Abstract
The deep ocean is one of the largest and least studied biomes on Earth. The microbes inhabiting these locales require physiological adaptations to handle the associated extreme environmental conditions, including high hydrostatic pressure, low temperatures, and low organic carbon. Few microbes have been successfully cultured that are capable of growth under in situ high-pressure conditions, especially at hadal depths, thanks to the relative inaccessibility of these sites, an inability to collect samples and maintain them under in situ conditions, and difficulties in culturing methodology. However, genome sequencing and high-throughput community analyses have provided insight into the prokaryotes which inhabit the deep sea and their lifestyles. This review discusses our current understanding of microbial adaptation to the deep-ocean through genomic comparisons of deep-ocean adapted microbial ecotypes and their shallow-water counterparts, including opportunistic heterotrophic microbes belonging to the Gammaproteobacteria and the fastidious taxa SAR11 and Thaumarchaea. These comparisons are addressed in the context of culture-independent metagenomics and community diversity analyses on deep, oligotrophic pelagic communities. Both culture-dependent and—independent analyses suggest the presence of bathytypes as both isolates and whole communities are distinct from those found above them. While these studies show many attributes indicative of deep-ocean genomes, including genes for particle-association, heavy-metal resistance, the loss of a UV photolyase, and increased abundances of mobile elements, they also suggest that high-pressure adaptation seems to arise from the accumulation of many small changes, such as differences in gene expression or the accumulation of compatible solutes. Genomic analyses on a larger dataset of samples and piezophilic isolates are necessary to distinguish attributes specific to deep-sea adaptation.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
2.1 Introduction
With an average depth of 3800 m the deep ocean is one of the largest biomes on Earth. Microorganisms dominate these deep-sea pelagic and benthic environments in terms of biomass, metabolic activity and turnover. The deep sea is characterized by low temperatures, high hydrostatic pressures, no sunlight, and low abundances and recalcitrant forms of organic carbon. These conditions lead to total microbial biomass decreases per milliliter of seawater by approximately three orders of magnitude from epipelagic to abyssal habitats (Nagata et al. 2010). Still, the pelagic ocean deeper than 200 m and its underlying subseafloor sediment contain ~6.5 × 1028 and 2.9 × 1029 cells respectively, values similar to those in the entire soil and terrestrial subsurface environments (Whitman et al. 1998; Kallmeyer et al. 2012). Therefore, a large fraction of the total microbes in the biosphere must be adapted to deep-ocean conditions. Hydrostatic pressure, which increases by 1 MPa (megapascal) for every 10 m of depth and can reach ~110 MPa (16,000 lbs/in2, 1100 atmospheres) in the deepest trenches, is one important selective parameter in the deep sea. Piezophiles, microorganisms that show optimal growth at hydrostatic pressures greater than 0.1 MPa (Yayanos 1995), have been successfully isolated from the deep ocean. They are closely related to shallow-water species but have adaptations to their in situ physical conditions mediated in part through changes in cell membrane composition (Allen et al. 1999; Allen and Bartlett 2000; Bartlett 2002), DNA replication and cell division (Yayanos and Pollard 1969; Welch et al. 1993; Ishii et al. 2002, 2004; El-Hajj et al. 2009, 2010) and protein synthesis (Kawano et al. 2004; Lauro et al. 2007; Lauro and Bartlett 2008). Advances over the past decade in cultivation-independent genomics have helped provide a more complete picture of deep-ocean microbial physiology and function by overcoming the small sample sizes and culturing challenges typically affiliated with the deep sea. Metagenomics and deep-sequencing community analysis, which shows community composition and the functional repertoire of a community, and single-cell genomics, which shows genomic content from one individual cell, can provide genomic insight into unculturable or rare members of a community. These studies, along with culture-dependent analyses, have revealed a number of distinct genomic and physiological features distinct to deep-ocean ecotypes. Here we briefly review genomic comparisons between cold, oligotrophic, deep-ocean microbial isolates and communities with their shallow-water counterparts.
