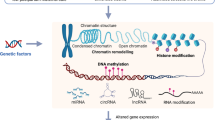
Abstract
Epigenetic mechanisms regulate gene expression, influencing protein levels and ultimately shaping phenotypes during life. However, both stochastic epigenetic variations and environmental reprogramming of the epigenome might influence neurodevelopment and ageing, and this may contribute to the origins of mental ill-health. Studying the role of epigenetic mechanisms is challenging, as genotype-, tissue- and cell type-dependent epigenetic changes have to be taken into account, while the nature of mental disorders also poses significant challenges for linking them with biological profiles. In this chapter, we summarise the current evidence suggesting the role of DNA methylation as a key epigenetic mechanism in major depressive disorder.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
Major depressive disorder (MDD) is a prevalent mood disorder characterised by persistent low mood accompanied by significant morbidity and mortality. The lifetime prevalence of MDD is 15–20% and women suffer from MDD about twice more often than men [1]. MDD is a complex multifactorial disorder, with both genetic and environmental factors playing an important role in its development. The heritability is estimated to be approximately 37%, and numerous links have been made between genetic variation and clinical depression [2]. However, DNA sequence variations cannot fully explain the susceptibility to MDD as detecting strong and replicable genetic associations with the development and course of clinical depression have proven difficult. Moreover, exposure to known environmental risk factors for MDD such as childhood adversities does not always generate the disorder. Therefore, the development and course of MDD are thought to be explained by gene-environment (GxE) interactions, where the effect of the environment depends on a person’s genotype or, equivalently, the effect of a person’s genotype depends on the environment [3]. Thus, it can be suggested that individuals genetically vulnerable to mental illness might undergo structural brain changes, imbalances in multiple neurotransmitter systems, alterations in neurotrophic signalling, and neuroendocrine abnormalities when exposed to harmful environmental factors at critical times during neurodevelopment.
A consistent line of evidence in human and rodent studies has shown that environmental factors regulate gene transcription and epigenetic mechanisms emerged as prime candidates for mediating GxE interactions in several brain regions [4]. In this chapter, we focus on DNA methylation, representing the most studied epigenetic mechanisms in psychiatric research. We will first summarise the current stage of knowledge within this field indicating the potential contribution of DNA methylation to MDD and discuss the results of the most replicated human studies on DNA methylation alterations in relation to environmental exposures associated with MDD. Finally, we will discuss the current challenges and perspectives in the field of epigenetic research on MDD.
1 DNA Methylation and MDD
Several methods have been applied to investigate MDD-associated and stress-induced alterations in DNA methylation. In the following sections, we will discuss the observed candidate and methylome-wide associations in MDD.
1.1 Candidate Gene Studies
Most DNA methylation studies thus far used candidate gene approaches. Impairments in stress response pathways, neurotrophic signalling and monoaminergic systems are well-known processes involved in the pathogenesis of MDD. Therefore, the focus of candidate gene methylation studies was predominantly on the promoter sequences of genes involved in these biological pathways. Here, we will address a selection of the main findings of the first studies on candidate genes implicated in MDD.
1.2 Stress Reactivity Genes
Given the role of the hypothalamic-pituitary-adrenal (HPA) axis and early life experiences and the aetiology of MDD, a series of studies investigated the DNA methylation patterns of the NR3C1 gene. NR3C1 encodes the glucocorticoid receptor (GR), which is known for its regulatory role in dampening the activity of the HPA axis. Environmental reprogramming of NR3C1 gene expression has been shown in both the brain and periphery. Both human and rodent studies suggested that early life trauma is associated with significant hypermethylation in the promoter region of the alternate exon 1F (humans) or 17 (rodents) within the hippocampus and, subsequently, reduced GR expression [5, 6]. Increased methylation levels of the NR3C1 promoter have been reported in lymphocytes of newborn prenatally exposed to maternal depression [7, 8]. One study examined a functional association between methylation statuses of NR3C1 gene promoter and cortisol as the end product of the stress pathway. A decreased cortisol response to the dexamethasone/corticotropin-releasing hormone (CRH) test was associated with increased methylation levels exon 1F NR3C1 gene promoter in leukocytes [9].
