Abstract
Plants belonging to family fabaceae play an imperative role in restoring soil fertility, with the remarkable ability to engage endosymbiotically with both rhizobia and arbuscular mycorrhiza (AM). Establishment of both symbioses is based on a finely regulated molecular dialogue between two partners. Plant roots secrete an assortment of flavonoids, competent to shape rhizosphere microflora by amplifying chemotactic surface motility of beneficial microorganisms while combating pathogenic ones. Flavonoids potentially regulate transcriptional activity of many microbial genes, e.g. nod genes, and fungal hyphal branching and initiate the production of microsymbiont signal molecules (Nod/Myc factor). The perception of these lipo-chito-oligosaccharides at epidermis stimulates partly analogous downstream signal transduction cascade to activate symbiosis-related genes and consequently enable successful penetration of both microsymbionts in the host. In response to host-specific microbe, selective accumulation of flavonoids drives suppression of plant innate immunity as well as cortical cell dedifferentiation into symbiosome. High degree of coordination between root cortical cell machinery and rhizobia/AM results in the formation of symbiotic interfaces—nodules/arbuscules respectively, where harboring bacteroids and arbuscules deliver macronutrients (nitrogen and phosphorus) to host in exchange for photosynthates. Flavonoids cross-link with plant proteins to form an O2− diffusion barrier in the symbiosome membrane and serve as a checkpoint for nitrogenase efficiency. Under nutrient-rich conditions, plants regulate flavonoid fluxes to prevent an excessive establishment of metabolically expensive symbioses. Therefore, understanding these selective forces that govern host selection of beneficial rhizomicrobiome, followed by underlying establishment and regulation of symbioses in legumes, is crucial for agrobiologists to achieve sustainable agriculture.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
8.1 Introduction
Among plant nutrients, nitrogen (N) and phosphorus (P) are the most limiting nutrients worldwide whose continued supply as fertilizers is necessary if world food needs are to be fulfilled. Modern agriculture has been highly reliant on industrial fertilizers, and the vast amount of resources spent on their production is leading to a substantial carbon footprint (exploiting ~50% of fossil fuel) of this industrial sector (Jensen et al. 2012). However, the rising fossil fuel cost is making chemical fertilizers dramatically expensive; CO2 emission during fossil fuel combustion is contributing to the greenhouse effect, and leaching of applied fertilizers (~30–50%) is leading to major environmental problems, e.g. eutrophication (Crutzen et al. 2007). Thus, efforts must be directed to enhance the exploitation of rhizospheric soil microorganisms which can effectively ensure substantial uptake of essential nutrients by plants and thus recuperate agricultural fields with nutrient-poor soils (Bonfante and Genre 2008; Parniske 2008; Vieira et al. 2010). Two of the most widespread rhizospheric interactions, the Rhizobium–legume (RL) and the arbuscular mycorrhiza (AM)–plant symbioses, have particular significance as natural mini-fertilizer factories in land ecosystems (Venturi and Keel 2016).
Rhizobium–legume (RL) symbiosis is almost entirely limited to economically important family Fabaceae which evolved only about 60 Mya (Manchanda and Garg 2007; Delaux et al. 2015). On the other hand, since 500 million years ago (Mya), ubiquitous soil AM fungi establish a symbiotic relationship with the roots of more than 90% of all higher plants (Smith and Read 2008; Wilde et al. 2009). RL interaction leads to the formation of symbiotic root nodules, where Rhizobium bacteria acquires vast fitness output from host carbon and energy (Peix et al. 2015) and legumes gain access to otherwise unavailable soil nitrogen (replenishing approximately 200 million tons of N2 annually) (Ferguson et al. 2010; Kondorosi et al. 2013). In mycorrhizal association, vegetative growth and reproductive spore production of biotropic fungal symbionts rely on reduced carbon of living plant tissue (4–20% of their photosynthate) and in return provide various benefits, including—but not limited to—nutrient and water uptake (Parniske 2008).
Legumes have the ability to host N2-fixing bacteria and AM at the same time (Antunes et al. 2006), and mycorrhizal symbiosis associated with legumes is an essential link for effective phosphorus (P) nutrition leading to enhanced N2 fixation that advocates a synergistic tripartite association (Geneva et al. 2006). In most cases investigated, especially when both nitrogen and phosphate are limiting factors, rhizobia and AM fungi appear to act synergistically since combined inoculation with rhizobia and mycorrhiza enhances plant growth and reproduction more than inoculation with either microsymbiont alone and also mutually increases each other’s establishment (Gould and Lister 2005). Striking similarities between two symbionts have been reported with respect to mutual recognition, infection process, and genetic and hormonal regulation (Mukherjee and Ané 2011). Exchange of molecular signals between the host plant and microsymbiont in the form of cell-to-cell inter-organismal communication is an important stage for the initiation of effective symbiosis (Mapope and Dakora 2013), and during N2 fixation, a wide variety of host FLAVONOIDS have been shown to attract compatible rhizobia for symbiosis, by their nod gene inducing activity (Shaw et al. 2006; Mandal et al. 2010). Apart from the function of flavonoids in RL interaction, they also act as signaling compounds in the host communication with AM (Catford et al. 2006; Steinkellner et al. 2007; Shaw and Hooker 2008). Moreover, plant flavonoids vary qualitatively and quantitatively with root endosymbiont colonization, indicating dependability of nodulation and mycorrhization on flavonoids beyond signaling (Carlsen et al. 2008; Zhuang et al. 2013). Thus, this prompts in-depth review on diverse role of flavonoids in the establishment of legume symbioses with rhizobia and AM.
8.2 Flavonoid: A Versatile Compound
8.2.1 Structural Diversity and Subcellular Distribution
Flavonoids (derived from the Latin word for yellow, favus), the low-molecular-weight secondary metabolites, are biologically active polyphenolic compounds and are widespread throughout the plant kingdom, ranging from mosses to angiosperms (Williams and Grayer 2004). Diverse flavonoid molecules share the same core carbon framework of phenyl-benzopyran functionality, the flavan nucleus (Fig. 8.1), consisting of two aromatic rings with six carbon atoms (rings A and B) interconnected by a heterocyclic benzopyrano (chromano) C ring with three carbon atoms (Saito et al. 2013; Cheng et al. 2014). The position of B aromatic ring linkage to the benzopyrano moiety allows a broad separation of these compounds into flavonoids (2-phenyl-benzopyrans, e.g., kaempferol, apigenin), isoflavonoids (3-phenyl-benzopyrans, e.g., genistein), and neoflavonoids (4-phenyl-benzopyrans) (Winkel-Shirley 2001; Dixon and Pasinetti 2010). These groups usually share a common chalcone precursor and therefore are biogenetically and structurally related (Marais et al. 2006). A number of divergent chalcones and flavonoid structures are formed from the extensive modification (rearrangement, alkylation, oxidation, and glycosylation) of the basic molecules (Halbwirth 2010). To date, more than 10,000 different flavonoids have been identified in plants (broadly classified into flavonols, flavones, flavan-3-ols, flavanones, anthocyanins, isoflavones) and the number is still increasing (D’haeseleer et al. 2010; Hassan and Mathesius 2012; Cheynier et al. 2013) and even within the same species a number of different flavonoids have been identified (Martens and Mithöfer 2005). Typically, flavonoids are stored in their glycoside forms (more stable than the free form); however, some flavones and flavonols can be found naturally as the aglycone (Birt and Jeffery 2013). The chemical diversity, size, three-dimensional shape, and physical and biochemical properties of flavonoids allow them to interact with targets in different subcellular locations to influence biological activity in plants, animals, and microbes (Taylor and Grotewold 2005; Buer et al. 2010).
Consistent with their diverse physiological functions, flavonoids are found in most plant cell compartments, including cytosol, vacuole (anthocyanins, flavonol and flavone glycosides), ER, chloroplast (quercetin and kaempferol glycosides), nucleus (isoflavonoids coumestrol and 4′,7-dihydroxyflavone), pollen surface and small vesicles, as well as the extracellular space (Saslowsky et al. 2005; Lepiniec et al. 2006; Hsieh and Huang 2007; Naoumkina et al. 2007; Hernández et al. 2009). In the plant, flavonoids function as developmental regulators, antioxidants, pigments, UV sunscreens, nutrient acquisitor, energy escape valve, auxin transport regulators, defense compounds against pathogens (signal for jasmonate-induced mobilization of vacuolar isoflavonoid glucosides for phytoalexin biosynthesis), and signals during symbiosis (Buer and Muday 2004; Naoumkina and Dixon 2008; Cheynier et al. 2013). However, their importance in plant biology goes beyond their specific functions within the plant (Dixon and Pasinetti 2010). Flavone and flavonolaglycones have been detected in root exudates from numerous species, where isoflavones are particularly secreted by legume roots into the rhizosphere (Zhao and Dixon 2010).
8.2.2 Flavonoids in the Rhizosphere
In response to elicitors, both aglycone and glycoside flavonoids are often exuded into the rhizosphere in order to fulfill some of their ecological roles as mediators of belowground interactions, for example, in order to attract compatible rhizosphere-dwelling rhizobia, stimulate or inhibit rhizobial nod gene expression, inhibit root pathogens, stimulate mycorrhizal spore germination and hyphal branching, affect quorum sensing, and chelate soil nutrients (Broughton et al. 2003; Cooper 2004; Martens and Mithöfer 2005). Exudation of flavonoids is ATP dependent and catalyzed by an ABC-type transporter as has been suggested by the study of isoflavonoid genistein exudation from soybean root plasma membrane vesicles (Sugiyama et al. 2007). This was also supported by the study of Badri et al. (2009) where ABC transporter mutants of Arabidopsis like abcg30 had altered root exudate profiles. Besides ABC-type transpoters, flavonoids can also be released passively in the rhizosphere from decomposing root cap and border cells (Shaw et al. 2006). Another important mechanism for releasing active flavonoid aglycones during root–microbe interactions is through apoplastic β-glucosidases, which release isoflavones from their conjugates in soybean roots (Suzuki et al. 2006). Recently, Sugiyama et al. (2016) reported that the expression of gene encoding isoflavone conjugates hydrolyzing beta-glucosidase (ICHG) coordinately peaked at vegetative stages with the higher secretion of daidzein. However, under nitrogen-deficient conditions, besides daidzein, genistein was also highly secreted, with no induction of ICHG. Thus, their study suggested that two pathways for isoflavone secretion in soybean roots are expected to have distinct physiological roles, i.e., ICHG-mediated secretion during vegetative growth and ATP-dependent transport during nitrogen deficiency.
Once in the rhizosphere, the fate of flavonoid persistence in the rhizosphere (likely to be from hours to days) varies with environmental conditions, flavonoid structure, and the presence of soil microbes, some of which can metabolize or modify flavonoids. Flavonoids can be absorbed to the cell wall and to soil particles with cationic binding sites, thus becoming unavailable (Shaw and Hooker 2008). Depending on their structural modifications, the solubility and mobility of flavonoids in the soil varies. For example, genistein is described as “practically insoluble in water,” whereas genistin, the glucoside, is “sparingly soluble in water” (O’Neil et al. 1996). Thus, conjugated forms are expected to be less adsorbed to the soil matrix, more mobile, and therefore more bioavailable than the free aglycone form. While glycosylation improves their solubility in water, it is likely that flavonoid glycosides are quickly deglycosylated by microorganisms and plant exoenzymes, leaving the more hydrophobic aglycone (Sosa et al. 2010; Weston and Mathesius 2013). Once present in the aglycone form, new flavonoid structures may be produced during biodegradation of a parent flavonoid, which can be more efficient inducers of nod genes than the flavonoids themselves (Anders and Huber 2010; Rose et al. 2012). Moderate bioavailablity and mobility of flavonoids (organic carbon normalized partition coefficients, i.e., logKoc of 3.12 for formononetin and 3.19 for naringenin), makes ecological sense, as it would be evolutionarily favorable for a plant root to produce a chemical signal that is sufficiently bioavailable to allow interaction with its intended microbial target, but, at the same time, not so mobile that it will diffuse rapidly to the outer reaches of the rhizosphere. Thus, bioavailability, mobility and persistence of a flavonoid will determine the degree and outcome of flavonoid–microbe interaction with important consequences for plant nutrition (Shaw and Hooker 2008; Weston and Mathesius 2013).
8.3 Flavonoids in Rhizobium–Legume (RL) Endosymbiosis
Among the wide range of bacteria that have the ability to reduce N2 to ammonia (NH3), the most important are soil-dwelling prokaryotic bacteria collectively called rhizobia, where each legume species has its own cognate Rhizobium partner(s) (Gibson et al. 2008; Soyano and Kawaguchi 2014). Legume genomes are at least 50 times larger than those of their microsymbionts; nevertheless, their respective contributions are probably not vastly different (Irving et al. 2000). Thus, RL symbioses are cross-kingdom collaboration between two vastly different genomes (Yang et al. 2010). The endosymbiosis generally commences within a specific “susceptible” root zone close to the root tip, where initial bacteria–host recognition takes place (Laloum et al. 2014). The bacteria invade the roots of compatible legume plants leading to the development of specialized root structures called nodules, providing a unique ecological niche in which bacteria differentiate into bacteroids and fix N2. Although the interaction is beneficial to both partners, it comes with rigid rules that are strictly enforced by both the partners (Oldroyd et al. 2011). Here, we review the various signaling pathways by which the plant allows bacterial infection and promotes the construction of the nodule as well as how intricate transaction of metabolites (especially flavonoids) directs the bacteria into a nitrogen-fixing organelle-like state.
8.3.1 Signaling in Rhizobium–Legume Symbiosis
The succession to the symbiotic affirmation by two originally autonomous, free-living partners is governed by reciprocal generation and perception of signals, which has been described as “molecular dialogue” (De’narie et al. 1993). At least three different sets of symbiotic signals are exchanged between legumes and rhizobia during nodule development: flavonoids, Nod factors, EPS, and extracellular proteins (NOPs = nodulation outer proteins) (Broughton et al. 2003).
8.3.1.1 Signals from the Host Plants
A diverse array of compounds is exuded into the rhizospere, including sugars, aliphatic as well as aromatic acids, amino acids, amines, and many other low-molecular-weight compounds such as flavonoids, isoflavonoids, steroids, alkaloids, vitamins, and growth regulators (Skorupska et al. 2010; Lareen et al. 2016). The “rhizosphere effect,” first described by Hiltner (1904), assumes that many microorganisms are attracted to sugars, acids, and amino acids exuded by plant roots, which serve as C and energy sources for microorganisms. However, in addition to providing a C-rich environment, plant roots initiate cross talk with soil microbes by producing signals that are recognized by the microbes, which in turn produce signals that initiate colonization (Bais et al. 2004). Among the myriad of rhizodepositions, normally and continuously exuded by plants into the rhizosphere are phenolic compounds especially flavonoids that mediate signal traffic between roots and beneficial rhizosphere-dwelling rhizobia (Gibson et al. 2008; Skorupska et al. 2010; McNear 2013; Mandal et al. 2016). Although small quantities are excreted continuously, flavonoid concentrations in the rhizosphere increase in response to compatible rhizobia. Niches in the legume rhizosphere are tailored to rhizobial inoculation, as exudation of flavonoids is mostly restricted to the elongating root hair zone from which most nodules later develop (Zuanazzi et al. 1998). The role of flavonoids as bacterial chemoattractant and transcriptional activator of bacterial nod genes points out their central position in modifying rhizobial phenotypes possibly in relation to plant–root association and then symbiotic interaction (Brencic and Winans 2005; Spini et al. 2016). Once exuded, flavonoids, especially aglycone forms, are recognized to diffuse into the rhizobial membrane (Kobayashi et al. 2004; Wang et al. 2012), possibly through porins (Taylor and Grotewold 2005). Further, it activates rhizobia nod (nodulation) D gene expression (Ferguson et al. 2010), and this successful liaison between flavonoids and NodD proteins signals the beginning of association (Broughton et al. 2000). The first flavonoid to be discovered to act as nod gene inducers was luteolin, isolated from Medicago sativa and 7,4′ dihydroxyflavone (DHF) from Trifolium repens (white clover) (Redmond et al. 1986). Since then, about 30 nod gene-inducing flavonoids (especially flavanones and isoflavonoids) have been isolated from nine legume genera (Limpens and Bisseling 2008; Gholami et al. 2014). Most of these flavonoids are active as nod gene inducers at nanomolar to low micromolar concentrations (Begum et al. 2001a; Gholami et al. 2014) and stimulate bacterial nod gene expression within minutes. The specific exudation of flavonoid (mixtures) from legume hosts together with the specific perception of flavonoids by NodD proteins of different rhizobia is partially responsible for the host specificity of the symbiosis (Gibson et al. 2008; Skorupska et al. 2010; Rose et al. 2012). Hence, point mutations in nodD affect recognition of inducing flavonoids and cause extension of host range (Broughton et al. 2000). Generally, the NodDs of broad host range rhizobia, e.g., NGR234, respond to a wider range of flavonoid species (including phenolics that are inhibitors in other rhizobia, e.g., vanillin, iso-vanillin, as well as several estrogenic compounds) than those present in restricted host range rhizobia (Peck et al. 2006; Wang et al. 2012). nodD products of various Rhizobium species respond in different ways to flavonoids, and NodD homologues from the same strain may have various flavonoid preferences. In R. meliloti, NodD1 was activated when cells were supplied with a complex plant seed extract or the flavonoid luteolin. NodD2 only derepressed transcription when supplied with the complex extract, not with purified luteolin, while NodD3 apparently modulates the expression of nod genes even in the absence of any plant factor (Smit et al. 1992). Recently, Peck et al. (2013) presented a structural model of wild-type NodD1 identifying residues important for inducer binding, protein multimerization, and interaction with RNA polymerase at nod gene promoters. Species-specific flavonoids interact with a class of transcriptional activators of the LysR family which have an N-terminal ligand-binding domain that regulates the activity of the associated C-terminal DNA-binding domain, and NodD ligand-binding domain is thought to function as a flavonoid receptor. The perception of flavonoids by rhizobia is linked to elevation in concentrations of intracellular calcium in rhizobia that subsequently induces NodD proteins for Nod factor expression (Moscatiello et al. 2010; Hassan and Mathesius 2012).