2.2 The Culturable Community
To date at least 41 bacterial and 12 archaeal strains showing maximum growth at elevated hydrostatic pressure have been isolated (Jebbar et al. 2015). Almost all psychrotolerant or psychrophilic obligate piezophiles are members of the genera Shewanella, Colwellia, Moritella, and Psychromonas within the Gammaproteobacteria (Jebbar 2015). In general, the psychrophilic piezophiles display a good correlation between their isolation depth and growth pressure optimum, a fact that has been used to infer that high pressure has selected for the evolution and distribution of distinct groups of microorganisms (Yayanos 1986). However, studies attempting to isolate microbes capable of growth above or below the pressures corresponding to their collection depth have not yet been reported. Genomes of some cultured deep-sea piezophiles, including Shewanella benthica KT99 (Lauro et al. 2013a), Colwellia sp. MT41 (Kyaw et al.; unpublished data), P. profundum SS9 (Vezzi et al. 2005), Psychromonas sp. CNPT3 (Lauro et al. 2013b), Moritella sp. PE36, Marinitoga piezophila KA3 (Lucas et al. 2012), Desulfovibrio piezophilus (Pradel et al. 2013), Desulfovibrio hydrothermalis AM13 (Ji et al. 2013), Carnobacterium sp. AT7 (Stratton 2008), Thermococcus barophilus (Vannier et al. 2011), and Pyrococcus yayanosii (Jun et al. 2011) have been sequenced to date. 16S rRNA gene phylogenies show that piezophiles of the same genera are typically closely related to one another (Lauro et al. 2007; Miyazaki and Nogi 2014; Urakawa 2014; Satomi and Fujii 2014). Piezophilic clusters have radiated out from within multiple genera, separately evolving many times. Piezophiles that show optimum growth at pressures exceeding 40 MPa are either psychrotolerant/psychrophilic or thermophilic/hyperthermophilic (DeLong et al. 1997; Lauro et al. 2007). This likely reflects the habitats sampled as much of the deep ocean is low temperature and hydrothermal vent sites have been intensively sampled. Some microbes show better, and sometimes optimum, growth at higher pressures when grown at higher temperatures (Yayanos 1995; Kaneko et al. 2000; Martini et al. 2013). This may be due to elevated pressure compensating for temperature effects (Yayanos et al. 1983). Future attempts at isolating more mesophilic piezophilic microbes, such as from the deep marine subsurface, could yield success.
Despite the major role of elevated pressure in structuring the distribution of piezophilic deep-sea life, many pressure-sensitive microbes have also been obtained from the deep sea. Analogous to the enrichments for piezophiles, a select group of heterotrophic taxa are consistently isolated from the deep-sea when incubated at atmospheric pressure, including the genera Pseudomonas, Pseudoalteromonas, Halomonas, Marinobacter, and Psychrobacter and the phyla Actinobacteria and Firmicutes (Takami et al. 1999; Biddle et al. 2005; Batzke et al. 2007; Kaye et al. 2011; Orcutt et al. 2011). These taxa do not seem to be location or depth-specific as they have been isolated from many different oceanic basins and depths, including trenches (Yanagibayashi et al. 1999; Takami et al. 1997; Pathom-aree et al. 2006). While many of these microbes are likely present in low abundance, some culture-indepent studies also indicate their presence or even dominance (Kato et al. 1997; Li et al. 1999; Xu et al. 2005; Nunoura et al. 2015; Salazar et al. 2015a, b). Extreme conditions, food scarcity, or an ability to adapt to broad environmental conditions may select for these microbes and result in their widespread distribution. While specific strains of Halomonas (Kaye et al. 2011) and Pseudomonas (Tamegai et al. 1997; Sikorski et al. 2002) may be adapted to the deep sea, the lack of culturable members of these taxa at high pressure suggests they may originate in more shallow waters but survive in a dormant state at depth. Dormancy, a microbial strategy to minimize energy requirements for long-term survival (Lennon and Jones 2011), can be accomplished in one manner through the formation of spores. Sporeformers are abundant in subseafloor sediments based on metagenome studies (Kawai et al. 2015), and this is even more apparent when spore levels are directly measured using the endospore-specific compound dipicolinic acid (Langerhuus et al. 2012; Lomstein et al. 2012).