FK506-binding protein 5 (FKBP5), a member of the immunophilin protein family, functionally interacts with GRs and is linked to environmental stress exposure and MDD. FKBP5 protein reduces glucocorticoid-binding affinity [10], while cortisol and its binding to GR induce FKBP5 expression [11]. Several studies have shown the interactive effects of polymorphisms in FKBP5 and early life adversities predicting MDD [12]. Decreased allele-specific methylation of FKBP5 has been observed in peripheral blood cells of subjects who have experienced childhood abuse, whereas a similar effect has been observed in a neuronal progenitor cell line after exposure to GR agonists [13].
1.3 Neurotrophic Signalling Genes
Brain-derived neurotrophic factor (BDNF) is a member of the neurotrophin family and has been shown to regulate the development, plasticity and survival of dopaminergic, cholinergic and serotonergic neurons. The neurotrophic hypothesis of depression suggests that low levels of BDNF, concomitant with reduced neuronal and synaptic plasticity, are associated with clinical depression [14]. The BDNF protein is abundant in the brain and periphery and known to be able to cross the blood-brain barrier [15]. Chronic stress-induced decreases in BDNF and, in particular, antidepressant-induced increases in BDNF have been extensively studied in relation to the development and course of MDD [16]. Therefore, the correlation between BDNF methylation status and MDD has already been a focus of interest for years [17]. Differential regulation of BDNF exons I, IV and IX expression has been reported repeatedly, accompanied by changes in (allele-specific) DNA methylation of the corresponding promoters within various brain regions as well in the blood of patients with MDD [18]. The majority of studies have found increased methylation levels at different loci within the BDNF gene in MDD patients compared to controls in both brain and the periphery [19]. However, the direction of the effects is not always congruent [20].
To explore the functional relevance of DNA methylation variation and MDD, some studies examined the association between DNA methylation at the BDNF gene and structural changes in the brain of MDD patients. As such, in patients with MDD, the prefrontal and occipital cortices have been indicated as regions in which BDNF promoter hypermethylation is associated with reduced cortical thickness [21]. Another study showed a correlation between BDNF promoter hypermethylation and reduced white-matter integrity in individuals with MDD [22]. Significant associations have been found between BDNF promoter VI hypermethylation and a history of suicidal attempts and suicidal ideation [23]. In two subsequent studies, the same group found a significant association between BDNF promoter VI methylation levels and late-life depression [24], as well as depression related to breast cancer [25].
Methylation status of BDNF promoters has also been studied as a predictor for antidepressant treatment. Differential methylation of BDNF promoter I was observed when comparing responders and nonresponders to electroconvulsive therapy (ECT) [26]. Likewise, the methylation state of a CpG site within the exon IV promoter region of BDNF has been suggested to predict responses to antidepressant pharmacotherapy [27].
1.4 Monoaminergic Transporter Genes
The monoamine hypothesis of depression highlights the importance of serotonin (5-HT) turnover and transmission in synaptic cleft in the pathophysiology of major depression [28]. In particular, genetic variation in SLC6A4, the gene encoding the serotonin transporter (5-HTT or SERT), is a well-known candidate gene studied in MDD. 5-HTT is responsible for the reuptake of 5-HT into the presynaptic neuron and the primary target of antidepressants to normalise 5-HT levels and, consequently, the clinical symptoms of depression. SLC6A4 polymorphisms and their interaction with environmental stress have been studied extensively in association with MDD [29]. However, studies on the link between genetic variation of SLC6A4, life events and MDD are characterised by inconsistent findings, which may be partly explained by the additional and possibly interdependent role of epigenetic variation. Hypermethylation in the promoter region of SLC6A4 in lymphoblast cell lines was accompanied by lower gene expression and associated with vulnerability to MDD, concomitant with a complex interaction with genetic variation in SLC6A4 [30]. Clearly, these findings await replication.