Of the large number of available flavonones, flavones, isoflavones, and other related compounds, capability of flavonoid to act as nod gene inducers varies with the variation in host varieties, bacterial strains, and/or signal compounds (Begum et al. 2001a, b). Within the variety of flavonoids, isoflavonoids, and other compounds secreted by Lupinus albus roots into the rhizosphere, major proportion is composed of aldonic acids which act as natural nod gene inducers of Rhizobium lupini, Mesorhizobium loti, and Sinorhizobium meliloti (Gagnon and Ibrahim 1998). Luteolin, genistein, naringenin, hesperetin, and apigenin are the principal flavonoids involved in nod gene expression in S. meliloti, Bradyrhizobium japonicum, Rhizobium, and R. tibeticum, respectively (Kapulnik et al. 1987; Graham 1991; Begum et al. 2001b; Belkheir et al. 2001; Novák et al. 2002; Tsvetkova et al. 2006; Brechenmacher et al. 2010; Abd-Alla et al. 2014). Genistein, coumestrol, and daidzein have been reported as important inducers of rhizobial nodulation genes in the early stages of symbiosis between soybean and B. japonicum (Antunes and Goss 2005; Miransari and Smith 2009; Tian et al. 2014; Sugiyama et al. 2016). Begum et al. (2001a) suggested that the attachment of the B-ring to C-2 of flavonoids, as found in flavones and flavanones, is of crucial importance for induction. In this regard, Zhang et al. (2007) provided genetic evidence that RNA interference-mediated suppression of MtFNSII genes in Medicago truncatula resulted in flavone-depleted roots and led to significantly reduced nodulation when inoculated with S. meliloti. In addition, hydroxylation at the C-4 and the C-7 positions of flavones is important for this activity (Brencic and Winans 2005; Subramanian et al. 2007).
Interestingly, some flavonoids also show nod gene repressing activity for certain rhizobia. Isoflavonoids, medicarpin and coumestrol, have been reported to negatively control Nod factor production in S. meliloti (Zuanazzi et al. 1998). Jain and Nainawatee (1999) studied that except quercetin, alfalfa exudates decreased growth and protein content of R. meliloti cells. Naringenin induces the expression of nod genes in R. leguminosarum–Pisum sativum; however, quercetin is an inhibitor of nodulation (Novák et al. 2002). Inducers in one species or strain of Rhizobium are frequently anti-inducers in another species; thus, one type of flavonoid can have opposing effects on different bacteria; for example, the isoflavone diadzein induces nod gene expression in B. japonicum (nodulating soybean), whereas it inhibits that of those from R. leguminosarum (nodulating clover or peas) and thus contributes to host specificity (Andersen and Markham 2006). It has been suggested that a mixture of flavonoids is more effective in inducing nod genes as opposed to a single compound (Mandal et al. 2010). Both functions (induction or anti-induction) can co-occur in the exudates of the same plant, where different flavonoids in root exudates can act synergistically as nod gene inducers, but also in an antagonistic manner as anti-inducers (Cooper 2007; Makarova et al. 2015). Luteolin and 7,40-dihydroxyflavone are inducers, whereas genistein inhibits expression of S. meliloti nod genes (Kosslak et al. 1987; Hartwig et al. 1990; Peck et al. 2006). Thus, the ratio of inducers to anti-inducers in root exudates may be involved in determination of host recognition, maintaining an optimal level of Nod factor production and preventing elicitation of defense responses by the plant (Zuanazzi et al. 1998; Cesco et al. 2010). Li et al. (2016) provided a previously unidentified mechanism by which flavonoids in exudates of one crop root can promote N2 fixation in another crop in a two-crop intercropping system. In their study, maize root exudates contained significant flavonoids and promoted flavonoid synthesis in faba bean, thus triggering N2 fixation.
8.3.1.2 Signals from the Microsymbiont
Second set of signals is synthesized when cytoplasmic membrane-bound NodD–flavonoid complexes activate transcription from conserved 49-bp DNA motifs, i.e., “nod-box” promoters found in the promoter regions of many nodulation loci (Gibson et al. 2008). Thus, NodD proteins act as both sensors of the plant signal and transcriptional activators of nod loci on symbiotic plasmids (Redmond et al. 1986; Downie 2010). The concerted transcriptional activation of common and host-specific nod genes leads to the synthesis of Nod factor (NF) which is a key to legume doors (Mulder et al. 2005; Cooper 2007; Remigi et al. 2016). NFs belong to lipo-chito-oligosaccharides (LCOs) family, having an oligosaccharide backbone of four or five β-1-4-linked N-acetyl-d-glucosamine units with a terminal nonreducing sugar N-acylated by a 16–18 carbon fatty acid (Fauvart and Michiels 2008; Hamel and Beaudoin 2010). Assembly of the chitin backbone is performed by an N-acetylglucosaminyltransferase encoded by nodC, and the deacetylase NodB removes the N-acetyl moiety from the nonreducing terminus of the N-acetylglucosamine oligosaccharides. NodG has the enzymatic activity of a 3-oxoacyl-acyl carrier protein reductase and is involved in fatty acid elongation (López-Lara and Geiger 2001). Finally, an acyltransferase coded by nodA links the acyl chain to the acetyl-free carbon C-2 of the nonreducing end of the oligosaccharide (Brencic and Winans 2005). In addition to the common nod-DABC genes that are essential for symbiosis, the rhizobia harbor different combinations of other nod, nol, and noe genes which may have been recruited from paralogues in the course of the evolution, allowing the diversification of NF structures and host ranges (Taurian et al. 2008; Vieira et al. 2010). For instance, nodV and nodW of B. japonicum are essential for the nodulation of Macroptilium atropurpureum, Vigna radiata, and V. unguiculata but contribute only marginally to the symbiosis with G. max. nodH encodes a sulfotransferase that transfers a sulfate group to the reducing end of NFs of R. meliloti and elicits Ca2+ spiking (Wais et al. 2002). A nodF mutant produces a NF with a modified N-acylation on the terminal nonreducing N-acetyl glucosamine residue (Demont et al. 1993). NF produced by nodL mutants lacks a C6-O-acetylation on the terminal nonreducing glucosamine (Ardourel et al. 1995). nodFL double mutants trigger Ca2+ spiking and root hair deformation but are unable to infect their hosts (Wais et al. 2002; Haney et al. 2011). Thus, a major determinant of host-symbiont specificity is attributed to the different NF substituents (sulphuryl, methyl, carbamoyl, acetyl, fucosyl, arabinosyl, and other groups) attached to the oligosaccharide backbone as well as differences in the structure of the acyl chain (Downie 2010; Ferguson et al. 2010; Kouchi et al. 2010). Nod factors can trigger plant responses like root hair deformation and calcium oscillations (called calcium spiking) at astonishingly low concentrations, i.e., as little as 10−13 M Nod factor (Rose et al. 2012). As Nod factors also stimulate the synthesis and release of flavonoids from legume roots, the response to inoculation is amplified (Broughton et al. 2003).
8.3.2 Nod Factor (NF) Signaling Pathway in the Root Epidermis
Symbiosis initiates if the above-stated chemical cross talk between the interacting partners successfully culminates in the production of NFs (Bek et al. 2010). The mechanism, by which plants regulate the intracellular uptake of symbiotic bacteria, depends on physiological and molecular reprogramming that is associated with the perception of NF and resultant downstream signaling (Xie et al. 2012; Liang et al. 2013). Rhizobia have two main ways of entering the plant root: via the root hair or through cracks in root epidermal tissue (Oldroyd and Downie 2008; Ribeiro et al. 2015); however, rhizobial entry along the infection threads in root hairs is largely common (Mathesius 2009; Downie 2010). Two receptor-like kinases (RLK) of chitin-binding LysM RLK family, located on epidermal cells, are involved in nod factor binding: LjNFR1 and LjNFR5 in L. japonicus, PsSYM2A and PsSYM10 in P. sativum, MtLYK3/MtLYK4 and MtNFP in M. truncatula, and GmNFR1α/β and GmNFR5α/β in soybean (Ferguson et al. 2010; Indrasumunar et al. 2010; Broghammer et al. 2012; Wang et al. 2012). These receptors consist of an intracellular kinase domain, a transmembrane domain, and an extracellular portion having LysM domains (Gough 2003; Mathesius 2009). Interestingly, LjNFR1/PsSYM2A/MtLYK3/MtLYK4/GmNFR1α/β has a typical serine/threonine kinase domain, while LjNFR5/PsSYM10/MtNFP/GmNFR5α/β lacks the activation loop (Indrasumunar et al. 2010). The absence of an activation loop in one of the kinase domains suggests that the two LysM RLKs may assemble into a heterodimeric receptor, with the active kinase (NFR kinase) domain triggering downstream signal transduction through phosphorylation (Markmann et al. 2008; Radutoiu et al. 2008; Hamel and Beaudoin 2010; Indrasumunar et al. 2015).
Rhizobial infection has many similarities with pathogenic infection and induction of defense responses accompanies both interactions, but defense responses are induced to a lesser extent during rhizobial infection. Recently, it was evidenced by Ivanova et al. (2015) that a range of plant defense responses like suberization, callose and unesterified pectin deposition, as well as activation of defense genes can be triggered by different single mutations in symbiotic genes (sym33, sym40, sym42) that cause perception of an otherwise beneficial strain of Rhizobium as a pathogen. Besides symbiosis, LysM-RLKs have a role in immune signaling, indicating that NF signaling and pathogen chitin-based immune signaling are intertwined. However, Nod factor signal, unlike microbe-associated molecular pattern (MAMP) derived from microbes, suppresses an innate immune response in the host (Tóth and Stacey 2015). Two putative models have been put forward to explain this evolutionarily conserved dual function: (a) perception of LCOs factors modulates the balance between different LysM-RK receptor complexes, favoring a symbiotic complex at the expense of complexes required for immune responses, or (b) tight regulation of the receptor complexes at the posttranslational level, involving rapid endocytotic turnover, subsequently prevents activation of defense responses (Limpens et al. 2015). NF perception leads to root hair deformation and to changes in the root hair cytoskeleton (within 3–5 min) that are required for root hair curling (the so-called Shepherd’s crooks within 1–3 h) and invasion (Weerasinghe et al. 2005; Yokota et al. 2009). Mutations in genes coding for the NF LRR RLK (Leucine-rich repeat receptor-like kinases), the putative ion channels, or the nucleoporins abolish Ca2+ spiking and continued nodule development events; however, they maintain the Ca2+ fluxes and root hair deformation events (Kanamori et al. 2006; Miwa et al. 2006; Saito et al. 2007; Capoen et al. 2011; Morieri et al. 2013). In contrast, mutations in genes encoding for a calcium and calmodulin-dependent kinase called CCaMK or DMI3 do not affect Ca2+ fluxes and Ca2+ spiking events but block continued nodule development (Lévy et al. 2004; Miwa et al. 2006). This suggests that the NF LRR RLK, the ion channels, and the nucleoporins act downstream of NF perception, but upstream of Ca2+ spiking, whereas the CCaMK acts downstream of Ca2+ spiking (Limpens and Bisseling 2008). Within the nucleus, this sustained calcium spiking is decoded by CCaMK, which phosphorylates a transcriptional regulator CYCLOPS/IPD3 (Yano et al. 2008; Kouchi et al. 2010; Singh et al. 2014). CYCLOPS, together with other TFs belonging to the GRAS (NSP1 and NSP2) (Smit et al. 2005; Oldroyd and Downie 2008), ERF (ERN1) (Cerri et al. 2012; Rose et al. 2012), and the nodule inception (NIN) activator (Marsh et al. 2007) families, modulates early symbiotic gene expression (like ENOD2, ENOD40, ENOD11) for infection thread (IT) formation and polar tip growth (Yano et al. 2008; Madsen et al. 2010). Downstream of DMI3 and NIN, members of Nuclear Factor Y family, i.e., NF-YA1 and NF-YA2 (a CCAAT-box-binding heterotrimeric TF complex), act as early symbiotic regulators of ENOD11 (Cerri et al. 2012; Laloum et al. 2014). The secondary induction of Nod signals by flavonoids inside the roots is thought to be responsible for an additional level of host specificity. Thus, flavonoids play a multitude of roles during the process of nodulation (Subramanian et al. 2007).
8.3.3 Formation of Nodules: The Conjugal Lodging
NF perception in the epidermis activates a series of events, including polarized root hair tip growth, invagination associated with bacterial infection, and the promotion of cell division in the cortex leading to the nodule meristem (Oldroyd et al. 2011; Gourion et al. 2015; Laplaze et al. 2015). Cytoskeletal rearrangements have been reported in pericycle cells of M. truncatula within just 16 h of rhizobia inoculation (Timmers et al. 1999), and ENOD40 expression is reported in cortical cells within just 24 h of rhizobia inoculation (Mulder et al. 2005; Murray 2011). Thus, coordination between epidermal and nodule organogenesis seems to be crucial for successful nitrogen-fixing nodule formation (Oldroyd and Downie 2008). Although initial bacterial infection events do not require nodule primordia formation, it is subsequently required to direct infection thread growth (Oldroyd et al. 2011; Rose et al. 2012). To achieve such rapid mitotic activity in the underlying cortical cells after exposing the outer root to rhizobia/NF, role of auxin and cytokinin is imperative (Ryu et al. 2012).
8.3.3.1 Rhizobial Invasion
The host plant permits rhizobium to enter root tissues through plasma membrane-derived conduits called infection threads (ITs) (Jones et al. 2007; Fournier et al. 2008). The new growth can result in the root hair curling/bending, which results in NF-producing bacteria becoming entrapped between appressed cell walls, forming so-called infection pocket (Murray 2011; Wang et al. 2012). The rhizobia entrapped in these infection pockets continue to divide, forming colonies that are referred to as infection foci from which root hair ITs start to develop (Oldroyd et al. 2011). This results in elevated Nod factor concentrations, which are thought to be required to reach a Nod factor threshold concentration (Oldroyd and Downie 2008). The invaginating plant cell wall, along with the extended plasma membrane, grows as a hollow tube within the root hair cell and the bacteria multiply within the polar centripetally growing infection threads, where new cell wall and membrane material are being synthesized at their tip (Xie et al. 2012; Haag et al. 2013).
A third set of signals are represented by other rhizobial products necessary for continued infection thread development and/or preventing defense mechanisms (López-Baena et al. 2016). Among them are extracellular lipopolysaccharides (LPS), extracellular polysaccharides (EPS) and related compounds, as well as proteins exported by the type III secretion system (T3SS) (Limpens and Bisseling 2008; Haag et al. 2013). EPS facilitate attachment of bacterial cells to both biotic and abiotic surfaces and biofilm formation due to hydrophobic interactions and heterogeneity of the envelope surface (Janczarek et al. 2015), affect different stages of the organogenesis of nodules (Kelly et al. 2012), and along with LPS also facilitate suppression of defense response (Dalla Via et al. 2016). Further, bacterial effector proteins delivered from rhizobia into the plant cytosol through a T3SS or T4SS can act to either negatively or positively modulate nodulation, i.e., alter the symbiotic state toward pathogenesis or vice versa (Nelson and Sadowsky 2015; Tóth and Stacey 2015). The suppression of the MAMP-triggered immunity through a T3SS constituted the first evolutionary step toward symbiosis (Gourion et al. 2015; Yamazaki and Hayashi 2015; Okazaki et al. 2016). Interestingly, rhizobial NF, T3SS, and T4SS depend on a common regulator activated by legume-secreted flavonoids (Janczarek and Skorupska 2011; Gourion et al. 2015; Smith et al. 2015). Role of flavonoids in protein secretion in ITs suggests that the same keys can unlock different doors (Broughton et al. 2003). Growth in the ITs is critical stage for selection of competitive rhizobia, and this stage provides checkpoint for the plant because bacteria mutated in cell wall components such as LPS, EPS, as well as BacA either fail to be released from ITs or fail to form mature symbiosomes (Gibson et al. 2008; Janczarek et al. 2015).
