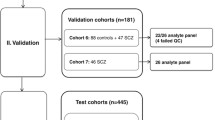
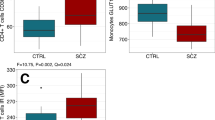
Abstract
An increasing number of studies are now exploring the potential of using blood-based biomarkers for prediction of antipsychotic treatment response in studies of schizophrenia patients. Here we describe the detailed setup of a clinical study to identify biomarker candidates for prediction of response of patients prior to receiving antipsychotics over a 6-week treatment period. The main emphasis is on study design, patient recruitment, sampling and outcome measures.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Approximately half of schizophrenia patients fail to respond favourably to an initial treatment with antipsychotic medications [1, 2]. Also, traditional treatment for schizophrenia sometimes involves the administration and switching of drugs multiple times until an adequate response is achieved [3]. Moreover, a high rate of non-compliance and relapse is a common occurrence [4, 5]. These issues are likely to be due to the fact that there is still insufficient understanding of the molecular pathways affected in this disease to guide treatment. Thus, the availability of objective biomarker tests to inform treatment selection is urgently needed, and a small number of studies have now been carried out in this area [6,7,8,9]. This is in line with the objectives of the Food and Drug Administration and other regulatory agencies to set out personalized medicine approaches for improved disease management [10]. However, studies to identify such biomarkers should be set up using a standardized design to minimize the effects of potential confounding factors on study outcome.
Here, we describe a clinical protocol involving 77 newly diagnosed schizophrenia patients to identify a baseline biomarker signature that could be used to predict response over a 6-week treatment period with the antipsychotics olanzapine, quetiapine, risperidone or others [6]. The main outcome measure was the Positive and Negative Syndrome Scale (PANSS) [11], and serum proteins were measured by using a multiplex immunoassay at Myriad RBM (Austin, TX, USA) as described previously [6, 7, 9]. Emphasis was placed on minimizing statistical effects of gender, age, body mass index (BMI), smoking and cannabis use on experimental outcomes.
2 Materials
-
1.
Approval obtained from the institutional ethical committee for the study protocols (see Note 1)
-
2.
Written consent obtained for all participants
-
3.
A study plan developed according to the Declaration of Helsinki [12]
-
4.
Patients diagnosed with paranoid schizophrenia according to the Diagnostic and Statistical Manual (DSM)-IV (Table 21.1) (see Note 2), with exclusion of psychosis resulting from other medical conditions or substance-induced psychosis by physical examination, routine blood analysis, screening for illegal drugs and magnetic resonance imaging of the brain
-
5.
S-Monovette 7.5 mL serum tubes (Sarstedt; Numbrecht, Germany)
-
6.
Low protein-binding Eppendorf tubes (Hamburg, Germany)
-
7.
Human DiscoveryMAP(R) multiplex immunoassay platform service (Myriad RBM; Austin, TX, USA) (see Note 3)
-
8.
Microsoft Office Excel software (Redmond, WA, USA) or equivalent
-
9.
Principal component analysis (SIMCA-P+ vs12.0, Umetrics, Umea, Sweden)
-
10.
Statistical software package R (http://www.r-project.org)
3 Methods
-
1.
Record all indicated parameters (and others as necessary) using those given in Table 21.1 as a guide.
-
2.
Carry out tests to determine whether or not statistical differences exist across the groups as appropriate, and exclude patients lying outside two standard deviations (see Note 4).
-
3.
From the selected patient group, exclude those individuals with other medical conditions (see Note 5).
-
4.
Treat participants as inpatients over the 6-week study period ( see Note 6).
-
5.
Collect blood from all subjects by venipuncture into serum tubes at the start of the study (T0) and after the 6-week treatment period (T6) (see Note 7).
-
6.
Prepare serum by placing samples at room temperature for 90 min to allow blood coagulation, followed by centrifugation at 4,000 × g for 5 min to recover the supernatants.
-
7.
Store serum at 80 °C in low protein-binding Eppendorf tubes prior to analysis.
-
8.
Randomize and blind samples to analysts using code numbers until all biochemical assays are complete.
-
9.
Analyse samples by multiplex immunoassay (see note 8).
-
10.
Record all values such as molecular levels of each analyte on an Excel spread sheet.
-
11.
Carry out data analyses using the statistical software package R (http://www.r-project.org).
-
12.
Preprocess the data by filtering out analytes which contain measurement values outside the linear assay range in more than 30% of samples (see Note 9).
-
13.
Assess data quality using principal component analysis (see Note 10).
-
14.
Determine significant associations using non-parametric Spearman’s correlation tests and adjust for false discovery rate [13] (see Note 11).
-
15.
To identify a molecular fingerprint for prediction patient responses, apply a Random Forests analysis [14] (Table 21.2) (see Note 12).
4 Notes
-
1.
In this study, approval was obtained from the University of Magdeburg.
-
2.
The focus on paranoid schizophrenia, which comprises the most prevalent subtype of the illness, was intended to minimize variability. In this study, 36 patients were in the first stage of illness and had not taken antipsychotics at the start of the study, and 41 patients had not taken antipsychotics for at least 6 weeks before the start of the study.
-
3.
This consists of approximately 200 immunoassays in multiplexed formats based on the Luminex technology (https://rbm.myriad.com).
-
4.
In this study, we also attempted to control for smoking and cannabis use, considering the increased consumption of these drugs by schizophrenia patients and known links to psychiatric illnesses.
-
5.
Clinicians had access to detailed clinical files of all patients including medical histories and referral letters from the general practitioners. Any patients with other conditions such as type 2 diabetes, hypertension, cardiovascular or autoimmune diseases were excluded to minimize potential confounding factors during data analysis.