Increased buccal cell methylation levels have been observed in short-allele carriers of the SLC6A4 gene in association with depressive symptoms [31]. In addition, increased SLC6A4 promoter methylation status was significantly associated with childhood adversities, a family history of depression, but not antidepressant treatment outcomes [32]. However, another study showed that lower average SLC6A4 CpG methylation was associated with an impaired antidepressant treatment response [33]. Thus far, all studies with regard to the association between DNA methylation within the SLC6A4 gene have been performed on peripheral blood tissues. A number of studies have examined the association between SLC6A4 promoter methylation and structural and functional changes in the brain. Using positron emission tomography (PET), decreased 5-HT synthesis in the orbitofrontal cortex was shown to be correlated to increased SLC6A4 promoter methylation [34]. In addition, increased methylation of the SLC6A4 promoter has been shown to be associated with increased hippocampal volume assessed by voxel-based morphometry [35]. Increased SLC6A4 methylation has furthermore been associated with childhood trauma and decreased hippocampal volume [36].
2 Methylome-Wide Association Studies
Methylome-wide association studies (MWAS) using different platforms have shown distinguished patterns of DNA methylation in MDD (see Table 10.1). A study comparing post-mortem frontal cortex tissue from MDD patients and healthy controls identified 224 differentially methylated regions (DMRs). These regions were highly enriched for genes involved in neuronal growth and development. However, the technique that has been used to measure the methylation levels for this study could not provide information at a single CpG resolution [37]. Another MWAS on the prefrontal cortex of suicidal completers with depression and sudden death controls showed 115 DMRs mainly related to astrocytic functioning [38]. Findings from MWAS performed on blood samples of individuals with a lifetime history of depression compared to nondepressed controls provided evidence for the involvement of inflammatory pathways previously implicated in depression [39]. Another genome-wide DNA methylation profiling of peripheral leukocytes of medication-free MDD patients identified 363 hypomethylated CpG sites [40]. In this study, three CpG sites residing in DGKH, GSK3B and SGK1 genes have been previously implicated in MDD, and they could show a significant inverse correlation between GSK3B promoter DNA methylation and expression. A methylome-wide study that incorporated an environmental risk factor reported an association between a history of childhood maltreatment and changes in saliva DNA methylation within three genes (ID3, GRIN1 and TPPP), of which the degree of methylation predicted depressive symptoms. These genes are known to be involved in stress-related neuroendocrine pathways and neural plasticity [41]. A recent genome-wide profiling of cortical brain regions (BA11 and BA25) from MDD suicide cases compared to controls identified a DMR, upstream of the PSORS1C3 noncoding gene, which is consistently hypomethylated across both cortical brain regions in MDD patients [42].
2.1 Twin Studies
Phenotypic discordance in monozygotic (MZ) twin pairs provides a valuable tool to differentiate genetic from non-genetic causes of diseases. Utilising MZ twin designs allows ruling out DNA sequence variations as a confounding source for epigenetic studies. While the genomic content in MZ twins is almost identical, the discordance in phenotypes can be attributed to non-shared environmental and stochastic factors. It has been suggested that different levels of DNA methylation at specific loci within MZ pairs measured as differentially methylated positions (DMPs) can be linked to the environmental causes of disease, while the changes in methylation variance, measured as variably methylated probes (VMPs), may be related to stochasticity. There is increasing evidence suggesting that epigenetic variation between MZ pairs plays a role in the aetiology of MDD. However, using buccal and blood cells, studying MZ pairs discordant for MDD has not identified any methylome-wide significant loci after correcting for multiple testing, which is most likely the result of a lack of statistical power. Interestingly, several of the most differentially methylated genes have previously been associated with the pathogenesis of MDD (see Table 10.1) [43, 44]. A meta-analysis of 50 MZ pairs, all female discordant for MDD, using 8.1 K human CpG island microarrays, identified 17 DMRs with genome-wide significance, and some of these genes have previously been associated with MDD (see Table 10.1) [45].
3 DNA Methylation and Antidepressant Treatment
There is increasing evidence suggesting that the effectiveness of antidepressants might be associated with DNA methylation status of certain genes. One example is the finding that decreased DNA methylation of SLC6A4 promotor region was associated with impaired antidepressant treatment response [33]. On the contrary, another study showed that less improvement in clinical symptoms of depression was correlated with higher methylation percentage at SLC6A4 gene promotor region [32].