Because so few distinct high pressure-adapted microbes are known and piezosensitive isolates are commonly obtained it is unclear what fraction of deep-ocean communities are adapted to high hydrostatic pressure. Measurements of microbial activity as a function of pressure, measured by comparing activity rates at both in situ and atmospheric pressure conditions, suggest that activity is dependent not only on depth but also collection location and sample type, with sediment, water column, and benthic boundary layer samples showing different levels of piezophily (Tamburini et al. 2013). These findings are likely due to the mixing of autochthonous communities with microbes attached to sinking particulate organic matter (POM) as less stratified and benthic boundary layer communities showed less in situ pressure adaptation than their counterparts. This issue could be addressed in future work by determining the taxonomic distribution of deep-sea microbial communities active at in situ pressures versus atmospheric pressure.
2.3 Insights from Deep Ecotypes
Ecotypes, or closely related microbial lineages adapted to specific environmental conditions, possess genomic adaptations that allow them to exploit different habitats or ecological roles. For example, different ecotypes of high- and low-light adapted Prochlorococcus strains show adaptive strategies unique to their respective depths (Ting et al. 2002; Rocap et al. 2003; Delong and Karl 2005; Martiny et al. 2009). Hundreds of coexisting subpopulations of Prochlorococcus have been identified, each with a distinct genomic backbone that provides differential fitness, leading to changes in relative abundances with varying environmental conditions (Kashtan et al. 2014). Similar ecotypes have been identified among deep-ocean microbes and their shallow-water counterparts. Depth-specific ecotypes, termed bathytypes, have been defined as a population of a species adapted to a certain water column depth (Lauro and Bartlett 2008). This classification of bathytypes has been expanded to include not only species but also distinct clades as many putative deep-ocean-specific microbes are members of groups with poorly defined phylogenies. Ubiquitous taxa with important ecological roles, such as the SAR11 clade (Field et al. 1997; Eloe et al. 2011c; Thrash et al. 2014) and the Thaumarchaea (Brochier-Armanet et al. 2008; Hu et al. 2011; Luo et al. 2014; Swan et al. 2014) show deep-specific clades distinct from those at shallower depths. Members of candidate bacterial phyla have also been suggested to be deep-specific. The ‘Gracilibacteria,’ ‘Microgenomates,’ and ‘Parcubacteria,’ members of the “candidate phyla radiation” that show relatively reduced genome size and metabolic potential (Brown et al. 2015), potentially include piezophiles because of their identification at depth within the East Pacific Rise (Hedlund et al. 2014). Other uncultured groups, including SAR406 (Nunoura et al. 2015) and SAR324 (Brown and Donachie 2007) show depth-dependent distributions and likely have members that are specifically adapted to the deep-ocean. Here we briefly review the findings of comparisons between ten sets of sequenced bathytypes.