Concomitantly, certain cortical cells divide to form nodule primordia, and it is toward these primordia that the infection thread grows. After initiating the symbiotic dialog, flavonoids function as positional signals for cell division and/or growth in nodulating roots. This function was inferred because both the induction of a chalcone synthase-GusA (CHS–GusA) fusion and the accumulation of flavonoids occurred at the site where either purified Nod factor or nodulating rhizobia strains (but not of non-nodulating strains) were applied (Mathesius et al. 1998). Genes encoding phenyl propanoid biosynthesis enzymes including Chalcone-O-Methyltransferase (required for the production of the potent nod gene inducer 4,4-dihydroxy-2-methoxychalcone) not only express in rhizobially infected root hairs but also in nodule infection zone (not in the nitrogen fixation zone) (Chen et al. 2015). Some intriguing effects of plant phenolics are the ones associated with long-distance polar auxin transport (PAT) streams (Taylor and Grotewold 2005; Peer et al. 2011). Evidence that flavonoids regulate auxin accumulation in vivo was obtained using the flavonoid-deficient mutant, tt4, where accumulation of [14C] indole-3-acetic acid in whole seedling was defective as a considerable amount of auxin escaped from the roots. Treatment of the tt4 mutant with the missing intermediate naringenin restored normal auxin distribution and accumulation by the root (Murphy et al. 2000). Silencing of lignin biosynthetic gene in Arabidopsis thaliana led to redirection of metabolic flux into flavonoid synthesis through chalcone synthase activity. The level of plant growth reduction of HCT-deficient plants was correlated with the inhibition of auxin transport, while suppression of flavonoid accumulation by chalcone synthase repression in HCT-deficient plants restored normal auxin transport and wild-type plant growth. Thus, reduced size phenotype of HCT-silenced plants is not due to the alteration of lignin synthesis but to flavonoid accumulation (Besseau et al. 2007). The molecular interplay of flavonoids and Nod factors is likely to occur at several stages during nodule ontogeny (Taylor and Grotewold 2005). Nod factors bring about an immediate and transient inhibition of PAT in a highly localized fashion, possibly through the action of specific flavonoids, resulting in a change in auxin concentrations and subsequent stimulation of cell divisions at the site of nodule initiation (de Billy et al. 2001; Laplaze et al. 2015). Wasson et al. (2006) identified that accumulation of auxin and nodule formation was restored in the naringenin and liquiritigenin-supplemented chalcone synthase (CHS)-silenced root cultures, thereby indicating that the ability of nodule-forming rhizobia to inhibit auxin transport was flavonoid dependent. Zhang et al. (2009) silenced different flavonoid biosynthesis enzymes to generate transgenic M. truncatula roots with different flavonoid profiles. Silencing of chalcone synthase led to flavonoid-deficient roots, while silencing of isoflavone synthase and flavone synthase led to roots deficient for a subset of flavonoids, isoflavonoids (formononetin and biochanin A), and flavones (7,4-dihydroxyflavone), respectively. When tested for nodulation by S. meliloti, flavonoid-deficient roots had a near complete loss of nodulation, whereas flavone-deficient roots had reduced nodulation. Isoflavone-deficient roots nodulated normally, suggesting that isoflavones might not play a critical role in M. truncatula nodulation, even though they are the most abundant root flavonoids. Supplementation of flavone-deficient roots with 7,4-dihydroxyflavone, a major inducer of S. meliloti nod genes, completely restored nodulation. However, the same treatment did not restore nodulation in flavonoid-deficient roots, suggesting that other non-nod gene-inducing flavonoid compounds are also critical to nodulation. Supplementation of roots with the flavonol kaempferol (an inhibitor of auxin transport), in combination with the use of flavone-pretreated S. meliloti cells, completely restored nodulation in flavonoid-deficient roots. These observations indicated that flavones might act as internal inducers of rhizobial nod genes and that flavonols might act as auxin transport regulators during nodulation. However, all flavonoids have not been shown to exhibit auxin transport inhibition. The flavonol subclass in particular, such as kaempferol and quercetin, shows the strongest inhibitory activity (Ng et al. 2015).
Several possible mechanisms by which flavonoids modulate auxin transport have been reported. Acropetal auxin transport in the root, as well as basipetal auxin transport in the inflorescence, hypocotyl, and root, is all elevated in the absence of flavonoids. Flavonoids, such as quercetin, apigenin, and kaempferol, do not directly compete with IAA but are implicated as inhibitors of IAA transport across the plasma membrane by binding to a plasma membrane protein known as NPA receptor (Cooke et al. 2002). Rhizobia-induced flavonoids could bind to the auxin transporters AtMDR (Multidrug resistance), AtAPM (Aminopeptidase M1), and AtPINs (Pin-formed) (Peer et al. 2004) and prevent their intracellular trafficking, i.e., negatively regulate PAT in vivo (Buer and Muday 2004; Taylor and Grotewold 2005). Another possible mechanism is that flavonoids alter either the amount of synthesis or localization of auxin transport proteins, perhaps by phosphorylation of transcription factors that control the synthesis of these auxin carriers (Buer and Muday 2004). In addition to the participation of flavonoids in the polar transport of auxins, flavonoids are involved in the inhibition of auxin breakdown by peroxidases. Monohydroxy B-ring flavonoids are suggested as cofactors of peroxidase functioning as an IAA oxidase (destroys the hormone), whereas dihydroxy B-ring forms act as inhibitors of IAA-degrading activity (Mathesius 2001). Similarly, Agati and Tattini (2010) postulated that the high light-induced biosynthesis of flavonoids may have a role in regulating whole-plant and individual organ architecture. Pollastri and Tattini (2011) confirmed this as high sunlight-induced synthesis of quercetin derivatives fine-tuned auxin gradients as well as local auxin concentrations which represent the actual determinants for different morphological responses.
As IT reaches the base of the root hair cell, the nucleus in the adjacent cortical cells starts to reposition itself and a centrally located cytoplasmic bridge forms to establish pre-IT in this cell (Timmers et al. 1999). This process is repeated at each cell junction, thereby extending transcellularly, through a tip growth-like mechanism, from the epidermis or subepidermal cortex toward a subtending region of dividing cortical cells that have initiated the formation of a nodule primordium (NP) (Fournier et al. 2008; Haag et al. 2013). NSP1/2 and NIN not only act downstream of CCaMK in the epidermis, but they also act downstream of CCaMK and the cytokinin receptor in the cortex (Marsh et al. 2007; Suzaki and Kawaguchi 2014; van Zeijl et al. 2015). ENOD11 and ENOD12 encode hydroxyproline-rich glycoproteins (HyPRPs), with relatively few tyrosine residues, which enhance cell wall plasticity or in components of the infection thread matrix. Hence, very rapidly and even before contact, legume root cells are paving the way for the accommodation of their symbiotic partner by remodeling the cell wall barrier (Rose et al. 2012; comprehensively reviewed by Rich et al. 2014). Cytokinin receptor LjLHK1/MtCRE1 is essential for Nod factor-induced gene expression and subsequent nodule primordium formation. Miri et al. (2016) postulated that cytokinin participates in orchestrating signaling events that promote rhizobial colonization of the root cortex and limit the extent of subsequent infection at the root epidermis by systemic autoregulation of nodulation (AON), thus maintaining homeostasis of the symbiotic interaction. Recent studies have revealed that exogenously applied cytokinin can bypass Rhizobium Nod factor signaling and the symbiotic phenotype of the Mtcre1 mutant can be complemented, at least in part, by the exogenous application of flavonoids, suggesting that flavonoids mimic cytokinin functioning or even act as small secondary molecules downstream of cytokinin (Ng et al. 2015).
8.3.3.2 Nodule Organogenesis into Symbiosome: The Unifying Feature of Endosymbioses
Eventually rhizobia are released from the growing tip of ITs into an infection droplet in the cytoplasm [a subset of nascent nodule primodia (NP) cells], where they undergo differentiation to bacteroids in confined facultative organelle-like compartments called symbiosomes (SMs) (Kereszt et al. 2011). The rapid growth of bacteria at the tip tends to select out a single bacterial strain even if more than one strain becomes entrapped initially. This selection even works with two near-identical rhizobial strains in the infection (Gage 2002) and can select against potential “cheaters,” which will attempt to coinfect the nodule niche without conferring any benefit to the plant (Downie 2010). Through a process resembling endocytosis, the bacteria are surrounded by a plant-derived membrane, called the peribacteroid membrane, which forms SMs (McNear 2013). To accommodate a high number of endosymbionts, extreme plant cell enlargement is the consequence of repeated endoreduplication (ER) of the genome without mitosis (Kondorosi and Kondorosi 2004; Ribeiro et al. 2015). The bacteroids multiply in the growing host nodule cells to a certain cell density, adapt to the endosymbiotic lifestyle and microaerobic conditions, and mature to nitrogen-fixing bacteroids which convert atmospheric nitrogen gas into ammonia, using the nitrogenase enzyme complex (Ferguson et al. 2010). The couple jointly needs to modify their house especially to insulate the microsymbiont from the plant cytoplasm and to lower oxygen tensions for nitrogen fixation. One essential modification to the corridor is seen as the peribacteroid or symbiosome membrane in infected cortical cells (Broughton et al. 2000). Thus, rhizobia remain outside the plant cytoplasm and are engulfed in a symbiosome membrane, which functions to regulate nutrient exchange between the partners (Mathesius 2009). In nodulating plants, flavonoids have been observed to delimit the border between symbiotic sites and non-symbiotic plant tissue. Another notable feature in these barrier regions is the accumulation of histidine-rich glycoproteins, such as ENOD2, which are capable of cross-linking with flavonoids (Stafford 1997). The existence of this barrier in legumes has been postulated to act as an O2− diffusion barrier that helps to prevent inactivation of nitrogenase in bacteroids (Cohen et al. 2001).
Two major morphological types of nodules are determined by the host legumes: determinate and indeterminate. Differences between the two nodule types are the site of first internal cell divisions, maintenance of a meristematic region, and the form of the mature nodules (Oldroyd et al. 2011; Hichri et al. 2016). Different roles of flavonoids in regulating auxin homeostasis for development of determinate and indeterminate nodules have been reported (Wasson et al. 2006; Subramanian et al. 2007; Zhang et al. 2009). Silencing the flavonoid pathway in M. Truncatula (forming indeterminate nodules) prevented localized auxin transport inhibition, and thus, flavonoid-deficient roots were unable to initiate nodules, even though normal root hair curling was observed (Wasson et al. 2006). However, Subramanian et al. (2005) silenced isoflavone synthase, the entry point enzyme for isoflavone biosynthesis in soybean (forming determinate nodules). Isoflavonoid-depleted roots of the determinate nodule forming legume soybean were also deficient in both auxin transport inhibition and nodulation. However, nodulation in isoflavone-deficient soybean roots could be restored by using isoflavone-hypersensitive rhizobia cells, suggesting that induction of Nod signal biosynthesis by isoflavones is crucial, whereas auxin transport inhibition by isoflavones is not. Consistently, during indeterminate nodule formation, flavonoid-regulated auxin transport inhibition occurs at the site of rhizobial infection, whereas during determinate nodule formation there is no significant auxin transport inhibition (Subramanian et al. 2007).
8.3.4 Role of Flavonoids in Autoregulation of Nodulation
At times of sufficient nitrogen supply to the legumes, rhizobial symbiosis can be limited by both local and systemic feedback regulatory mechanisms (Mathesius 2009). Since nodulation and the subsequent nitrogen fixation are energy-intensive processes, host plant maintains a balance between cost and benefit by tightly controlling the number of nodules it forms, via a complex root-to-shoot-to-root signaling loop called AON (Herder and Parniske 2009; Magori and Kawaguchi 2009). There are various root [e.g., CLAVATA3/ESR-related (CLE) peptide] and shoot (e.g., SDI) derived inhibitors identified in autoregulation of nodulation (Reid et al. 2011). Rossen et al. (1987) recognized four classes of mutations in the nodD gene of Rhizobium leguminosarum biovar viciae that can affect its ability to autoregulate and/or activate other nod genes in the presence of flavonoid inducers. Class I mutations led to defects in both autoregulation and activation of other nod genes in the presence of inducers, class II mutations left activation of the transcription of nodABC and nodEF unaffected but virtually abolished autoregulation, and class III mutations affected the induction of other nod genes but not autoregulation. More adversely, class IV mutations showed the activation of other nod genes even in the absence of inducer molecules. Some of the anti-inducers also acted as inducers in the strains with class IV nodD mutations, thereby pointing toward the direct interaction of nodD proteins with the flavonoids. All the mutations led to decrease in nodule number; however, class IV mutations even failed to fix N2 (even when a wild-type copy of nodD was present in the strain). This suggests that continued transcription of nodABCIJ and/or nodEF by bacteria in the nodule interferes with their ability to develop in N2-fixing bacteroids (Rossen et al. 1987). Catford et al. (2006) identified that levels of formononetin and its 7-O-glucoside ononin systemically suppressed in response to S. meliloti and NFs, suggesting that these isoflavonoids are probably involved in the systemic suppression of nodulation as their application to autoregulated roots again promoted nodulation (Fig. 8.2).
8.4 Flavonoids in Arbuscular Mycorrhizal Symbiosis
Phosphorus (P) is the second most important macronutrient, next to nitrogen; however, P is the least accessible and hence most frequently deficient macronutrient limiting growth of crop plants (Balemi and Negisho 2012). Further, excess P supply in the soil, as chemical fertilizers, is another major environmental concern as it increases the risk of P movement to surface and groundwaters (Grant et al. 2005). To this end, arbuscular mycorrhiza (AM) fungi could be of great benefit in enhancing soil P utilization efficiency of the crop and improve nutritional status of the crop (Aggarwal et al. 2011). Unlike rhizobial symbiosis, mycorrhizae are the rule in nature, not the exception. In this mutualistic relationship, mycorrhizal hyphae (up to 100 times longer than root hairs) act as an extension of the root system, extending to soil beyond the P depletion zone (Mohammadi et al. 2011; Balemi and Negisho 2012). Thus, mycorrhization aids roots by providing a more widespread nutrient foraging system to plants, and this foraging provides fungus, an avenue to find new hosts (Denison and Kiers 2011; Bücking et al. 2012). A better understanding of this microbial interaction is therefore a potential key to determine the profitability and sustainability of agricultural systems (Mohammadi et al. 2011). Thus, here we review the mechanisms by which the plant signals (especially flavonoids) allow fungal infection and promote mutualistic symbiosis.
8.4.1 Unfolding Role of Flavonoids in Signaling Imputed to Arbuscular Mycorrhiza (AM) Development
Round-shaped thick cell-walled multinucleate fungal spores germinate to form haploid coenocytic hyphae (Hijri and Sanders 2005), when environmental soil conditions such as matric potential, temperature, and CO2 levels are favorable (Mandal et al. 2010). This phase of growth in the absence of signal from the plant is known as asymbiotic stage (Requena et al. 2007). During this time the fungal colonies, living mainly on its triacylglyceride reserves, lengthen a few centimeters showing a characteristic growth pattern with marked apical dominance and sporadic hyphal branching. AM spores are surprisingly dynamic, and in the absence of a host root, growth ceases where hyphal septation from the apex arrests development and retracts protoplasm back into the spore within 8–20 days, becoming dormant again before consumption of the spore reserves (Bonfante and Genre 2010; Giovannetti et al. 2010). Therefore, even though spores are competent to germinate without the presence of a host, but a switch from the aymbiotic stage of development to an active presymbiotic growth phase triggers only in response to initial recognition by multifaceted fine-tuned reciprocal signaling events (Gachomo et al. 2009). Prior to colonization, significant developmental step in the life cycle of mycorrhizal fungi is host signal interceded increase in hyphal branching and metabolic activity, which ensures directional growth of hyhal branches toward host root (Pinior et al. 1999).
Germ tubes, with limited growth potential, respond to the presence of “branching factor” in host root exudates in their vicinity, where fungal morphology shifts toward enhanced hyphal growth and extensive hyphal branching (Buee et al. 2000). The branch-inducing factor is present in root exudates of all the mycotrophic plants but absent in those of nonhost plants. Although no directional growth has been observed toward the root, several experiments showed that host-derived signals intensify hyphal ramification, thereby increasing the probability of contact with a host root (Requena et al. 2007). By analogy to the Rhizobium–legume symbiosis, some of the potential host exudates that stimulate spore germination, hyphal branching in the soil, and fungal invasion and arbuscule formation inside the root, often in a symbiont-specific manner, have been identified as flavonoids (Scervino et al. 2007; Steinkellner et al. 2007). These chemical signals exuded by the plant and the thigmotropic signals from the rhizodermis are possibly recognized by receptor proteins associated with the fungal plasma membrane (Requena et al. 2007). Interestingly, Siqueira et al. (1991b) suggested that flavonoids can stimulate fungal growth directly or remove AM fungal self-inhibition. Pyranoisoflavones produced by white lupin, which is not a host for mycorrhizal fungi, inhibited hyphal branching of mycorrhizal fungi, suggesting that flavonoids could play both stimulating and inhibitory roles on fungal symbionts in the soil (Akiyama et al. 2010). Martens and Mithöfer (2005) considered the absence of flavone biosynthesis pathway in Brassicaceae as one of the reasons of this family lacking mycorrhizae. Thus, it is likely that host and nonhost plants can modulate the establishment of symbiosis by altering the profile of flavonoid exudates (Hassan and Mathesius 2012).