Epigenetic mechanisms have also been proposed to mediate the mechanism of action of antidepressants. Alterations in DNA methylation and enzymes catalysing the methylation processes have been suggested to be involved in therapeutic effects of tricyclic antidepressants (TCA), selective serotonin reuptake inhibitor (SSRI) and valproate (VPA) [46, 47].
A number of MWA studies investigated the methylation changes in newborns exposed to maternal antidepressant use during pregnancy. Exposure to antidepressants, regardless of the medication family, was shown to be associated with differential methylation of CpG sites in the TNFRSF21 and CHRNA2 genes [48]. Another genome-wide methylation study identified increased methylation levels at CpG sites in CYP2E1, EVA1 and SLMAP in SSRIs exposed neonates [49]. Moreover, various studies compared the methylation changes associated with maternal use of antidepressants in candidate genes such as NR3C1, BDNF and SLC6A4 [27, 50]. However, results were not conclusive and none have been replicated in genome-wide array-based approaches.
4 Discussion and Future Perspectives
Over the past few years, a considerable number of epigenome-wide and candidate gene studies have been performed to identify a relation between DNA methylation and the pathogenesis of MDD. Current state of evidence on the link between DNA methylation and MDD indicates limited overlap between the identified genes in different studies. Moreover, the significant associations identified in candidate gene studies have not been observed in MWAS. These inconsistent findings can be explained by a broad range of methodological concerns in the field of epigenetic psychiatry. Evidently, limited access to human brain tissue is a major challenge in studying the specific epigenetic patterns in mental illnesses. Moreover, DNA methylation is characterised by the addition of a methyl or hydroxymethyl group to the C5 position of cytosine. DNA hydroxymethylation can both act as a stable functional epigenetic mark and may also serve as an intermediate mark in the active process of DNA demethylation. However, the large majority of current commercially available techniques that have been used to study epigenetic variation in MDD do not distinguish between DNA methylation and other related epigenetic marks, e.g. DNA hydroxymethylation, limiting the power of such approaches. Furthermore, true epigenetic variations can be influenced by state- or disease-specific differences in the cellular composition of both brain and peripheral blood tissues. In addition, significant findings remain vulnerable to confounding factors such as age, gender and smoking, which might cause false association discoveries between MDD and DNA methylation marks. Similarly, disease-related factors like medication, heterogeneity in diagnostic tools, the onset and course of MDD in different studies might explain the observed inconsistency in findings. Moreover, thus far, no MWAS of MDD has been integrated with genomic data. As such, genetic variations known to predict variations in the methylome should be taken into account in future approaches. Larger sample sizes and meta-analytical approaches can be utilised to boost statistical power in order to identify methylome-wide significant genes in association with MDD or the effects of antidepressants. Evidently, heterogeneity between studies, though strengthening the value of observed converging evidence, at the same time challenges the consolidation of less robust findings within individual datasets. Replication studies of epigenetic findings are therefore highly necessary. In addition, it will be crucial to move from associations and correlations towards examining causality. For instance, in vivo epigenetic editing of identified targets, as well as using longitudinal study designs in human cohorts would significantly improve our understanding about the causal relationship between changes in DNA methylation and the development and course of MDD.
References
Kessler RC, Bromet EJ. The epidemiology of depression across cultures. Annu Rev Public Health. 2013;34:119–38. doi:10.1146/annurev-publhealth-031912-114409.
Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry. 2000;157(10):1552–62. doi:10.1176/appi.ajp.157.10.1552.
Duncan LE, Keller MC. A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. Am J Psychiatry. 2011;168(10):1041–9. doi:10.1176/appi.ajp.2011.11020191.
Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–54. doi:10.1038/ng1089.
McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonte B, Szyf M, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12(3):342–8. doi:10.1038/nn.2270.
Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7(8):847–54. doi:10.1038/nn1276.
Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3(2):97–106.