2.3.1 Bacteria: Alphaproteobacteria: SAR11
The SAR11 clade within the Alphaproteobacteria is one of the most numerically dominant bacterial lineages in the ocean (Morris et al. 2002). The clade’s success has been attributed to a number of traits, including minimal cell size, genome streamlining (Giovanonni et al. 2005; Grote et al. 2012), and large population sizes to evade viral attack (Brown and Fuhrman 2005; Zhao et al. 2013). The SAR11 clade has been separated into subgroups, with some confined to warmer, temperate, or polar locations while others show a cosmopolitan distribution (Brown and Fuhrman 2005; Rusch et al. 2007; Brown et al. 2012), and many clades show variable responses to seasonal changes, suggesting ecological differentiation (Brown and Furhman 2005; Morris et al. 2005; Carlson et al. 2009; Brown et al. 2012). Deep populations have also been identified (Field et al. 1997; García-Martínez and Rodríguez-Valera 2000), including at abyssal and hadal depths (DeLong et al. 2006; Martin-Cuadrado et al. 2007; Konstantindis et al. 2009; Eloe et al. 2011c; Léon-Zayas et al. 2015). Single-cell amplified genomes (SAGs) of members of subclade Ic, a deep bathytype, were obtained from 770 m at Station ALOHA and were compared to surface SAR11 genomes, including those from subclade Ia, and metagenomic datasets (Thrash et al. 2014). Subclades Ia and Ic showed a rRNA sequence identity of 95% and an amino acid identity of 62%, suggesting they may be different genera. Relative abundances of subclades Ia and Ic estimated by read recruitment from metagenomic datasets showed that subclade Ia was dominant in the upper surface waters while subclade Ic represented more than half the reads taken from aphotic depths. The metabolism of many of the deep genomes suggested a metabolism similar to surface sucblade Ia strains, focused on the oxidation of organic acids, amino acids, and C1 and methylated compounds, but with the capacity for nitrogen salvage and sulfite oxidation. COG distribution was conserved except for enrichment in genes for cell wall/membrane/envelope biogenesis (M) and inorganic ion transport (P) in subclade Ic. Unique phage related genes were also identified, including a CRISPR region that showed preferential recruitment to the mesopelagic at Station ALOHA. Adaptations of this SAR11 bathytype are thought to be reflected in subtle differences, such as increases in genome size, larger intergenic spacer regions, and preferential amino acid-substitutions. Populations of Pelagibacter (a major genus within the SAR11 group) identified using metagenomics at 4000 m at Station ALOHA had nonsynonymous to synonymous (Dn/Ds) ratios two times higher when compared to shallow-water representatives, suggesting decreased purifying selection (Konstantinidis et al. 2009). One gene conspicuously missing is that encoding deoxyribodipyrimidine photolyase (phr) (Vezzi et al. 2005). This gene, which uses blue-light energy to repair UV-mediated DNA damage (Todo 1999), is expected to be missing from microbes not exposed to light and may be a diagnostic tool to identify obligate deep-ocean microbes (Lauro and Bartlett 2008). Despite relatively good genome sequence coverage for the SAGs, ranging from 55 to 86%, none of the subclade Ic genomes had a DNA photolyase. However, genes for proteorhodopsins, which are light-driven proton pumps, were identified in two SAGs. Thrash et al. suggest that members of subclade Ic may occasionally circulate to the euphotic zone where proteorhodopsins would provide an energetic advantage. The work by Thrash et al. (2014) provides a framework for genomic adaptation to the deep ocean in one of the most-abundant microbes in the ocean.