A variety of flavanones (hesperetin and naringenin), flavones (apigenin), and isoflavones (formononetin) have been reported to stimulate spore germination and hyphal growth of AM fungi in vitro (Gianinazzi-Pearson et al. 1989; Tsai and Phillips 1991; Nair et al. 1991). Effect of these compounds on AM fungi is flavonoid type and concentration, AM fungal species and genera, and host specific (reviewed by Vierheilig et al. 1998). At low concentration, different flavonoids can increase AM fungal spore germination, hyphal growth, and hyphal branching, while at high concentration, the same flavonoid turns inhibitory (Nair et al. 1991; Baptista and Siqueira 1994). Siqueira et al. (1991b) reported that isoflavonoids (formononetin and biochanin A) at concentrations of 5 mg l−1 stimulated AM colonization in T. repens L., while flavone chrysin increased root colonization at concentration 40–60 mg l−1. Morandi et al. (1992) tested the effect of 2 isoflavonoids (glyceollin I and coumestrol at concentration 0, 0.05, 0.5, 5, and 50 μM) and 1 flavonoid (quercetin at concentration 0, 0.1, 1, and 10 μM) on in vitro spore germination of Gigaspora margarita. After 5 and 7 days, number of germ tubes per spore was slightly increased by glyceollin I; mycelium length from germinated spores was increased by low concentrations of glyceollin I but was significantly decreased at the highest concentration. A positive correlation was found between coumestrol concentration and mycelium length, while vesicle number decreased by coumestrol, quercetin, and the highest concentration of glyceollin (but was increased by glyceollin at 0.5 μM). Thus, similar to the case of nod gene induction, it is probably not simply the absence or presence of a specific flavonoid in root exudates which determines the signal properties, but rather a tightly controlled concentration-specific release pattern of inducers and anti-inducers.
Bécard et al. (1992) reported that only the flavonols stimulated in vitro growth of germinated spores of Gi. margarita, while the flavones, flavanones, and isoflavones tested were generally inhibitory. Quercetin (10 μM) prolonged hyphal growth from germinated spores of Gi. margarita, while the glycosides of quercetin, rutin and quercitrin, were not stimulatory, thereby indicating the role of structure of flavonoid in AM colonization. In general, at least one hydroxyl group on the B aromatic ring has been found to be necessary for stimulatory effect. Besides this, a hydroxyl on position 3 is also essential to confer stimulatory activity to the flavonol molecule, as the flavones luteolin and apigenin, lacking this hydroxyl, showed no effect (Bécard et al. 1992). Glycosylation at position 3 also promoted the loss of the stimulatory activity, as in quercitrin and rutin. Moreover, it has been suggested that saturation of the 2,3 double bond in flavonols, as seen in flavanones, promoted the loss of activity on hyphal growth of Gi. margarita (Chabot et al. 1992). This loss of activity was attributed to the loss of the planar configuration of the flavonol molecule when the double bond disappears; e.g., dihydroquercetin and dihydrokaempferol showed no effect on hyphal growth on Gi. margarita. However, these results are in contrast with those of Gianinazzi-Pearson et al. (1989) and Baptista and Siqueira (1994). Naringenin, lacking the 2,3 double bond, was stimulatory with Gi. margarita and Gi. gigantea, whereas apigenin, differing only by the presence of the 2,3 double bond, showed no effect with Gi. gigantea. In general, Glomus spp. are stimulated by flavonols as well as by isoflavones (Vierheilig et al. 1998). Whereas some flavonoids such as quercetin exhibit a general stimulatory effect on the hyphal growth of different AMF genera (Chabot et al. 1992, Baptista and Siqueira 1994), data with biochanin A showed a stimulation of Glomus (Nair et al. 1991; Vierheilig et al. 1998) but not Gigaspora species (Chabot et al. 1992, Baptista and Siqueira 1994).
8.4.2 Myc Factor Signaling Pathway in the Root Epidermis
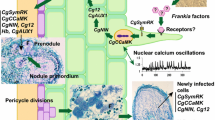
Branched fungal hyphae secrete fungal chitin elicitor to be called “Myc factor” that drive various morphological and physiological changes in the hosts (Requena and Breuninger 2004), to orchestrate the AM infection process by counteracting the plant immune program and upregulating the expression of symbiosis-related genes (Kloppholz et al. 2011). At an early stage of mycorrhizal formation, plant constitutive chitinases may partially cleave these elicitors and thus inducing transient plant defense response (Antunes and Goss 2005; Maillet et al. 2011). However, unlike fungal pathogens, diffusible Myc factors from AM increase lateral root formation, elicit a transient cytosolic calcium elevation within a few minutes, and induce expression of specific genes in only specific root cells in direct contact with the penetrating fungus (Navazio et al. 2007), while a suppressor activity is induced in non-colonized neighboring cells (Kosuta et al. 2003; Genre et al. 2005).
Several mutants in legumes like M. truncatula, Lotus japonicus, and pea are deficient for nodulation and AM, indicating the existence of a conserved symbiotic (Sym) pathway required for the establishment of both symbioses (Mitra et al. 2004b; Kistner et al. 2005). One common signaling component is the receptor-like leucine-rich kinase SymRK (MtDMI2/MtSYM2) that is involved in the direct or indirect transduction of fungal or rhizobial signals through its intracellular kinase domain to the cytoplasm (Stracke et al. 2002). Evolutionarily more recent Nod factors also overlap with Myc symbiotic signals, both structurally and functionally (Laparre et al. 2014; Limpens et al. 2015). Three nucleopore-associated proteins: NUP85, NUP133, and NENA (Saito et al. 2007) act downstream and could be involved in the transport of CASTOR and/or DMI1/POLLUX (ion channels) to the inner nuclear envelope (Riely et al. 2007). In both the symbioses, these channels lead to calcium oscillations in the nucleus and perinuclear cytoplasm for activating calcium-calmodulin-dependent protein kinase MtSYMI3/DMI3/CCAMK (Mitra et al. 2004a). CCAMK is known to phosphorylate the last identified SYM gene CYCLOPS, encoding IPD3/CYCLOPS protein which interacts with DMI3 (Messinese et al. 2007). In order to activate the appropriate symbiotic program, legumes have to discriminate the two types of symbiotic signals. CYCLOPS represents a branch point in the common SYM pathway, as infection threat formation and arbuscular development are CYCLOPS dependent, but nodule organogenesis is CYCLOPS independent (Yano et al. 2008). Microbial activation of the common symbiosis signaling pathway can also modulate innate immune responses through its effect on the hormonal landscape, where Limpens et al. (2015) speculated a central role for DELLA proteins, in part by influencing the salicylic acid–jasmonic acid balance. However, profound mycorrhization in roots of cre1 mutant of M. truncatula suggested that MtCRE1 (Cytokinin Response 1 required for N2 fixation) does not belong to the ancestral common symbiotic pathway (Laffont et al. 2015). In addition to the Sym pathway, a parallel pathway exists and mediates AM signaling in non-nodulating eudicots and monocots (Mukherjee and Ané 2011).
Further, hyphal contact with host roots has been reported to induce the flavonoid pathway in a number of host species, in particular in infected cells (Steinkellner et al. 2007; Abdel-Lateif et al. 2012). Harrison and Dixon (1993) reported that in M. truncatula + G.versiforme interactions, the most striking changes in identified root metabolites included the overall accumulation of formononetin malonyl glucoside (FGM), medicarpin malonyl glucoside (MGM), and daidzein and a transient increase in free medicarpin (the major phytoalexin from MGM) in the early stages of the interaction, as a defense response to fungal elicitors. In mycorrhizal state, either the fungal symbiont may be consuming theses phytoalexin precursors as carbon precursors or conjugation of medicarpin may inactivate or remove a potentially toxic metabolite. In contrast to the levels of phenylalanine ammonia lyase (PAL) and CHS transcripts, which remained elevated throughout the interaction, the level of Isoflavone reductase (IFR) transcripts decreased in later stages of the interaction to 2.5-fold below the level in uninoculated control roots, suggesting that the established mycorrhizal association in M. truncatula brought about a specific suppression of this transcript to decline medicarpin levels in these roots. However, in the interaction between myc− line and G. versiforme the levels of medicarpin and related transcripts remained elevated throughout the interaction suggesting that a defense response was occurring. Further, the inability of myc− M. sativa line to form a complete interaction may be attributed to the absence of coumestrol and 4′,7-dihydroxyflavone (Harrison and Dixon 1993). Studies of Guenoune et al. (2001) and Akiyama et al. (2002) on flavonoids and AM reopened the discussion about the involvement of flavonoids as regulatory compounds during signaling of mycorrhization. Guenoune et al. (2001) demonstrated that medicarpin-3-O-glucoside, an isoflavonoid phytoalexin, accumulated both in roots colonized by the pathogenic fungus and in AM-treated roots receiving high P and prevented these roots from being colonized by AMF. Increases in the steady-state levels of chalcone isomerase and IFR mRNAs, as defense responses of alfalfa roots to the pathogenic fungus Rhizoctonia solani, were reduced significantly in roots simultaneously infected with the arbuscular mycorrhizal (AM) fungus Glomus intraradices. These data suggested that during early stages of colonization by G. intraradices, suppression of defense-related properties is associated with the successful establishment of AM symbiosis. Akiyama et al. (2002) detected the C-glycosylflavone—isovitexin 2″-O-β-glucoside in non-mycorrhizal, P-deficient melon roots but not in roots with a high P status or in mycorrhizal roots (inoculated with G. caledonium). Application of this flavonoid (at concentrations of 20 and 50 μM) to AM plants not only enhanced root colonization in plants grown under low P conditions, but also in plants grown under high P conditions, thus clearly showing that a high P status or the mycorrhizal status of a plant can reduce the accumulation of a flavonoid in roots which stimulates mycorrhization.
Tsai and Phillips (1991) reported that after 21 days of application, 1.0–2.5 μM quercetin-3-d-galactoside, 4′,7-dihydroxyflavone, and 4′,7-dihydroxyflavanone promoted spore germination, hyphal elongation, and hyphal branching of Glomus etunicatum and Glomus macrocarpum in vitro. On the other hand, formononetin, an isoflavone that is released from stressed alfalfa roots, inhibited germination of both Glomus species. However, flavonoids tested in this study were identified as products from non-AM alfalfa seedlings only, strongly advocating that AM fungi alter flavonoid metabolism in plants and compounds produced as a result of infection have a role in differentiation of vesicles, arbuscules, and/or spores (Tsai and Phillips 1991). Again, application of biochanin A and formononetin in the rhizosphere of AM nonhost plants—Lupinus polyphyllus and Spinacia oleracea (grown in the presence of G. mosseae)—resulted in hyphal attachment and more hyphae around these roots (Vierheilig and Piché 1995). These results could indicate a simple hyphal growth stimulation to be responsible for the enhanced root colonization in AM host plants; however, as soil application also resulted in a colonization of L. polyphyllus roots by G. mosseae (Vierheilig and Piché 1995; Vierheilig et al. 1996), a role of these compounds in the plant–fungus communication during the colonization stage is suggested (Vierheilig et al. 1998). Vierheilig and Piché (2002) presented a complex model for the role of flavonoids during the AM association. They proposed that a fundamental molecular dialogue occurs that regulates not only the early development of AM symbiosis but also subsequent colonization which has to be balanced for establishment of genuine mycorrhizal symbiosis.
8.4.3 AM Hyphal Invasion and Differentiation of Intraradical Hyphae: The Conjugal Lodging
After hyphal docking to cell surface by means of an appresorium, the fungus enters into root epidermis via AM-specific prepenetration apparatus (Bonfante and Genre 2010). AM fungi first grow intercellularly or cross outer cells with linear or simple coiled hyphae (Gianinazzi and Gianinazzi-Pearson 1988). Reaching the inner cortex the fungal symbionts switch to different mode of colonization and penetrate the plant cell, where between cell wall and plant plasma membrane fungal hyphae extensively ramify to form a dichotomously branched haustorium, called “arbuscule” with a high surface to volume area (Gutjahr and Parniske 2013). Although the arbuscule eventually expands to largely fill the cortical cell, the hyphal tree does not enter the plant symplast. This plant–fungal interface consists of the fungal cell wall, the perihyphal or periarbuscular membrane (PAM), and a matrix between the two organisms—periarbuscular space (Pumplin and Harrison 2009; Rich et al. 2014). Hofferek et al. (2014) reported that in the root cortex nodulation and mycorrhization are regulated by NSP2 whose spatiotemporal expression negatively regulates both types of root endosymbioses through perception of the nutritional status (such as high Pi or N) of the host plant. VAPYRIN also promotes intracellular accommodation of endosymbionts by interacting with membranes/cytoskeleton and is thus required for intracellular accommodation of AM fungi and rhizobia in epidermal as well as cortical cells (Feddermann et al. 2010; Gobbato 2015). Shtark et al. (2016) indicated that as compared to wild type (wt), though non-nodulating pea mutants (sym7, sym11, sym14) had considerable increase in root surface colonization by Rhizophagus irregularis, a substantial decrease in internal colonization (especially arbuscule formation) was recorded. sym34 mutants displayed strongly reduced root surface colonization, but 10 days later internal colonization did not differ from wt. While sym38 mutant did not differ from wt indicating that except SYM38, all analyzed genes were essential for both nodule and AM development (Shtark et al. 2016). Various homologues of MtCBS1/2 gene, to encode protein containing a cystathionine-b-synthase (CBS) domain in ITs, act downstream NIN in rhizobial infection, in nodule organogenesis, as well as in mycorrhization, again suggesting common infection mechanisms (Sinharoy et al. 2016).
Harrison and Dixon (1993) reported that roots of the mycorrhizal resistant M. sativa mutant (Myc−) do not become colonized, but hyphae grew attached to the root surface forming appresoria. In the presence of G. versiforme, levels of several flavonoids (diadzein, formononetin, FGM, medicarpin, MGM) increased in the M. sativa (Myc−) roots in the same way as in colonized root of the wild-type (Myc+) M. sativa (Harrison and Dixon 1993). Volpin et al. (1994) also found an increase of formononetin in inoculated but still uncolonized roots of M. sativa (Myc+). Thus, these two researches suggested that these flavonoids play a role in the plant–fungus signaling during precolonization and cell-to-cell stage. Further, Harrison and Dixon (1993) also demonstrated that some flavonoids are newly induced after AM fungal root colonization, where coumestrol and 4′,7-dihydroxyflavone were only present in colonized roots of (Myc+) M. sativa and M. truncatula, but were not detected in uncolonized roots of the two plants and in roots of inoculated (Myc−) M. sativa. This further suggested that these flavonoids play a signaling role during the intraradical phase of the hyphae in the AM symbiosis and might be an indication for its role in the formation of highly ramified arbuscules. Naringenin found in root exudates of bean (Hungria et al. 1991) enhanced in axenic culture of Gi. margarita, where the number of vesicle clusters per germinated spore increased. Earlier in vitro experiments by Gianinazzi-Pearson et al. (1989) evidenced similar effects with apigenin and hesperetin. Luteolin, found in the seed rinse of alfalfa (Hartwig et al. 1990), although showing no stimulation of hyphal growth, stimulated the production of auxiliary cells in Gi. margarita (Bécard et al. 1992). Thus, hyphal differentiation possibly requires a different stimulatory mechanism or induction than do hyphal growth. Some flavonoids may be involved in only one of these mechanisms and not in the other. However, quercetin and myricetin, which exhibit a hyphal growth stimulating effect on different AM fungal genera, also enhanced the formation of auxiliary cells (Bécard et al. 1992), suggesting again a more general role of these two compounds compared to other flavonoids. Sometimes similar changes could be observed within a plant family. The level of coumestrol and daidzein increased in colonized roots of Glycine max, M. sativa, and M. truncatula (Morandi et al. 1984; Harrison and Dixon 1993), all belonging to the leguminosae family, and the level of blumenin was enhanced in colonized roots of several members of the Gramineae (Maier et al. 1995). Change of flavonoids level also depends on the AM fungal species, i.e., different AM fungi have different requirements for their development and thus induce different levels of these compounds or that the plant recognizes the two fungi differently. Ponce et al. (2004) reported that 5,6,7,8,9-hydroxy chalcone (NM7), 3,7-hydroxy-4′-methoxy flavone, 5,6,7,8-hydroxy-4′-methoxy flavones (RR4), and 3,5,6,7,4′-hydroxy flavones (RR4-2) could be detected only in non-mycorrhizal roots of white clover, whereas the flavonoids acacetin, quercetin, and rhamnetin were only present in roots inoculated with G. intraradices.
Arbuscules act as the epicenter of mutualism as they represent an extreme form of intimacy and compatibility and are site of nutrient transfer from the fungus to the host plant (Hughes et al. 2008; Denison and Kiers 2011; Mohammadi et al. 2011). In return, host-derived carbon is transferred to the fungi and stored in energy-rich vesicles to support vegetative growth or spore formation (Genre and Bonfante 2010). Whereas a high frequency of arbuscules usually indicates cost-effective nutrient exchange in both directions, high vesicular colonization is a potential indicator of fungal resource hoarding prominently under high external nutrient conditions, when hosts are less dependent on fungal partners (Johnson 2010; Nijjer et al. 2010). Further, host molecules like lysophosphatidylcholine (LPc) potentially allow them to evaluate the amount of P delivered via the mycorrhizal pathway (Bucher et al. 2009). Host will decrease C provision or directly digest arbuscules when there is insufficient P being transferred to the host across the colonized cell (Kobae and Hata 2010). Resource exchange is followed by rapid arbuscule collapse in 4–5 days, with structures degenerating within 2.5–5.5 h (Bonfante and Genre 2010), much more rapidly than the decline in N2 fixation in nodules (Wong and Evans 1971). The fungal life cycle is completed after formation of asexual chlamydospores on the external mycelium, which allow them to propagate and survive in the absence of a host, for more than 10 years (Giovannetti et al. 2010) (Fig. 8.3).