Palma-Gudiel H, Cordova-Palomera A, Eixarch E, Deuschle M, Fananas L. Maternal psychosocial stress during pregnancy alters the epigenetic signature of the glucocorticoid receptor gene promoter in their offspring: a meta-analysis. Epigenetics. 2015;10(10):893–902. doi:10.1080/15592294.2015.1088630.
Tyrka AR, Parade SH, Welch ES, Ridout KK, Price LH, Marsit C, et al. Methylation of the leukocyte glucocorticoid receptor gene promoter in adults: associations with early adversity and depressive, anxiety and substance-use disorders. Transl Psychiatry. 2016;6(7):e848. doi:10.1038/tp.2016.112.
Denny WB, Valentine DL, Reynolds PD, Smith DF, Scammell JG. Squirrel monkey immunophilin FKBP51 is a potent inhibitor of glucocorticoid receptor binding. Endocrinology. 2000;141(11):4107–13. doi:10.1210/endo.141.11.7785.
Binder EB. The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology. 2009;34(Suppl 1):S186–95. doi:10.1016/j.psyneuen.2009.05.021.
Zannas AS, Binder EB. Gene-environment interactions at the FKBP5 locus: sensitive periods, mechanisms and pleiotropism. Genes Brain Behav. 2014;13(1):25–37. doi:10.1111/gbb.12104.
Klengel T, Binder EB. Gene × environment interactions in the prediction of response to antidepressant treatment. Int J Neuropsychopharmacol. 2013;16(3):701–11. doi:10.1017/S1461145712001459.
Duman RS. Role of neurotrophic factors in the etiology and treatment of mood disorders. Neuromol Med. 2004;5(1):11–25. doi:10.1385/NMM:5:1:011.
Pan W, Banks WA, Fasold MB, Bluth J, Kastin AJ. Transport of brain-derived neurotrophic factor across the blood–brain barrier. Neuropharmacology. 1998;37(12):1553–61.
Lee BH, Kim YK. The roles of BDNF in the pathophysiology of major depression and in antidepressant treatment. Psychiatry Investig. 2010;7(4):231–5. doi:10.4306/pi.2010.7.4.231.
Boulle F, van den Hove DL, Jakob SB, Rutten BP, Hamon M, van Os J, et al. Epigenetic regulation of the BDNF gene: implications for psychiatric disorders. Mol Psychiatry. 2012;17(6):584–96. doi:10.1038/mp.2011.107.
Januar V, Ancelin ML, Ritchie K, Saffery R, Ryan J. BDNF promoter methylation and genetic variation in late-life depression. Transl Psychiatry. 2015;5:e619. doi:10.1038/tp.2015.114.
Keller S, Sarchiapone M, Zarrilli F, Videtic A, Ferraro A, Carli V, et al. Increased BDNF promoter methylation in the Wernicke area of suicide subjects. Arch Gen Psychiatry. 2010;67(3):258–67. doi:10.1001/archgenpsychiatry.2010.9.
Fuchikami M, Morinobu S, Segawa M, Okamoto Y, Yamawaki S, Ozaki N, et al. DNA methylation profiles of the brain-derived neurotrophic factor (BDNF) gene as a potent diagnostic biomarker in major depression. PLoS One. 2011;6(8):e23881. doi:10.1371/journal.pone.0023881.
Na KS, Won E, Kang J, Chang HS, Yoon HK, Tae WS, et al. Brain-derived neurotrophic factor promoter methylation and cortical thickness in recurrent major depressive disorder. Sci Rep. 2016;6:21089. doi:10.1038/srep21089.
Choi S, Han KM, Won E, Yoon BJ, Lee MS, Ham BJ. Association of brain-derived neurotrophic factor DNA methylation and reduced white matter integrity in the anterior corona radiata in major depression. J Affect Disord. 2015;172:74–80. doi:10.1016/j.jad.2014.09.042.
Kang HJ, Kim JM, Lee JY, Kim SY, Bae KY, Kim SW, et al. BDNF promoter methylation and suicidal behavior in depressive patients. J Affect Disord. 2013;151(2):679–85.
Kang HJ, Kim JM, Bae KY, Kim SW, Shin IS, Kim HR, et al. Longitudinal associations between BDNF promoter methylation and late-life depression. Neurobiol Aging. 2015;36(4):1764.e1–7. doi:10.1016/j.neurobiolaging.2014.12.035.