-
6.
Participants were treated as inpatients as they were acutely ill. This also had the added benefit of monitoring patients more closely to better control the study.
-
7.
In addition to providing the material for biomarker analysis, a serum was used for therapeutic drug monitoring to assess compliance.
-
8.
In this study, all samples were shipped to Myriad RBM (Austin, TX, USA) for multiplex immunoassay using the HumanMAP® panel comprised of approximately 200 analytes. The screening was carried out in a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory as described previously [6, 7]. Assays were calibrated using standards, raw intensity measurements converted to absolute protein concentrations by comparison to the standards and performance verified using quality controls.
-
9.
Out of 191 analytes, 168 remained in the data set after this procedure, and only 6.8% of the values were missing.
-
10.
This was used to detect any strong effects on the overall data variance. In this case no effects were identified across the first ten principal components. Outliers in each analyte as well as response variables were excluded from analysis if these differed by more than two standard deviations from the mean. This resulted in removal of less than three subjects per analyte.
-
11.
Comparisons between the groups are based on non-parametric Wilcoxon rank-sum tests or paired Wilcoxon rank-sum tests for within-subject comparisons. Analysis of covariance (ANCOVA) was used to account for ‘regression to the mean’ effects between baseline and follow-up measurements [15]. This is a statistical phenomenon that can make natural variation in repeated data look like real change and can occur when large or small measurements tend to be followed by measurements that are closer to the mean. In addition, ANCOVA was used to estimate effects of BMI on analyte differences between the groups.
-
12.
This method builds multiple decision trees and uses a majority decision of classification outputs across trees to assign patients to one out of two groups based on the levels of the serum molecules. Random Forests was also used to select the most important molecules for prediction by permuting the measured values of each molecule sequentially and determining the importance based on the impact of this randomization on the output (results not presented here).
-
13.
No proteins were identified at baseline (T0) associated with improvements in PANSS positive or general scores. However, one protein (insulin) was associated with improvements in PANSS negative symptom scores. The negative correlation means that lower levels of insulin at T0 were associated with greater improvements [6].
References
Buckley PF, Friedman L, Krowinski AC, Eaton Y, Tronetti M, Miller DD (2001) Clinical and biochemical correlates of high-dose clozapine therapy for treatment-refractory schizophrenia. Schizophr Res 49:225–227
Heres S, Cirjaliu DM, Dehelean L, Matei VP, Podea DM, Sima D et al (2016) The SWITCH study: rationale and design of the trial. Eur Arch Psychiatry Clin Neurosci 266:513–521
Hashimoto N, Toyomaki A, Honda M, Miyano S, Nitta N, Sawayama H et al (2015) Long-term efficacy and tolerability of quetiapine in patients with schizophrenia who switched from other antipsychotics because of inadequate therapeutic response-a prospective open-label study. Ann Gen Psychiatry 14:1. doi:10.1186/s12991-014-0039-6
Kulkarni J, Reeve-Parker K (2015) Psychiatrists' awareness of partial- and non-adherence to antipsychotic medication in schizophrenia: results from the Australian ADHES survey. Australas Psychiatry 23:258–264
McIlwain ME, Harrison J, Wheeler AJ, Russell BR (2011) Pharmacotherapy for treatment resistant schizophrenia. Neuropsychiatr Dis Treat 7:135–149
Schwarz E, Guest PC, Steiner J, Bogerts B, Bahn S (2012) Identification of blood based molecular signatures for prediction of response and relapse in schizophrenia patients. Transl Psychiatry 2:e82
Schwarz E, Steiner J, Guest PC, Bogerts B, Bahn S (2015) Investigation of molecular serum profiles associated with predisposition to antipsychotic-induced weight gain. World J Biol Psychiatry 16:22–30
Mondelli V, Ciufolini S, Belvederi Murri M, Bonaccorso S, Di Forti M, Giordano A et al (2015) Cortisol and inflammatory biomarkers predict poor treatment response in first episode psychosis. Schizophr Bull 4:1162–1170
Tomasik J, Schwarz E, Lago SG, Rothermundt M, Leweke FM, van Beveren NJ et al (2016) Pretreatment levels of the fatty acid handling proteins H-FABP and CD36 predict response to olanzapine in recent-onset schizophrenia patients. Brain Behav Immun 52:178–186
Butterfield LH, Palucka AK, Britten CM, Dhodapkar MV, Hakansson L, Janetzki S et al (2011) Recommendations from the iSBTc-SITC/FDA/NCI Workshop on Immunotherapy Biomarkers. Clin Cancer Res 17:3064–3076
Kay SR, Fiszbein A, Opler LA (1987) The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr Bull 13:261–276
Rickham PP (1964) Human experimentation. code of ethics of the world medical association. declaration of Helsinki. Br Med J 2:177
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B 57:289–300
Breiman L (2001) Random forests. Machine Learning 45:5–32
Barnett AG, van der Pols JC, Dobson AJ (2005) Regression to the mean: what it is and how to deal with it. Int J Epidemiol 34:215–220
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Steiner, J., Guest, P.C. (2017). A Clinical Study Protocol to Identify Serum Biomarkers Predictive of Response to Antipsychotics in Schizophrenia Patients. In: Guest, P. (eds) Proteomic Methods in Neuropsychiatric Research. Advances in Experimental Medicine and Biology(), vol 974. Springer, Cham. https://doi.org/10.1007/978-3-319-52479-5_21
Download citation
DOI: https://doi.org/10.1007/978-3-319-52479-5_21
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-52478-8
Online ISBN: 978-3-319-52479-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)