8.4.4 Role of Flavonoids in Autoregulation of Mycorrhization
An already mycorrhizal plant develops one mechanism to repulse further colonization by fungi, i.e., to not differentiate between AMF and soilborne pathogenic fungi (Vierheilig and Piché 2002; Mechri et al. 2015). Flavonoids again play a role in autoregulating mycorrhization, as depending on the flavonoids and their concentration, they can exhibit an inhibitory and/or a stimulatory effect on fungi (Vierheilig et al. 1998; Subramanian et al. 2007; reviewed by Lira et al. 2015). Larose et al. (2002) found that after an application of a mixture of spores and extraradical hyphae of G. intraradices to M. sativa roots, coumestrol, ononin, and daidzein levels were increased, whereas formononetin was detected at lower levels, indicating that at this stage of symbiosis, i.e., during appressoria formation, fungal partner, with signals released by its mycelium, is actually asking for a change of the concentration of certain flavonoids, which exhibit stimulatory effects on fungi. The accumulation of medicarpin (defense phytoalexin) only at the stage of appressoria formation, when the fungus has formed its first structures in the root, indicates that once the fungal partner has formed its appressoria it was perceived in a similar way as a pathogen. Later the perception of fungi by the plant changed and at a stage of intense AM root colonization, no altered medicarpin accumulation was observed, while coumestrol accumulation further increased. Interestingly, formononetin/daidzein levels were linked inversely, as at the beginning of root colonization formononetin was reduced in mycorrhizal roots whereas daidzein increased and at the end of the experiment, when formononetin increased daidzein levels seemed to decrease. Besides coumestrol, medicarpin again accumulated at the end of colonization, thereby pointing toward the role of coumestrol at any stage of root colonization; medicarpin probably plays a role at the beginning of root colonization and at a later stage. There was a positive link between the accumulation of coumestrol, medicarpin, and/or formononetin at the later stages and the autoregulation of mycorrhization (Vierheilig and Piché 2002). Some species-specific quantitative changes were observed, as ononin was highly accumulated in roots colonized by G. mosseae, but only weakly accumulated in roots colonized by G. intradices while coumestrol was high in roots colonized by G. intradices but lower in roots colonized by G. mosseae. The only compound that accumulated to similar levels in all AMF-colonized roots was genistein, and the similar accumulation pattern of genistein in mycorrhizal roots independent of the root-colonizing AMF indicated a general, non-species- or non-genus-specific regulatory role of this compound during mycorrhization (Larose et al. 2002). Later on, Scervino et al. (2005a) reported that acacetin and rhamnetin inhibited the formation of AM penetration structures as well as colonization and pointed toward a possible involvement of these two flavonoids in the autoregulation process of the AM symbiosis. Even, flavonoid NM7 inhibited the number of entry points and the percentage of root length colonization of the test plant by Gigaspora and Glomus species. The presence of 5,6,7,8,9-hydroxy chalcone in non-AM colonized clover roots and the disappearance of this flavonoid in mycorrhizal clover roots indicated that the AM fungus G. intraradices induced a mechanism, which reduces this inhibitory molecule in the mycorrhizal root. Quercetin, 5,6,7,8-hydroxy-4′-methoxy flavones, and 3,5,6,7,4′-hydroxy flavone increased the number of entry points and the AM colonization of tomato roots by Gigaspora (not Glomus). Further, Scervino et al. (2005b) linked flavonoids changes in mycorrhizal roots directly to regulatory processes of the AM symbiosis for the first time. Interestingly, RR4 and RR4-2 stimulated most of the presymbiotic stages of Gigaspora (but not of the two Glomus species), supporting the implication of Akiyama et al. (2002) that a flavonoid, stimulating root colonization by AM fungi, was only present in non-mycorrhizal but not in mycorrhizal roots. They further suggested that the compound might be involved in one stage of the fungal regulation, but not in another, as RR4 increased the percentage of spore germination of Gi. margarita, however, inhibited the hyphal growth, and showed no effect on hyphal branching and the cluster formation of auxiliary cells. NM7 was detectable only in non-mycorrhizal white clover roots and inhibited the presymbiotic development of all AM fungi tested. There was stimulatory effect of newly synthesized quercetin on spore germination and/or hyphal growth of Gigaspora species. However, acacetin and rhamnetin exhibited an inhibitory effect on nearly all fungal parameters of both AM genera (Gigaspora and Glomus) tested, pointing toward an implication in the autoregulation of mycorrhization (Scervino et al. 2005b). Scervino et al. (2009) reported that 2 μM concentration of 3-methoxy-5,6,7,8-hydroxy-4′hydroxy flavone (NMHTV) isolated from shoots of non-arbuscular mycorrhizal (AM) inoculated clover did not affect the percentage of germination of spores but significantly increased the formation of entry points and symbiotic stage of Gi. margarita spores on colonization of tomato (Lycopersicum esculentum), while at higher concentration of 8 μM concentration inhibited the hyphal length of Gi. rosea. The absence of stimulation of the AM presymbiotic and symbiotic stages in tomato by exogenous application of the newly synthesized flavonoids 5,6,7,8-hydroxy-3-methoxy flavone (MH-1), 5,6,7,8-hydroxy-4′-hydroxy flavone (MH-2), and 5,7-hydroxy-3,4′-methoxy flavone (MH-3), isolated from AM clover (T. repens) shoots, indicated that the autoregulation of the AM symbiosis can be, at least partially, due to the disappearance of flavonoids in AM-colonized plants that stimulated the AM symbiosis.
Working with split-root systems of barley, alfalfa, and soybean, it was reported that root colonization of one side of a split-root system strongly suppressed mycorrhization of “autoregulated roots” on the other side (Vierheilig 2004b; Meixner et al. 2005). Meixner et al. (2005) reported that the soybean supernodulating mutant nts1007 [mutated in the receptor kinase gene GmNARK (Searle et al. 2003)], which lacks the autoregulatory mechanism to control nodulation, did not autoregulate AMF root colonization. Thus, these reports indicated a similar mechanism of autoregulation in both symbioses. Alike Rhizobium–legume symbiosis, flavonoids have also been suggested to be involved in the regulation of mycorrhization as roots treated with certain flavonoids exhibited increased AM fungal root colonization (Vierheilig and Piché 2002; Vierheilig 2004a; Scervino et al. 2005a, b). Catford et al. (2006) studied split-root systems (SRS) of alfalfa plants, where one side of SRS was inoculated with S. meliloti or Glomus mosseae or was treated with Nod factor. All the three applications resulted in accumulation of daidzein but reduced levels of formononetin, ononin, and medicarpin. These treatments given on one side altered isoflavonoids of the medicarpin synthesis pathway on the other side, thereby suggesting that levels of formononetin and its glycoside ononin were not only locally reduced but also systemically downregulated in parts of the root system that were not in contact with the invading symbionts. Thus, Catford et al. (2006) suggested that systemic downregulation of formononetin and ononin levels in non-treated root parts is a metabolic reponse to the putative autoregulation signals that mediate suppression of symbioses. Exogenous application of isoflavonoids to roots inactivated or compensated the symbiosis-suppressing effects induced by long-distance autoregulation signals and promoted nodule formation and stimulated establishment of the AM fungal symbiosis. Ononin (but not formononetin) promoted AMF root colonization, whereas formononetin stimulated nodulation in autoregulated parts of the root system. Hence, although autoregulation of nodulation and mycorrhization seem to share some common signaling events (Vierheilig 2004a; Meixner et al. 2005; Vierheilig et al. 2008), the effects of systemically regulated isoflavonoids on establishment of symbiosis were different (Fig. 8.4).
Many legume plants benefit from the tripartite symbiosis of arbuscular mycorrhizal fungi and rhizobia. Siqueira et al. (1991b) reported that in isoflavonoid formononetin and biochanin A (considered “anti-nod inducers”) treated plants, Rhizobium nodulation was enhanced in the presence of AM fungi which might have resulted from improved mycorrhizal colonization and plant growth. Soil applications of these compounds had also shown to stimulate mycorrhizal formation and growth of non-nodulated plants in the presence of AM fungi (Siqueira et al. 1991a), thus indicating that isoflavonoid growth stimulation was not nodulation mediated. However, Xie et al. (1995) found that the inoculation of G. max with B. japonicum increased the colonization by G. mosseae, and an increased flavonoid concentration in root exudates was suggested to be responsible for this effect. Xie et al. (1997) reported the increase in mycorrhizal colonization (from <30 to 65%) and sporocarp formation of G. mosseae (Nicol. & Gerd.) Gerdemann & Trappin in roots of Lablab purpureus (L.) Sweet treated with Nod factors purified from Rhizobium sp. NGR234. Antunes et al. (2006) found that the concentration of daidzein was at least four times greater in soybean root than in the seed, whereas coumestrol, which was absent in the seed, was newly synthesized. The significant increase in diadzein and coumestrol appeared to result mainly from the development of AM, which might have helped in bradyrhizobial symbiosis (diadzein and coumestrol are involved in bradyrhizobial symbiosis) and thus played a key role in the early stages of the tripartite symbiosis. Morandi et al. (2009) reported a new M. truncatula mutant B9, defective for nodulation but was hypermycorrhizal. It represented a new tool for the study of plant metabolites differentially regulating mycorrhiza and nodulation symbioses, in particular those related to autoregulation mechanisms. In contrast to wild A17, mutant was characterized by considerably higher root concentrations of the phytoestrogen coumestrol (nod gene inhibitor for S. meliloti and hyphal growth inducer of Gi. margarita) and coumestrol conjugate malonyl glycoside, thus leading to low and late nodulation but high mycorrhizal colonization in B9 mutant. Fokom et al. (2010) pointed out significant interaction between arbuscular mycorrhizal fungi and 3-o-glucoside kaempferol in enhancing P nutrition, N2 fixation, as well as phenolic metabolism in Vigna unguiculata (L.) Walp growing in low-P soil of southern Cameroon. Many flavonoids like quercetin, luteolin, and other substituted flavones and flavanones are released by germinating seeds and living roots of legume crops over time and their interactions with fungal pathogens suggest that while stimulating mycorrhizal fungi they may be equally important for plant protection against soilborne pathogens (Hassan and Mathesius 2012). Cordeiro et al. (2015) evidenced from field experiment that formononetin can improve mycorrhizal colonization, nodules number, and reduce the negative effects of fungicides Carbendazim + Thiram in soybean production. Thus, utilizing flavonoids can be an excellent opportunity to utilize and manipulate Rhizobium and AM fungi in order to enhance crop productivity in a cost-effective manner and with reduced agricultural chemical inputs. Currently, seed exudates, which are mixture of flavonoids, are being used commercially to promote Rhizobium–legume symbiosis and N2 fixation in agricultural practices (Skorupska et al. 2010). A commercial product “SoyaSignal” (mixture of genistein and daidzein) in Northern America is being applied either directly to the seed or in furrows in soils that contain adequate populations of Bradyrhizobium (Smith and Zhang 1999). SoyaSignal significantly improved nodulation and nitrogen fixation which resulted in an average 7% increase in grain yield (Leibovitch et al. 2001; Broughton et al. 2003). This study linked the function of different flavonoids to the establishment of the tripartite symbiosis and suggested that these compounds are produced and released into the rhizosphere as a function of the colonization process.
8.5 Conclusion
Thus, this chapter highlighted the significance of flavonoids as common architects in the establishment of two agriculturally important root symbioses—nodulation and mycorrhizal interactions. Information attained from these studies illustrated that besides stimulating microbial swarming motility toward host and activating symbiosis-related microbial genes, multifaceted flavonoids contribute in various stages of symbiosome developmental program. Their differential endogenous accumulation regulates modulation of plant innate defense reactions, infection thread/hyphal growth, intraradical hyphal differentiation, cortical cell morphogenesis for providing optimal niche to the microbes, and prevention of metabolically inefficient symbioses. Deciphering the complexity of flavonoids induced responses and their interplays with other major regulators of symbioses such as hormones make flavonoids a vital player in plant–microbe symbioses. However, gaps still remain in the knowledge base and further researches involving the endogenous regulation of flavonoid biosynthesis during these mutualistic associations as well as use of flavonoids as potential soil amendments are needed to optimize their implementation in improving rhizobial and mycorrhizal symbioses.
References
Abd-Alla MH, El-enany A-WE, Bagy MK, Bashandy SR (2014) Alleviating the inhibitory effect of salinity stress on nod gene expression in Rhizobium tibeticum—fenugreek (Trigonella foenum graecum) symbiosis by isoflavonoids treatment. J Plant Interact 9:275–284
Abdel-Lateif K, Bogusz D, Hocher V (2012) The role of flavonoids in the establishment of plant roots endosymbioses with arbuscular mycorrhiza fungi, rhizobia and Frankia bacteria. Plant Signal Behav 7:636–641
Agati G, Tattini M (2010) Multiple functional roles of flavonoids in photoprotection. New Phytol 186:786–793
Aggarwal A, Kadian N, Tanwar A, Yadav A, Gupta KK (2011) Role of arbuscular mycorrhizal fungi (AMF) in global sustainable development. J Appl Nat Sci 3:340–351
Akiyama K, Matsuoka H, Hayashi H (2002) Isolation and identification of a phosphate deficiency-induced C-glycosyl-flavonoid that stimulates arbuscular mycorrhiza formation in melon roots. Mol Plant Microbe Interact 15:334–340
Akiyama K, Tanigawa F, Kashihara T, Hayashi H (2010) Lupin pyranoisoflavones inhibiting hyphal development in arbuscular mycorrhizal fungi. Phytochemistry 71:1865–1871
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106
Andersen OM, Markham KR (2006) Flavonoids: chemistry, biochemistry and applications. CRC Press, Boca Raton
Antunes PM, Goss MJ (2005) Communication in the tripartite symbiosis formed by arbuscular mycorrhizal fungi, Rhizobia and legume plants: a review. Am Soc Agron 48:199–221
Antunes PM, de Variennes A, Rajcan I, Goss MJ (2006) Accumulation of specific flavonoids in soybean as a function of the early tripartite symbiosis with arbuscular mycorrhizal fungi and Bradyrhizobium japonicum. Soil Biol Biochem 38:1234–1242
Ardourel M, Lortet G, Maillet F, Roche P, Truchet G, Promé JC, Rosenberg C (1995) In Rhizobium meliloti, the operon associated with the nod box n5 comprises nodL, noeA and noeB, three host-range genes specifically required for the nodulation of particular Medicago species. Mol Microbiol 17:687–699
Badri DV, Quintana N, El Kassis EG, Kim HK, Choi YH, Sugiyama A, Verpoorte R, Martinoia E, Manter DK, Vivanco JM (2009) An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol 151:2006–2017
Bais HP, Park SW, Weir TL, Callaway RM, Vivanco JM (2004) How plants communicate using the underground information superhighway. Trends Plant Sci 9:26–32