Kang HJ, Kim JM, Kim SY, Kim SW, Shin IS, Kim HR, et al. A longitudinal study of BDNF promoter methylation and depression in breast cancer. Psychiatry Investig. 2015;12(4):523–31. doi:10.4306/pi.2015.12.4.523.
Kleimann A, Kotsiari A, Sperling W, Groschl M, Heberlein A, Kahl KG, et al. BDNF serum levels and promoter methylation of BDNF exon I, IV and VI in depressed patients receiving electroconvulsive therapy. J Neural Transm (Vienna). 2015;122(6):925–8. doi:10.1007/s00702-014-1336-6.
Tadic A, Muller-Engling L, Schlicht KF, Kotsiari A, Dreimuller N, Kleimann A, et al. Methylation of the promoter of brain-derived neurotrophic factor exon IV and antidepressant response in major depression. Mol Psychiatry. 2014;19(3):281–3. doi:10.1038/mp.2013.58.
Owens MJ, Nemeroff CB. Role of serotonin in the pathophysiology of depression: focus on the serotonin transporter. Clin Chem. 1994;40(2):288–95.
Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301(5631):386–9. doi:10.1126/science.1083968.
Philibert RA, Sandhu H, Hollenbeck N, Gunter T, Adams W, Madan A. The relationship of 5HTT (SLC6A4) methylation and genotype on mRNA expression and liability to major depression and alcohol dependence in subjects from the Iowa Adoption Studies. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(5):543–9. doi:10.1002/ajmg.b.30657.
Olsson CA, Foley DL, Parkinson-Bates M, Byrnes G, McKenzie M, Patton GC, et al. Prospects for epigenetic research within cohort studies of psychological disorder: a pilot investigation of a peripheral cell marker of epigenetic risk for depression. Biol Psychol. 2010;83(2):159–65. doi:10.1016/j.biopsycho.2009.12.003.
Kang HJ, Kim JM, Stewart R, Kim SY, Bae KY, Kim SW, et al. Association of SLC6A4 methylation with early adversity, characteristics and outcomes in depression. Prog Neuropsychopharmacol Biol Psychiatry. 2013;44:23–8. doi:10.1016/j.pnpbp.2013.01.006.
Domschke K, Tidow N, Schwarte K, Deckert J, Lesch KP, Arolt V, et al. Serotonin transporter gene hypomethylation predicts impaired antidepressant treatment response. Int J Neuropsychopharmacol. 2014;17(8):1167–76. doi:10.1017/S146114571400039x.
Wang D, Szyf M, Benkelfat C, Provencal N, Turecki G, Caramaschi D, et al. Peripheral SLC6A4 DNA methylation is associated with in vivo measures of human brain serotonin synthesis and childhood physical aggression. PLoS One. 2012;7(6):e39501. doi:10.1371/journal.pone.0039501.
Dannlowski U, Kugel H, Redlich R, Halik A, Schneider I, Opel N, et al. Serotonin transporter gene methylation is associated with hippocampal gray matter volume. Hum Brain Mapp. 2014;35(11):5356–67. doi:10.1002/hbm.22555.
Booij L, Szyf M, Carballedo A, Frey EM, Morris D, Dymov S, et al. DNA methylation of the serotonin transporter gene in peripheral cells and stress-related changes in hippocampal volume: a study in depressed patients and healthy controls. PLoS One. 2015;10(3):e0119061.
Sabunciyan S, Aryee MJ, Irizarry RA, Rongione M, Webster MJ, Kaufman WE, et al. Genome-wide DNA methylation scan in major depressive disorder. PLoS One. 2012;7(4):e34451. doi:10.1371/journal.pone.0034451.
Nagy C, Suderman M, Yang J, Szyf M, Mechawar N, Ernst C, et al. Astrocytic abnormalities and global DNA methylation patterns in depression and suicide. Mol Psychiatry. 2015;20(3):320–8. doi:10.1038/mp.2014.21.