Balemi T, Negisho K (2012) Management of soil phosphorus and plant adaptation mechanisms to phosphorus stress for sustainable crop production: a review. J Soil Sci Plant Nutr 12:547–562
Baptista MJ, Siqueira JO (1994) Efeito de flavonoides na germinação de esporos e no crescimento assimbiótico do fungo micorrízico arbuscular Gigaspora gigantea. Rev Bras Fisiol Veg 6:127–134
Bécard G, Douds DD, Pfeffer PE (1992) Extensive in vitro hyphal growth of vesicular arbuscular mycorrhizal fungi in the presence of CO2 and flavonols. Appl Environ Microbiol 58:821–825
Begum AA, Leibovitch S, Migner P, Zhang F (2001a) Specific flavonoids induced nod gene expression and pre-activated nod genes of Rhizobium leguminosarum increased pea (Pisum sativum L.) and lentil (Lens culinaris L.) nodulation in controlled growth chamber environments. J Exp Bot 52:1537–1543
Begum AA, Leibovitch S, Migner P, Zhang F (2001b) Inoculation of pea (Pisum sativum L.) by Rhizobium leguminosarum bv. viceae preincubated with naringenin and hesperetin or application of naringenin and hesperetin directly into soil increased pea nodulation under short season conditions. Plant Soil 237:71–80
Bek AS, Sauer J, Thygesen MB, Duus JO, Petersen BO, Thirup S, James E, Jensen KJ, Stougaard J, Radutoiu S (2010) Improved characterization of nod factors and genetically based variations in LysM receptors domain identify amino acids expendable for nod factor recognition in lotus spp. Mol Plant Microbe 23:58–66
Belkheir AM, Zhou X, Smith DL (2001) Variability in yield and yield component responses to genistein pre-incubated Bradyrhizobium japonicum by soybean [Glycine max (L.) Merr] cultivars. Plant Soil 229:41–46
Besseau S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legranda M (2007) Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19:148–162
Birt DF, Jeffery E (2013) Flavonoids. American Society for Nutrition. Adv Nutr 4:576–577
Bonfante P, Genre A (2008) Plants and arbuscular mycorrhizal fungi: an evolutionary-developmental perspective. Trends Plant Sci 13:492–498
Bonfante P, Genre A (2010) Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosis. Nat Commun 1:48. doi:10.1038/ncomms1046
Brechenmacher L, Lei Z, Libault M, Findley S, Sugawara M, Sadowsky MJ, Sumner LW, Stacey G (2010) Soybean metabolites regulated in root hairs in response to the symbiotic bacterium Bradyrhizobium japonicum. Plant Physiol 153:1808–1822
Brencic A, Winans SC (2005) Detection of and response to signals involved in host-microbe interactions by plant-associated bacteria. Microbiol Mol Biol Rev 69:155–194
Broghammer A, Krusell L, Blaise M, Sauer J, Sullivan JT, Maolanon N, Vinther M, Lorentzen A, Madsen EB, Jensen KJ, Roepstorff P, Thirup S, Ronson CW, Thygesen MB, Stougaard J (2012) Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc Natl Acad Sci USA 109:13859–13864
Broughton WJ, Jabbouri S, Perret X (2000) Keys to symbiotic harmony. J Bacteriol 182:5641–5652
Broughton WJ, Zhang F, Perret X, Staehelin C (2003) Signals exchanged between legumes and Rhizobium: agricultural uses and perspectives. Plant Soil 252:129–137
Bucher M, Wegmuller S, Drissner D (2009) Chasing the structures of small molecules in arbuscular mycorrhizal signaling. Curr Opin Plant Biol 12:500–507
Bücking H, Liepold E, Ambilwade P (2012) The role of the mycorrhizal symbiosis in nutrient uptake of plants and the regulatory mechanisms underlying these transport processes. In: Dhal NK, Sahu SC (eds) Plant science. Intech, Janeza Trdine, p 107
Buee M, Rossignol M, Jauneau A, Ranjeva R, Becard G (2000) The pre-symbiotic growth of arbuscular mycorrhizal fungi is induced by a branching factor partially purified from plant root exudates. Mol Plant Microbe Interact 13:693–698
Buer CS, Muday GK (2004) The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 16:1191–1205
Buer CS, Imin N, Djordjevic MA (2010) Flavonoids: new roles for old molecules. J Integr Plant Biol 52:98–111
Capoen W, Sun J, Wysham D, Otegui MS, Venkateshwaran M, Hirsch S, Miwa H, Downie JA, Morris RJ, Ane JM, Oldroyd GE (2011) Nuclear membranes control symbiotic calcium signaling of legumes. Proc Natl Acad Sci USA 108:14348–14353
Carlsen SCK, Understrup A, Fomsgaard IS, Mortensen AG, Ravnskov S (2008) Flavonoids in roots of white clover: interaction of arbuscular mycorrhizal fungi and a pathogenic fungus. Plant Soil 302:33–43
Catford JG, Stehelin C, Larose G, Piché Y, Vierheilig H (2006) Systemically suppressed isoflavonoids and their stimulating effects on nodulation and mycorrhization in alfalfa split-root systems. Plant Soil 285:257–266
Cerri MR, Frances L, Laloum T, Auriac MC, Niebel A, Oldroyd GE, Barker DG, Fournier J, de Carvalho-Niebel F (2012) Medicago truncatula ERN transcription factors: regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol 160:2155–2172
Cesco S, Neumann G, Tomasi N, Pinton R, Weisskopf L (2010) Release of plant-borne flavonoids into the rhizosphere and their role in plant nutrition. Plant Soil 329:1–25
Chabot S, Bel-Rhlid R, Chênevert R, Piché Y (1992) Hyphal growth promotion in vitro of the VA mycorrhizal fungus, Gigaspora margarita Becker et Hall, by the activity of structurally specific flavonoid compounds under CO2-enriched conditions. New Phytol 122:461–467
Chen D-S, Liu C-W, Roy S, Cousins D, Stacey N, Murray JD (2015) Identification of a core set of rhizobial infection genes using data from single cell-types. Front Plant Sci 6:575. doi:10.3389/fpls.2015.00575
Cheng A-X, Han X-J, Wu Y-F, Lou H-X (2014) The function and catalysis of 2-oxoglutarate-dependent oxygenases involved in plant flavonoid biosynthesis. Int J Mol Sci 15:1080–1095
Cheynier V, Comte G, Davies KM, Lattanzio V, Martens S (2013) Plant phenolics: recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiol Biochem 72:1–20
Cohen MF, Sakihama Y, Yamasaki H (2001) Roles of plant flavonoids in interactions with microbes: from protection against pathogens to the mediation of mutualism. In: Pandalai SG (ed) Recent research developments in plant physiology 2. Research Signpost, Trivandrum, pp 157–173
Cooke TJ, Poli D, Sztein AE, Cohen JD (2002) Evolutionary patterns in auxin action. Plant Mol Biol 49:319–338
Cooper JE (2004) Multiple responses of rhizobia to flavonoids during legume root infection. Adv Bot Res 41:1–62
Cooper JE (2007) Early interactions between legumes and rhizobia: disclosing complexity in a molecular dialogue. J Appl Microbiol 103:1355–1365
Cordeiro MAS, Ferreira DA, Paulino HB, Souza CRF, Siqueira JO, Carneiro MAC (2015) Mycorrhization stimulant based in formononetin associated to fungicide and doses of phosphorus in soybean in the Cerrado. Biosci J 31:1062–1070
Crutzen P, Mosier AR, Smith KA, Winiwarter W (2007) N2O release from agro-fuel production negates global warming reduction by replacing fossil fuels. Atmos Chem Phys Discuss 7:11191–11205
D’haeseleer K, Keyser AD, Goormachtig S, Holsters M (2010) Transcription factor MtATB2: about nodulation, sucrose, and senescence. Plant Cell Physiol 51:1416–1424
Dalla Via V, Zanetti ME, Blanco F (2016) How legumes recognize rhizobia. Plant Signal Behav 11(2):e1120396. doi:10.1080/15592324.2015.1120396
de Billy F, Grosjean C, May S, Bennett M, Cullimore JV (2001) Expression studies on AUX1-like genes in Medicago truncatula suggest that auxin is required at two steps in early nodule development. Mol Plant Microbe Interact 14:267–277
De’narie J, Debelle F, Truchet G, Prome JC (1993) Rhizobium and legume nodulation: a molecular dialogue. In: Palacios R, Moira J, Newton WE (eds) New horizons in nitrogen fixation. Kluwer, Dordrecht, pp 19–30
Delaux P-M, Radhakrishnan G, Oldroyd G (2015) Tracing the evolutionary path to nitrogen-fixing crops. Curr Opin Plant Biol 26:95–99
Demont N, Debellé F, Aurelle H, Dénarié J, Promé JC (1993) Role of the Rhizobium meliloti nodF and nodE genes in the biosynthesis of lipo-oligosaccharidic nodulation factors. J Biol Chem 268:20134–20142
Denison RF, Kiers ET (2011) Life histories of symbiotic rhizobia and mycorrhizal fungi. Curr Biol 21:775–785
Dixon RA, Pasinetti GM (2010) Flavonoids and Isoflavonoids: from plant biology to agriculture and neuroscience. Plant Physiol 154:453–457
Downie JA (2010) The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol Rev 34:150–170
Fauvart M, Michiels J (2008) Rhizobial secreted proteins as determinants of host specificity in the Rhizobium-legume symbiosis. FEMS Microbiol Lett 285:1–9
Feddermann N, Finlaya R, Bollerb T, Elfstrand M (2010) Functional diversity in arbuscular mycorrhiza—the role of gene expression, phosphorous nutrition and symbiotic efficiency. Fungal Ecol 3:1–8
Ferguson BJ, Indrasumunar A, Hayashi S, Lin M-L, Lin Y-H, Reid DE, Gresshoff PM (2010) Molecular analysis of legume nodule development and autoregulation. J Integr Plant Biol 52:61–76
Fokom R, Nana WL, Tchameni S, Nwaga D (2010) Arbuscular mycorrhizal fungi (AMF) colonisation and rhizobia nodulation of cowpea as affected by flavonoid application. Res J Agric Biol Sci 6:1015–1021
Fournier J, Timmers AC, Sieberer BJ, Jauneau A, Chabaud M, Barker DG (2008) Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiol 148:1985–1995
Gachomo E, Allen JW, Pfeffer PE, Govindarajulu M, Douds DD, Jin HR, Nagahashi G, Lammers PJ, Shachar-Hill Y, Bücking H (2009) Germinating spores of Glomus intraradices can use internal and exogenous nitrogen sources for de novo biosynthesis of amino acids. New Phytol 184:399–411
Gage DJ (2002) Analysis of infection thread development using Gfp- and DsRed-expressing Sinorhizobium meliloti. J Bacteriol 184:7042–7046
Gagnon H, Ibrahim RK (1998) Aldonic acids: a novel family of nod gene inducers of Mesorhizobium loti, Rhizobium lupini, and Sinorhizobium meliloti. Mol Plant Microbe Interact 11:988–998
Geneva M, Zehirov G, Djonova E, Kaloyanova N, Georgiev G, Stancheva I (2006) The effect of inoculation of pea plants with mycorrhizal fungi and Rhizobium on nitrogen and phosphorus assimilation. Plant Soil Environ 52:435–440
Genre A, Bonfante P (2010) The making of symbiotic cells in arbuscular mycorrhizal roots. In: Koltai H, Kapulnik Y (eds) Arbuscular mycorrhizas: physiology and function. Springer, New York, pp 57–71. doi:10.1007/978-90-481-9489-6_3
Genre A, Chabaud M, Timmers T, Bonfante P, Barker DG (2005) Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 17:3489–3499
Gholami A, Geyter ND, Pollier J, Goormachtig S, Goossens A (2014) Natural product biosynthesis in Medicago species. Nat Prod Rep 31:356–380
Gianinazzi S, Gianinazzi-Pearson V (1988) Mycorrhizae: a plant’s health insurance. Chim Oggi 10:56–68
Gianinazzi-Pearson V, Branzanti B, Gianinazzi S (1989) In vitro enhancement of spore germination and early hyphal growth of a vesicular-arbuscular mycorrhizal fungus by host root exudates and plant flavonoids. Symbiosis 7:243–255
Gibson KE, Kobayashi H, Walker GC (2008) Molecular determinants of a symbiotic chronic infection. Annu Rev Genet 42:413–441
Giovannetti M, Avio L, Sbrana C (2010) Fungal spore germination and presymbiotic mycelial growth—physiological and genetic aspects. In: Koltai H, Kapulnik Y (eds) Arbuscular mycorrhizas: physiology and function. Springer, New York, pp 3–32
Gobbato E (2015) Recent developments in arbuscular mycorrhizal signaling. Curr Opin Plant Biol 26:1–7
Gough C (2003) Rhizobium symbiosis: insight into Nod factor receptors. Curr Biol 13:973–975
Gould KS, Lister C (2005) Flavonoid functions in plants. In: Anderson OM, Markham KR (eds) Flavonoids: chemistry, biochemistry, and applications. CRC, Boca Raton, pp 397–441
Gourion B, Berrabah F, Ratet P, Stacey G (2015) Rhizobium-legume symbioses: the crucial role of plant immunity. Trends Plant Sci 20:186–194
Graham TL (1991) Flavonoid and isoflavonoid distribution in developing soybean seedling tissues and in seed and root exudates. Plant Physiol 95:594–603
Grant C, Bittman S, Montreal M, Plenchette C, Morel C (2005) Soil and fertilizer phosphorus: effects on plant P supply and mycorrhizal development. Can J Plant Sci 85:3–14
Guenoune D, Galili S, Phillips DA, Volpin H, Chet I, Okon Y, Kapulnik Y (2001) The defense response elicited by the pathogen Rhizoctonia solani is suppressed by colonization of the AM-fungus Glomus intraradices. Plant Sci 160:925–932
Gutjahr C, Parniske M (2013) Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu Rev Cell Dev Biol 29:593–617
Haag AF, Arnold MFF, Myka KK, Kerscher B, Dall’Angelo S, Zanda M, Mergaert P, Ferguson GP (2013) Molecular insights into bacteroid development during Rhizobium-legume symbiosis. FEMS Microbiol Rev 37:364–383
Halbwirth H (2010) The creation and physiological relevance of divergent hydroxylation patterns in the flavonoid pathway. Int J Mol Sci 11:595–621
Hamel LP, Beaudoin N (2010) Chitooligosaccharide sensing and downstream signaling: contrasted outcomes in pathogenic and beneficial plant-microbe interactions. Planta 232:787–806
Haney CH, Riely BK, Tricoli DM, Cook DR, Ehrhardt DW, Long SR (2011) Symbiotic rhizobia bacteria trigger a change in localization and dynamics of the Medicago truncatula receptor kinase LYK3. Plant Cell 23:2774–2787
Harrison M, Dixon RA (1993) Isoflavonoid accumulation and expression of defense gene transcripts during the establishment of vesicular-arbuscular mycorrhizal associations in roots of Medicago truncatula. Mol Plant Microbe Interact 6:643–654
Hartwig UA, Maxwell CA, Joseph CM, Phillips DA (1990) Chrysoeriol and luteolin released from alfalfa seeds induce nod genes in Rhizobium meliloti. Plant Physiol 92:116–122
Hassan S, Mathesius U (2012) The role of flavonoids in root-rhizosphere signalling: opportunities and challenges for improving plant-microbe interactions. J Exp Bot 63:3429–3444
Herder GD, Parniske M (2009) The unbearable naivety of legumes in symbiosis. Curr Opin Plant Biol 12:491–499
Hernández I, Alegre L, Van Breusegem F, Munné-Bosch S (2009) How relevant are flavonoids as antioxidants in plants? Trends Plant Sci 14:125–132
Hichri I, Meilhoc E, Boscari A, Bruand C, Frendo P, Brouquisse R (2016) Nitric oxide: jack-of-all-trades of the nitrogen-fixing symbiosis? Adv Bot Res 77. doi:10.1016/bs.abr.2015.10.014
Hijri M, Sanders IR (2005) Low gene copy number shows that arbuscular mycorrhizal fungi inherit genetically different nuclei. Nature 433:160–163
Hiltner L (1904) Uber neure erfahrungen und probleme auf dem gebeit der bodenbackteriologie und unter besonderer berucksichtigung der grundungung und brache. Arb Deut Landwirsch Ges 98:59–78
Hofferek V, Mendrinna A, Gaude N, Krajinski F, Devers EA (2014) MiR171h restricts root symbioses and shows like its target NSP2a complex transcriptional regulation in Medicago truncatula. BMC Plant Biol 14:199. doi:10.1186/s12870-014-0199-1
Hsieh K, Huang AH (2007) Tapetosomes in Brassica tapetum accumulate endoplasmic reticulum-derived flavonoids and alkanes for delivery to the pollen surface. Plant Cell 19:582–596
Hughes JK, Hodge A, Fitter AH, Atkin OK (2008) Mycorrhizal respiration: implications for global scaling relationships. Trends Plant Sci 13:583–588
Hungria M, Joseph CM, Phillips DA (1991) Rhizobium nod-gene inducers exuded naturally from roots of common bean (Phaseolus vulgaris L.). Plant Physiol 97:759–764
Indrasumunar A, Kereszt A, Searle I, Miyagi M, Li D, Nguyen CD, Men A, Carroll BJ, Gresshoff PM (2010) Inactivation of duplicated nod factor receptor 5 (NFR5) genes in recessive loss-of-function non-nodulation mutants of allotetraploid soybean (Glycine max L. Merr.) Plant Cell Physiol 51:201–214
Indrasumunar A, Wilde J, Hayashi S, Li D, Gresshoff PM (2015) Functional analysis of duplicated symbiosis receptor kinase (SymRK) genes during nodulation and mycorrhizal infection in soybean (Glycine max). J Plant Physiol 176:157–168
Irving HR, Boukli NM, Kelly MN, Broughton WJ (2000) Nod-factors in symbiotic development of root hairs. In: Ridge RW, Emons AMC (eds) Root hairs: cell and molecular biology. Springer, Tokyo, pp 241–265
Ivanova KA, Tsyganova AV, Brewin NJ, Tikhonovich IA, Tsyganov VE (2015) Induction of host defences by Rhizobium during ineffective nodulation of pea (Pisum sativum L.) carrying symbiotically defective mutations sym40 (PsEFD), sym33 (PsIPD3/PsCYCLOPS) and sym42. Protoplasma 252:1505–1517
Jain V, Nainawatee HS (1999) Flavonoids influence growth and saccharide metabolism of Rhizobium meliloti. Folia Microbiol 44:311–316
Janczarek M, Skorupska A (2011) Modulation of rosR expression and exopolysaccharide production in Rhizobium leguminosarum bv. trifolii by phosphate and clover root exudates. Int J Mol Sci 12:4132–4155
Janczarek M, Rachwał K, Cieśla J, Ginalska G, Bieganowski A (2015) Production of exopolysaccharide by Rhizobium leguminosarum bv. trifolii and its role in bacterial attachment and surface properties. Plant Soil 388:211–227
Jensen ES, Peoples MB, Boddey RM, Gresshoff PM, Hauggaard-Nielsen H, Alves BJR, Morrison MJ (2012) Legumes for mitigation of climate change and feedstock in a bio-based economy—a review. Agron Sustain Dev 32:329–364
Johnson NC (2010) Resource stoichiometry elucidates the structure and function of arbuscular mycorrhizas across scales. New Phytol 185:631–647
Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC (2007) How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol 5:619–633
Kanamori N, Madsen LH, Radutoiu S, Frantescu M, Quistgaard EM, Miwa H, Downie JA, James EK, Felle HH, Haaning LL, Jensen TH, Sato S, Nakamura Y, Tabata S, Sandal N, Stougaard J (2006) A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc Natl Acad Sci USA 103:359–364
Kapulnik Y, Joseph CM, Phillips DA (1987) Flavone limitation to root nodulation and symbiotic nitrogen fixation in alfalfa. Plant Physiol 84:1193–1196
Kelly SJ, Muszynski A, Kawaharada Y, Hubber AM, Sullivan JT, Sandal N, Carlson RW, Stougaard J, Ronson CW (2012) Conditional requirement for exopolysaccharide in the Mesorhizobium–Lotus symbiosis. Mol Plant Microbe Interact 26:319–329
Kereszt A, Mergaert P, Kondorosi E (2011) Bacteroid development in legume nodules: evolution of mutual benefit or of sacrificial victims? Mol Plant Microbe Interact 24:1300–1309
Kistner C, Winzer T, Pitzschke A, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Webb KJ, Szczyglowski K, Parniske M (2005) Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17:2217–2229
Kloppholz S, Kuhn H, Requena N (2011) A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr Biol 21:1204–1209
Kobae Y, Hata S (2010) Dynamics of periarbuscular membranes visualized with a fluorescent phosphate transporter in arbuscular mycorrhizal roots of rice. Plant Cell Physiol 51:341–353
Kobayashi H, Naciri-Graven Y, Broughton WJ, Perret X (2004) Flavanoids induce temporal shifts in gene-expression of nod-box controlled loci in Rhizobium sp. NGR234. Mol Microbiol 51:335–347
Kondorosi E, Kondorosi E (2004) Endoreduplication and activation of the anaphase-promoting complex during symbiotic cell development. FEBS Lett 567:152–157
Kondorosi E, Mergaert P, Kereszt A (2013) A paradigm for endosymbiotic life: cell differentiation of rhizobium bacteria provoked by host plant factors. Annu Rev Microbiol 67:611–628
Kosslak RM, Bookland R, Barkei J, Paaren EH, Appelbaum ER (1987) Induction of Bradyrhizobium japonicum common nod genes by isoflavones isolated from Glycine max. Proc Natl Acad Sci USA 84:7428–7432
Kosuta S, Chabaud M, Lougnon G, Gough C, Dénarie J, Barker DG, Bécard G (2003) A diffusible factor from arbuscular mycorrhizal fungi induces symbiosis-specific MtENOD11 expression in roots of Medicago truncatula. Plant Physiol 131:1–11
Kouchi H, Imaizumi-Anraku H, Hayashi M, Hakoyama T, Nakagawa T, Umehara Y, Suganuma N, Kawaguchi M (2010) How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol 51:1381–1397
Laffont C, Rey T, André O, Novero M, Kazmierczak T, Debellé F, Bonfante P, Jacquet C, Frugier F (2015) The CRE1 cytokinin pathway is differentially recruited depending on Medicago truncatula root environments and negatively regulates resistance to a pathogen. PLoS One 10(1):e0116819. doi:10.1371/journal.pone.0116819
Laloum T, Baudin M, Frances L, Lepage A, Billault-Penneteau B, Cerri MR, Ariel F, Jardinaud M-F, Gamas P, de Carvalho-Niebel F, Niebel A (2014) Two CCAAT-box-binding transcription factors redundantly regulate early steps of the legume-rhizobia endosymbiosis. Plant J 79:757–768
Laparre J, Malbreila M, Letissec F, Portaisc JC, Rouxa C, Bécarda G, Puech-Pagèsa V (2014) Combining metabolomics and gene expression analysis reveals that propionyl- and butyryl carnitines are involved in late stages of arbuscular mycorrhizal symbiosis. Mol Plant 7:554–566
Laplaze L, Lucas M, Champion A (2015) Rhizobial root hair infection requires auxin signaling. Trends Plant Sci 20:332–334
Lareen A, Burton F, Schäfer P (2016) Plant root-microbe communication in shaping root microbiomes. Plant Mol Biol 90:575–587
Larose G, Chênevert R, Moutoglis P, Gagné S, Piché Y, Vierheilig H (2002) Flavonoid levels in roots of Medicago sativa are modulated by the developmental stage of the symbiosis and the root colonizing arbuscular mycorrhizal fungus. J Plant Physiol 159:1329–1339
Leibovitch S, Migner P, Zhang F, Smith DL (2001) Evaluation of the effect of Soyasignal technology on soybean yield [Glycine max (L.)] Merr under field conditions over 6 years in Eastern Canada and the Northern United States. J Agron Crop Sci 187:281–292
Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57:405–430
Lévy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ané JM, Lauber E, Bisseling T, Dénarié J, Rosenberg C, Debellé F (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303:1361–1364
Li B, Li Y-Y, Wu H-M, Zhang F-F, Lia C-J, Li X-X, Lambers H, Li L (2016) Root exudates drive interspecific facilitation by enhancing nodulation and N2 fixation. PNAS. doi:10.1073/pnas.1523580113
Liang Y, Cao Y, Tanaka K, Wan J, Choi J, ho ho Kang C, Qui J, Stacey G (2013) Nonlegumes respond to rhizobial nod factors by suppressing the innate immune response. Science 34:1384-1387
Limpens E, Bisseling T (2008) Nod factor signal transduction in the Rhizobium-legume symbiosis. In: AMC E, Ketelaar T (eds) Root hairs, Plant cell monographs, vol 12. Springer, Berlin, pp 249–276
Limpens E, van Zeijl A, Geurts R (2015) Lipochitooligosaccharides modulate plant host immunity to enable endosymbioses. Annu Rev Phytopathol 53:311–334
Lira MA Jr, Nascimento LRS, Fracetto GGM (2015) Legume-rhizobia signal exchange: promiscuity and environmental effects. Front Microbiol 6:945. doi:10.3389/fmicb.2015.00945
López-Baena FJ, Ruiz-Sainz JE, Rodríguez-Carvajal MA, Vinardell JM (2016) Bacterial molecular signals in the Sinorhizobium fredii-soybean symbiosis. Int J Mol Sci 17:755. doi:10.3390/ijms17050755
López-Lara IM, Geiger O (2001) The nodulation protein NodG shows the enzymatic activity of an 3-oxoacyl-acyl carrier protein reductase. Mol Plant Microbe Interact 14:349–357
Madsen LH, Tirichine L, Jurkiwicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J (2010) The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun 1:1–12
Magori S, Kawaguchi M (2009) Long-distance control of nodulation: molecules and models. Mol Cells 27:129–134
Maier W, Peipp H, Schmidt J, Wray V, Strack D (1995) Levels of a terpenoid glycoside (blumenin) and cell wallbound phenolics in some cereal mycorrhizas. Plant Physiol 109:465–470
Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, Martinez EA, Driguez H, Bécard G, Dénarié J (2011) Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469:58–63
Makarova LE, Dudareva LV, Petrova IG (2015) The content of phenolic compounds in the pea seedling root exudates depends on the size of their roots and inoculation of bacteria mutualistic and antagonistic type of interactions. J Stress Physiol Biochem 11:94–103
Manchanda G, Garg N (2007) Endomycorrhizal and rhizobial symbiosis: how much do they share? J Plant Interact 2:79–88
Mandal SM, Chakraborty D, Dey S (2010) Phenolic acids act as signaling molecules in plant-microbe symbioses. Plant Signal Behav 5:359–368
Mandal SM, Chakraborty D, Dutta SR, Ghosh AK, Pati BR, Korpole S, Paul D (2016) Induction of nodD gene in a Betarhizobium isolate, Cupriavidus sp. of Mimosa pudica, by root nodule phenolic acids. Curr Microbiol 72:733–737
Mapope N, Dakora FD (2013) Rrole of flavonoid and isoflavonoid molecules in symbiotic functioning and host-plant defence in the leguminosae. In: Gurib-Fakim A, Eloff JN (eds) Chemistry for sustainable development in Africa. Springer, Berlin, Heidelberg, pp 33–48
Marais JPJ, Deavours B, Dixon RA, Ferreira D (2006) The stereochemistry of flavonoids. In: Grotewold E (ed) The science of flavonoids. The Ohio State University, Columbus, OH, pp 1–46
Markmann K, Giczey G, Parniske M (2008) Functional adaptation of a plant receptorkinase paved the way for the evolution of intracellular root symbioses with bacteria. PLoS Biol 6:e68
Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GE (2007) Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol 144:324–335
Martens S, Mithöfer A (2005) Flavones and flavone synthases. Phytochemistry 66:2399–2407
Mathesius U (2001) Flavonoids induced in cells undergoing nodule organogenesis in white clover are regulators of auxin breakdown by peroxidase. J Exp Bot 52:419–426
Mathesius U (2009) Comparative proteomic studies of root-microbe interactions. J Proteomics 72:353–366
Mathesius U, Bayliss C, Weinman JJ, Schlaman HRM, Spaink HP, Rolfe BG, Mccully ME, Djordjevic MA (1998) Flavonoids synthesized in cortical cells during nodule initiation are early developmental markers in white clover. Mol Plant Microbe Interact 11:1223–1232
McNear DH Jr (2013) The rhizosphere—roots, soil and everything in between. Nat Educ Knowl 4:1
Mechri B, Tekaya M, Cheheb H, Attia F, Hammami M (2015) Accumulation of flavonoids and phenolic compounds in olive tree roots in response to mycorrhizal colonization: a possible mechanism for regulation of defense molecules. J Plant Physiol 185:40–43
Meixner C, Ludwig-Müller J, Miersch O, Gresshoff P, Staehelin C, Vierheilig H (2005) Lack of mycorrhizal autoregulation and phytohormonal changes in the supernodulating soybean mutant nts1007. Planta 222:709–715
Messinese E, Mun JH, Yeun LH, Jayaraman D, Rougé P, Barre A, Lougnon G, Schornack S, Bono JJ, Cook DR, Ané JM (2007) A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol Plant Microbe Interact 20:912–921
Miransari M, Smith DL (2009) Alleviating salt stress on soybean (Glycine max (L.) Merr.)-Bradyrhizobium japonicum symbiosis, using signal molecule genistein. Eur J Soil Biol 45:146–152
Miri M, Janakirama P, Held M, Ross L, Szczyglowski K (2016) Into the root: how cytokinin controls rhizobial infection. Trends Plant Sci. doi:10.1016/j.tplants.2015.09.003
Mitra RM, Gleason CA, Edwards A, Hadfield J, Downie JA, Oldroyd GED, Long SR (2004a) A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proc Natl Acad Sci USA 101:4701–4705
Mitra RM, Shaw SL, Long SR (2004b) Six nonnodulating plant mutants defective for Nod factor-induced transcriptional changes associated with the legume-rhizobia symbiosis. Proc Natl Acad Sci USA 101:10217–10222
Miwa H, Sun J, Oldroyd GE, Downie JA (2006) Analysis of Nod-factor-induced calcium signaling in root hairs of symbiotically defective mutants of Lotus japonicus. Mol Plant Microbe Interact 19:914–923
Mohammadi K, Khalesro S, Sohrabi Y, Heidari G (2011) A review: beneficial effects of the mycorrhizal fungi for plant growth. J Appl Environ Biol Sci 1:310–319
Morandi D, Bailey JA, Gianinazzi-Pearson V (1984) Isoflavonoid accumulation in soybean roots infected with vesicular-arbuscular mycorrhizal fungi. Physiol Plant Pathol 24:357–364
Morandi D, Branzanti B, Gianinazzi-Pearson V (1992) Effect of some plant flavonoid on in vitro behavior of an arbuscular mycorrhizal. Agronomie 12:811–816
Morandi D, Le Signor C, Gianinazzi-Pearson V, Duc G (2009) A Medicago truncatula mutant hyper-responsive to mycorrhiza and defective for nodulation. Mycorrhiza 19:435–441
Morieri G, Martinez EA, Jarynowski A, Driguez H, Morris R, Oldroyd GED, Downie JA (2013) Host-specific nod-factors associated with Medicago truncatula nodule infection differentially induce calcium influx and calcium spiking in root hairs. New Phytol 200:656–662
Moscatiello R, Squartini A, Mariani P, Navazio L (2010) Flavonoid-induced calcium signalling in Rhizobium leguminosarum bv. viciae. New Phytol 188:814–823
Mukherjee A, Ané JM (2011) Germinating spore exudates from arbuscular mycorrhizal fungi: molecular and developmental responses in plants and their regulation by ethylene. Mol Plant Microbe Interact 24:260–270
Mulder L, Hogg B, Bersoult A, Cullimore JV (2005) Integration of signalling pathways in the establishment of the legume-rhizobia symbiosis. Physiol Plant 123:207–218
Murphy A, Peer WA, Taiz L (2000) Regulation of auxin transport by aminopeptidases and endogenous flavonoids. Planta 211:315–324
Murray JD (2011) Invasion by invitation: rhizobial infection in legumes. Mol Plant Microbe Interact 24:631–639
Nair MG, Safir GR, Siqueira JO (1991) Isolation and identification of vesicular-arbuscular mycorrhiza-stimulatory compounds from clover (Trifolium repens) roots. Appl Environ Microbiol 57:434–439
Naoumkina M, Dixon RA (2008) Subcellular localization of flavonid natural products: a signalling function? Plant Signal Behav 3:573–575
Naoumkina M, Farag MA, Sumner LW, Tang Y, Liu C-J, Dixon RA (2007) Different mechanisms for phytoalexin induction by pathogen and wound signals in Medicago truncatula. Proc Natl Acad Sci USA 104:17909–17915
Navazio L, Moscatiello R, Genre A, Novero M, Baldan B, Bonfante P, Mariani P (2007) A diffusible signal from arbuscular mycorrhizal fungi elicits a transient cytosolic calcium elevation in host plant cells. Plant Physiol 144:673–681
Nelson MS, Sadowsky MJ (2015) Secretion systems and signal exchange between nitrogen-fixing rhizobia and legumes. Front Plant Sci 6:491. doi:10.3389/fpls.2015.00491
Ng JLP, Perrine-Walker F, Wasson AP, Mathesius U (2015) Flavonoids and auxin transport inhibitors rescue symbiotic nodulation in the Medicago truncatula cytokinin perception mutant cre1. Plant Cell 27:2210–2226
Nijjer S, Rogers WE, Siemann E (2010) The impacts of fertilization on mycorrhizal production and investment in western gulf coast grasslands. Am Midl Nat 163:124–133
Novák K, Chovanec P, Skrdleta V, Kropacova M, Lisa L, Nemcova M (2002) Effect of exogenous flavonoids on nodulation of pea (Pisum sativum L.) J Exp Bot 53:1735–1745
O’Neil M, Heckelman P, Koch C, Roman K, Kenny C, D’Arecca M (1996) The merck index: encyclopedia of chemicals, drugs and biological. Merck Publishing, Rahway, NJ
Okazaki S, Tittabutr P, Teulet A, Thouin J, Fardoux J, Chaintreuil C, Gully D, Arrighi J-F, Furuta N, Miwa H, Yasuda M, Nouwen N, Teaumroong N, Giraud E (2016) Rhizobium-legume symbiosis in the absence of Nod factors: two possible scenarios with or without the T3SS. ISME J 10:64–74
Oldroyd GE, Downie JA (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59:519–546
Oldroyd GED, Murray JD, Poole PS, Downie JA (2011) The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45:119–144
Parniske M (2008) Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol 6:763–775
Peck MC, Fisher RF, Long SR (2006) Diverse flavonoids stimulate NodD1 binding to nod gene promoters in Sinorhizobium meliloti. J Bacteriol 188:5417–5427
Peck MC, Fisher RF, Bliss R, Long SR (2013) Isolation and characterization of mutant Sinorhizobium meliloti NodD1 proteins with altered responses to luteolin. J Bacteriol 195:3714–3723
Peer WA, Bandyopadhyay A, Blakeslee JJ, Makam SN, Chen RJ, Masson PH, Murphy AS (2004) Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16:1898–1911
Peer WA, Blakeslee JJ, Yang HB, Murphy AS (2011) Seven things we think we know about auxin transport. Mol Plant 4:487–504
Peix A, Ramírez-Bahena MH, Velázquez E, Bedmar EJ (2015) Bacterial associations with legumes. Crit Rev Plant Sci 34:17–42
Pinior A, Wyss U, Piché Y, Vierheilig H (1999) Plants colonized by AM fungi regulate further root colonization by AM fungi through altered root exudation. Can J Bot 77:891–897
Pollastri S, Tattini M (2011) Flavonols: old compounds for old roles. Ann Bot 108:1225–1233
Ponce M, Cervino M, Erra-Balsells R, Ocampo JA, Godeas A (2004) Flavonoids from shoots and roots of Trifolium repens (white clover) grown in presence or absence of the arbuscular mycorrhizal fungus Glomus intraradices. Phytochemistry 65:3131–3134