Uddin M, Koenen KC, Aiello AE, Wildman DE, de los Santos R, Galea S. Epigenetic and inflammatory marker profiles associated with depression in a community-based epidemiologic sample. Psychol Med. 2011;41(5):997–1007. doi:10.1017/S0033291710001674.
Numata S, Ishii K, Tajima A, Iga J, Kinoshita M, Watanabe S, et al. Blood diagnostic biomarkers for major depressive disorder using multiplex DNA methylation profiles: discovery and validation. Epigenetics. 2015;10(2):135–41. doi:10.1080/15592294.2014.1003743.
Weder N, Zhang H, Jensen K, Yang BZ, Simen A, Jackowski A, et al. Child abuse, depression, and methylation in genes involved with stress, neural plasticity, and brain circuitry. J Am Acad Child Adolesc Psychiatry. 2014;53(4):417–24.e5. doi:10.1016/j.jaac.2013.12.025.
Murphy TM, Crawford B, Dempster EL, Hannon E, Burrage J, Turecki G, et al. Methylomic profiling of cortex samples from completed suicide cases implicates a role for PSORS1C3 in major depression and suicide. Transl Psychiatry. 2017;7(1):e989. doi:10.1038/tp.2016.249.
Dempster EL, Wong CC, Lester KJ, Burrage J, Gregory AM, Mill J, et al. Genome-wide methylomic analysis of monozygotic twins discordant for adolescent depression. Biol Psychiatry. 2014;76(12):977–83. doi:10.1016/j.biopsych.2014.04.013.
Cordova-Palomera A, Fatjo-Vilas M, Gasto C, Navarro V, Krebs MO, Fananas L. Genome-wide methylation study on depression: differential methylation and variable methylation in monozygotic twins. Transl Psychiatry. 2015;5:e557. doi:10.1038/tp.2015.49.
Davies MN, Krause L, Bell JT, Gao F, Ward KJ, Wu HL, et al. Hypermethylation in the ZBTB20 gene is associated with major depressive disorder. Genome Biol. 2014;15(4):R56. doi:10.1186/gb-2014-15-4-r56.
Zimmermann N, Zschocke J, Perisic T, Yu S, Holsboer F, Rein T. Antidepressants inhibit DNA methyltransferase 1 through reducing G9a levels. Biochem J. 2012;448(1):93–102. doi:10.1042/BJ20120674.
Detich N, Bovenzi V, Szyf M. Valproate induces replication-independent active DNA demethylation. J Biol Chem. 2003;278(30):27586–92. doi:10.1074/jbc.M303740200.
Schroeder JW, Smith AK, Brennan PA, Conneely KN, Kilaru V, Knight BT, et al. DNA methylation in neonates born to women receiving psychiatric care. Epigenetics. 2012;7(4):409–14. doi:10.4161/epi.19551.
Gurnot C, Martin-Subero I, Mah SM, Weikum W, Goodman SJ, Brain U, et al. Prenatal antidepressant exposure associated with CYP2E1 DNA methylation change in neonates. Epigenetics. 2015;10(5):361–72. doi:10.1080/15592294.2015.1026031.
Zhao JY, Goldberg J, Bremner JD, Vaccarino V. Association between promoter methylation of serotonin transporter gene and depressive symptoms: a monozygotic twin study. Psychosom Med. 2013;75(6):523–9. doi:10.1097/PSY.0b013e3182924cf4.
Oh G, Wang SC, Pal M, Chen ZF, Khare T, Tochigi M, et al. DNA modification study of major depressive disorder: beyond locus-by-locus comparisons. Biol Psychiatry. 2015;77(3):246–55. doi:10.1016/j.biopsych.2014.06.016.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Pishva, E., Rutten, B.P.F., van den Hove, D. (2017). DNA Methylation in Major Depressive Disorder. In: Delgado-Morales, R. (eds) Neuroepigenomics in Aging and Disease. Advances in Experimental Medicine and Biology(), vol 978. Springer, Cham. https://doi.org/10.1007/978-3-319-53889-1_10
Download citation
DOI: https://doi.org/10.1007/978-3-319-53889-1_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-53888-4
Online ISBN: 978-3-319-53889-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)