Pumplin N, Harrison MJ (2009) Live-cell imaging reveals periarbuscular membrane domains and organelle location in Medicago truncatula roots during arbuscular mycorrhizal symbiosis. Plant Physiol 151:809–819
Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, Stougaard J (2008) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425:585–592
Redmond JW, Batley M, Djordjevic MA, Innes RW, Kuempel PL, Rolfe BG (1986) Flavones induce expression of nodulation genes in Rhizobium. Nature 323:632–635
Reid DE, Ferguson BJ, Hayashi S, Lin Y-H, Gresshoff PM (2011) Molecular mechanisms controlling legume autoregulation of nodulation. Ann Bot 108:789–795
Remigi P, Zhu J, Young JPW, Masson-Boivin C (2016) Symbiosis within symbiosis: evolving nitrogen-fixing legume symbionts. Trends Microbiol 24:63–75
Requena N, Breuninger M (2004) The old arbuscular mycorrhizal symbiosis in the light of the molecular era. In: Esser K, Lüttge U, Beyschlag W, Murata J (eds) Progress in botany. Springer, Berlin, Heidelberg, pp 323–356
Requena N, Serrano E, Ocón A, Breuninger M (2007) Plant signals and fungal perception during arbuscular mycorrhizal establishment. Phytochemistry 68:33–40
Ribeiro CW, Alloing G, Mandon K, Frendo P (2015) Redox regulation of differentiation in symbiotic nitrogen fixation. Biochim Biophys Acta 1850:1469–1478
Rich MK, Schorderet M, Reinhardt D (2014) The role of the cell wall compartment in mutualistic symbioses of plants. Front Plant Sci 5:238. doi:10.3389/fpls.2014.00238
Riely BK, Lougnon G, Ané JM, Cook DR (2007) The symbiotic ion channel homolog DMI1 is localized in the nuclear membrane of Medicago truncatula roots. Plant J 49:208–216
Rose CM, Venkateshwaran M, Volkening JD, Grimsrud PA, Maeda J, Bailey DJ, Park K, Howes-Podoll M, den Os D, Yeun LH, Westphall MS, Sussman MR, Ane J-H, Coon JJ (2012) Rapid phosphoproteomic and transcriptomic changes in the rhizobia-legume symbiosis. Mol Cell Proteomics 11:724–744
Rossen L, Davis EO, Johnston AWB (1987) Plant-induced expression of Rhizobium genes involved in host specificity and early stages of nodulation. Trends Biochem Sci 12:430–433
Ryu H, Cho H, Choi D, Hwang I (2012) Plant hormonal regulation of nitrogen-fixing nodule organogenesis. Mol Cells 34:117–126
Saito A, Keda S, Ezura H, Minamisawa K (2007) Microbial community analysis of the phytosphere using culture-independent methodologies. Microbes Environ 22:93–105
Saito K, Yonekura-Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, Fernie AR (2013) The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiol Biochem 72:21–34
Saslowsky DE, Warek U, Winkel BSJ (2005) Nuclear localization of flavonoid enzymes in Arabidopsis. J Biol Chem 280:23735–23740
Scervino JM, Ponce MA, Erra-Bassells R, Vierheilig H, Ocampo JA, Godeas A (2005a) Arbuscular mycorrhizal colonization of tomato by Gigaspora and Glomus species in the presence of root flavonoids. J Plant Physiol 162:625–633
Scervino JM, Ponce MA, Erra-Bassells R, Vierheilig H, Ocampo JA, Godeas A (2005b) Flavonoids exhibit fungal species and genus specific effects on the presymbiotic growth of Gigaspora and Glomus. Mycol Res 109:789–794
Scervino JM, Ponce MA, Erra-Bassells R, Bornpadre J, Vierheilig H, Ocampo JA, Godeas A (2007) The effect of flavones and flavonols on colonization of tomato plants by arbuscular mycorrhizal fungi of the genera Gigaspora and Glomus. Can J Microbiol 53:702–709
Scervino JM, Ponce MA, Monica ID, Vierheilig H, Ocampo JA, Godeas A (2009) Development of arbuscular mycorrhizal fungi in the presence of different patterns of Trifolium repens shoot flavonoids. J Soil Sci Plant Nutr 9:102–115
Searle IR, Men AE, Laniya TS, Buzas DM, Iturbe-Ormaetxe I, Carroll BJ, Gresshoff PM (2003) Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299:109–112
Shaw LJ, Hooker JE (2008) The fate and toxicity of the flavonoids naringenin and formononetin in soil. Soil Biol Biochem 40:528–536
Shaw LJ, Morris P, Hooker JE (2006) Perception and modification of plant flavonoid signals by rhizosphere microorganisms. Environ Microbiol 8:1867–1880
Shtark OY, Sulima AS, Zhernakov AI, Kliukova MS, Fedorina JV, Pinaev AG, Kryukov AA, Akhtemova GA, Tikhonovich IA, Zhukov VA (2016) Arbuscular mycorrhiza development in pea (Pisum sativum L.) mutants impaired in five early nodulation genes including putative orthologs of NSP1 and NSP2. Symbiosis 68:129–144
Singh DP, Prabha R, Meena KK, Sharma L, Sharma AK (2014) Induced accumulation of polyphenolics and flavonoids in cyanobacteria under salt stress protects organisms through enhanced antioxidant activity. Am J Plant Sci 5(5):43916. doi:10.4236/ajps.2014.55087
Sinharoy S, Liu C, Breakspear A, Guan D, Shailes S, Nakashima J, Zhang S, Wen J, Torres-Jerez I, Oldroyd G, Murray JD, Udvardi MK (2016) A Medicago truncatula cystathionine-b-synthase-like domain-containing protein is required for rhizobial infection and symbiotic nitrogen fixation. Plant Physiol 170:2204–2217
Siqueira JO, Safir GR, Nair MG (1991a) Significance of phenolic compounds in plant-soil-microbial systems. Crit Rev Plant Sci 10:63–121
Siqueira JO, Safir GR, Nair MG (1991b) Stimulation of vesicular arbuscular mycorrhiza formation and growth of white clover by flavonoid compounds. New Phytol 118:87–93
Skorupska A, Wielbo J, Kidaj D, Marek-Kozaczuk M (2010) Enhancing Rhizobium-legume symbiosis using signaling factors. In: Khan MS (ed) Microbes for legume improvement. Springer, Wien, pp 27–54
Smit G, Puvanesarajah V, Carlson RW, Barbour WM, Stacey G (1992) Bradyrhizobium japonicum nodD1 can be specifically induced by soybean flavonoids that do not induce the nodYABCSUIJ operon. J Biol Chem 267:310–318
Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial nod factor-induced transcription. Science 308:1789–1791
Smith SE, Read DJ (2008) Mycorrhizal symbiosis. Academic Press, Inc., San Diego, CA
Smith DL, Zhang F (1999) Composition for enhancing grain yield and protein yield of legumes grown under environmental conditions that inhibit or delay nodulation thereof. US Patent-5922316
Smith DL, Praslickova D, Ilangumaran G (2015) Inter-organismal signaling and management of the phytomicrobiome. Front Plant Sci 6:722. doi:10.3389/fpls.2015.00722
Sosa T, Valares C, Alias JC, Lobon NC (2010) Persistence of flavonoids in Cictus landanifer soils. Plant Soil 337:51–63
Soyano T, Kawaguchi M (2014) Systemic regulation of root nodule formation. Division of Symbiotic Systems, National Institute for Basic Biology, Okazaki, Aichi. Available via doi:10.5772/56991
Spini G, Decorosi F, Cerboneschi M, Tegli S, Mengoni A, Viti C, Giovannetti L (2016) Effect of the plant flavonoid luteolin on Ensifer meliloti 3001 phenotypic responses. Plant Soil 399:159–178
Stafford HA (1997) Roles of flavonoids in symbiotic and defense functions in legume roots. Bot Rev 63:27–39
Steinkellner S, Lendzemo V, Langer I, Schweiger I, Khaosaad T, Toussaint J-P, Vierheilig H (2007) Flavonoids and strigolactones in root exudates as signals in symbiotic and pathogenic plant-fungus interactions. Molecules 12:1290–1306
Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, Parniske M (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417:959–962
Subramanian S, Graham MY, Yu O, Graham TL (2005) RNA interference of soybean isoflavone synthase genes leads to silencing in tissues distal to the transformation site and to enhanced susceptibility to phytophthora sojae. Plant Physiol 137:1345–1353
Subramanian S, Stacey G, Yu O (2007) Distinct, crucial roles of flavonoids during legume nodulation. Trends Plant Sci 12:282–285
Sugiyama A, Shitan N, Yazaki K (2007) Involvement of a soybean ATP-binding cassette-type transporter in the secretion of genistein, a signal flavonoid in legume-Rhizobium symbiosis. Plant Physiol 144:2000–2008
Sugiyama A, Yamazaki Y, Yamashita K, Takahashi S, Nakayama T, Yazaki K (2016) Developmental and nutritional regulation of isoflavone secretion from soybean roots. Biosci Biotechnol Biochem 80:89–94
Suzaki T, Kawaguchi M (2014) Root nodulation: a developmental program involving cell fate conversion triggered by symbiotic bacterial infection. Curr Opin Plant Biol 21:16–22
Suzuki H, Takahashi S, Watanabe R, Fukushima Y, Fujita N, Noguchi A, Yokoyama R, Nishitani K, Nishino T, Nakayama T (2006) An isoflavone conjugate-hydrolyzing beta-glucosidase from the roots of soybean (Glycine max) seedlings. Purification, gene cloning, phylogenetics, and cellular localization. J Biol Chem 281:30251–30259
Taurian T, Morón B, Soria-Díaz ME, Angelini JG, Tejero-Mateo P, Gil-Serrano A, Megías M, Fabra A (2008) Signal molecules in the groundnut-Bradyrhizobia interaction. Arch Microbiol 189:345–356
Taylor LP, Grotewold E (2005) Flavonoids as developmental regulators. Curr Opin Plant Biol 8:317–323
Tian F, Jia T, Yu B (2014) Physiological regulation of seed soaking with soybean isoflavones on drought tolerance of Glycine max and Glycine soja. Plant Growth Regul 74:229–237
Timmers ACJ, Auriac MC, Truchet G (1999) Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126:3617–3628
Tóth K, Stacey G (2015) Does plant immunity play a critical role during initiation of the legume-Rhizobium symbiosis? Front Plant Sci 6:401. doi:10.3389/fpls.2015.00401
Tsai SM, Phillips DA (1991) Flavonoids released naturally from alfalfa promote development of symbiotic Glomus spores in vitro. Appl Environ Microbiol 57:1485–1488
Tsvetkova G, Teofilova T, Georgiev GI (2006) Effect of naringein and quercetin on activity of nodabc genes of strain d293 and following nodulation and nitrogen fixation response of inoculated pea plants (Pisum sativum l.). Gen Appl Plant Physiol Special Issue:67–71
van Zeijl A, Op den Camp RHM, Deinum EE, Charnikhova T, Franssen H, Op den Camp HJM, Bouwmeester H, Kohlen W, Bisseling T, Geurts R (2015) Rhizobium lipo-chitooligosaccharide signaling triggers accumulation of cytokinins in Medicago truncatula roots. Mol Plant 8:1213–1226
Venturi V, Keel C (2016) Signaling in the rhizosphere. Trends Plant Sci 21:187–198
Vieira RF, Mendes IC, Reis-Junior FB, Hungria M (2010) Symbiotic nitrogen fixation in tropical food grain legumes: current status. In: Khan MS, Musarrat J, Zaidi A (eds) Microbes for legume improvement. Springer, Dordercht, Heidelberg, London, New York, pp 427–472. doi:10.1007/978-3-211-99753-6
Vierheilig H (2004a) Regulatory mechanisms during the plant-arbuscular mycorrhizal fungus interaction. Can J Bot 82:1166–1176
Vierheilig H (2004b) Further root colonization by arbuscular mycorrhizal fungi in already mycorrhizal plants is suppressed after a critical level of root colonization. J Plant Physiol 161:339–341
Vierheilig H, Piché Y (1995) Facteurs biochimiques potentiellement impliqués dans les interactions entre les champignons endomycorhiziens et leurs plantes non-hôtes. In: Fortin JA, Charest C, Picheé Y (eds) La symbiose mycorhizienne. Orbis Frelighsburg, Québec, pp 109–124
Vierheilig H, Piché Y (2002) Signalling in arbuscular mycorrhiza: facts and hypotheses. In: Buslig B, Manthey J (eds) Flavonoids in cell functions. Kluwer/Plenum, New York, pp 23–39
Vierheilig H, Albrecht C, Bago B, Piché Y (1996) Do flavonoids play a role in root colonization by AM fungi? First International Conference on Mycorrhizae, Berkeley, CA
Vierheilig H, Bago B, Albrecht C, Poulin MJ, Piché Y (1998) Flavonoids and arbuscular mycorrhizal fungi. In: Manthey J, Buslig B (eds) Flavonoids in the living system. Plenum Press, New York, pp 9–33
Vierheilig H, Steinkellner S, Khaosaad T, Garcia-Garrido JM (2008) The biocontrol effect of mycorrhization on soilborne fungal pathogens and the autoregulation of the AM symbiosis: one mechanism, two effects? In: Varma A (ed) Mycorrhiza: state of the art, genetics and molecular biology, eco-function, biotechnology, eco-physiology, structure and systematics. Springer, Berlin, Heidelberg, pp 307–320
Volpin H, Elkind Y, Okon Y, Kapulnik Y (1994) A vesicular-arbuscular mycorrhizal fungus (Glomus intraradix) induces a defense response in alfalfa roots. Plant Physiol 104:683–689
Wais RJ, Keating DH, Long SR (2002) Structure-function analysis of nod factor-induced root hair calcium spiking in Rhizobium-legume symbiosis. Plant Physiol 129:211–224
Wang D, Yang S, Tang F, Zhu H (2012) Symbiosis specificity in the legume-rhizobial mutualism. Cell Miocrobiol 14:334–342
Wasson AP, Pellerone FI, Mathesius U (2006) Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell 18:1617–1629
Weerasinghe RR, Bird DMK, Allen NS (2005) Root-knot nematodes and bacterial Nod factors elicit common signal transduction events in Lotus japonicus. Proc Natl Acad Sci USA 102:3147–3152
Weston LA, Mathesius U (2013) Flavonoids: their structure, biosynthesis and role in the rhizosphere, including allelopathy. J Chem Ecol 39:283–297
Wilde P, Manal A, Stodden M, Sieverding E, Hilderbrandt U, Bothe H (2009) Biodiversity of arbuscular mycorrhizal fungi in roots and soils of two salt marshes. Environ Microbiol 11:1548–1561
Williams CA, Grayer RJ (2004) Anthocyanins and other flavonoids. Nat Prod Rep 21:539–573
Winkel-Shirley B (2001) It takes a garden. How work on diverse plant species has contributed to an understanding of flavonoid metabolism. Plant Physiol 127:1399–1404
Wong PP, Evans HJ (1971) Poly-ß-hydroxybutyrate utilization by soybean (Glycine max Merr.) nodules and assessment of its role in maintenance of nitrogenase activity. Plant Physiol 47:750–755
Xie Z-P, Staehelin C, Vierheilig H, Wiemken A, Jabbouri S, Broughton WJ, Vögeli-Lange R, Boller T (1995) Rhizobial nodulation factors stimulate mycorrhizal colonization of nodulating and nonnodulating soybeans. Plant Physiol 108:1519–1525
Xie ZP, Muller J, Wiemken A, Broughton WJ, Boller T (1997) Nod factors and tri-iodobenzoic acid stimulate mycorrhizal colonization and affect carbohydrate partitioning in mycorrhizal roots of Lablab purpureus. New Phytol 139:361–366
Xie F, Murray JD, Kim J, Heckmann AB, Edwards A, Oldroyd GE, Downie JA (2012) Legume pectate lyase required for root infection by rhizobia. Proc Natl Acad Sci USA 109:633–638
Yamazaki A, Hayashi M (2015) Building the interaction interfaces: host responses upon infection with microorganisms. Curr Opin Plant Biol 23:132–139
Yang S, Tang F, Gao M, Krishnan HB, Zhu H (2010) R gene-controlled host specificity in the legume-rhizobia symbiosis. Proc Natl Acad Sci USA 107:18735–18740
Yano K, Yoshida S, Muller J, Singh S, Banba M, Vickers K, Markmann K, White C, Schuller B, Sato S, Asamizu E, Tabata S, Murooka Y, Perry J, Wang TL, Kawaguchi M, Imaizumi-Anraku H, Hayashi M, Parniske M (2008) CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci USA 105:20540–20545
Yokota S, Mizuo K, Moriya N, Oshio S, Sugawara I, Takeda K (2009) Effect of prenatal exposure to diesel exhaust on dopaminergic system in mice. Neurosci Lett 449:38–41
Zhang JA, Subramanian S, Zhang YS, Yu O (2007) Flavone synthases from Medicago truncatula are flavanone-2-hydroxylases and are important for nodulation. Plant Physiol 144:741–751
Zhang L, Tian LH, Zhao JF, Song Y, Zhang CJ, Guo Y (2009) Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol 149:916–928
Zhao J, Dixon RA (2010) The ‘ins’ and ‘outs’ of flavonoid transport. Trends Plant Sci 15:72–80
Zhuang X, Gao J, Ma A, Fu S, Zhuang G (2013) Bioactive molecules in soil ecosystems: masters of the underground. Int J Mol Sci 14:8841–8868
Zuanazzi JAS, Clergeot PH, Quirion JC, Husson HP, Kondorosi A, Ratet P (1998) Production of Sinorhizobium meliloti nod gene activator and repressor flavonoids from Medicago sativa roots. Mol Plant Microbe Interact 11:784–794
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Singla, P., Garg, N. (2017). Plant Flavonoids: Key Players in Signaling, Establishment, and Regulation of Rhizobial and Mycorrhizal Endosymbioses. In: Varma, A., Prasad, R., Tuteja, N. (eds) Mycorrhiza - Function, Diversity, State of the Art. Springer, Cham. https://doi.org/10.1007/978-3-319-53064-2_8
Download citation
DOI: https://doi.org/10.1007/978-3-319-53064-2_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-53063-5
Online ISBN: 978-3-319-53064-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)