2.3.2 Gammaproteobacteria: Alteromonadales: Alteromonas
Members of the genus Alteromonas are heterotrophic r-strategists typically found in nutrient-rich niches (López-López et al. 2005; Shi et al. 2012). One species, A. macleodii, is readily cultivable and has surface and deep-specific clades (López-López et al. 2005; Ivars-Martínez et al. 2008a), with members identified at high abundance in some deep-ocean samples (López-López et al. 2005; Quaiser et al. 2011; Smedile et al. 2013). When the genome sequence from a representative deep isolate, termed A. macleodii AltDE (Ivars-Martínez et al. 2008b), was compared to that of the shallow ecotype A. macleodii ATCC 27126 (Baumann et al. 1972), many similarities with other deep-sea genomic comparisons were found. 65 transposable elements and 63 insertion sequences (IS) were identified in AltDE while only three and one, respectively, were found in ATCC 27126. Transposases are abundant in the genomes of particle-attached microbial communities and those that are associated with surfaces (Burke et al. 2011; Ganesh et al. 2014). Genes involved in phage interaction, such as integrases and a CRISPR region, were also overrepresented in AltDE. These findings may be due to a reduced capacity to selectively purge mobile elements, such as within species of reduced effective population sizes, greater cell-cell interactions leading to increased mobile element transmission, or as a result of the adaptive value of greater transposon-mediated lateral gene transfer occurring in microbes encountering environmental change in association with sinking particles (Ganesh et al. 2014). AltDE also encodes for a cytochrome BD complex, which may function as an alternative respiratory chain during hypoxic conditions, such as those that could exist on POM. The piezophile Shewanella violacea DSS12 contains a gene for this same cytochrome and upregulates its expression at high pressure (Chikuma et al. 2007). The deep bathytype is also enriched in genes for extracellular polysaccharide biosynthesis and export, consistent with the increased mucosity of AltDE and other deep Alteromonas isolates (Raguenes et al. 1996). While ATCC 27126 is enriched in genes for signal transduction and transcriptional regulation, environmental sensing and sugar and amino acid utilization, which may reflect a capacity for making use of a broader array of substrates, AltDE contains more dioxygenases and genes involved in urea use and transport, suggesting it is more adept at using recalcitrant organic matter.
Deep isolates of A. macleodii were experimentally shown to be more resistant to zinc, mercury and lead than their shallower strains, consistent with the identification of large numbers of genes for heavy metal resistance and detoxification in its genome. Heavy metals are thought to be associated with particulate matter (Puig et al. 1999) and therefore particle-associated microbes may require genes to deal with high, inhibitory concentrations of these metals. Based on these findings and the distribution of AltDE and ATCC 27126 relatives it was hypothesized by Ivars-Martínez et al. (2008b) that niche separation of these ecotypes is achieved by the preference of AltDE for large, fast-sinking POM present in both the deep sea and the upper water column, whereas ATCC 27126 associates with smaller, more slowly sinking POM present in shallow environments.
2.3.3 Gammaproteobacteria: Alteromonadales: Pseudoalteromonas
Thanks to versatile metabolic capabilities the genus Pseudoalteromonas shows widespread oceanic distribution and adaptability to dissimilar ecological habitats (Ivanova et al. 2014). While no piezophilic Pseudoalteromonas spp. are known, members of this genus are consistently isolated from the deep ocean, suggesting they may be important participants in these environments. One deep-sea Pseudoalteromonas that has been studied is the psychrophile Pseudoalteromonas sp. SM9913. SM9913 produces exopolysaccharides (Liu et al. 2013), a function typical of other Pseudoalteromonas spp. (Nichols et al. 2005; Zhou et al. 2009), which may facilitate colonization of particles and stabilize cold-adapted proteases for the degradation of POM (Chen et al. 2003; Qin et al. 2007, 2011). To identify the deep-sea adaptations possessed by Pseudoalteromonas sp. SM9913 its genome was sequenced (Qin et al. 2011) and compared to Pseudoalteromonas haloplanktis TAC125, a psychrophilic isolate from Antarctic coastal seawater (Médigue et al. 2005). Like A. macleodii AltDE and other deep-ocean microbes, SM9913 is enriched in transposases and integrases. A large number of SM9913-specific genes belong to COG categories involved in cell motility (N) and signal transduction (T), which are suggested to be involved in sensing and movement towards POM. Indeed, SM9913 has three gene clusters for flagellar biosynthesis, with one encoding a lateral flagellum similar to that found in P. profundum SS9 (see Sect. 2.3.6; Eloe et al. 2008). SM9913 also has more genes involved in heavy metal resistance and is more resistant to zinc than TAC125. These findings, along with the presence of a complete glycolysis pathway and TCA cycle, suggest Pseudoalteromonas sp. SM9913 is adept at colonizing labile particulate organic carbon in the deep ocean.
2.3.4 Gammaproteobacteria: Alteromonadales: Shewanella
Members of the genus Shewanella are some of the most common deep-sea isolates under both high and low hydrostatic pressure conditions. The Shewanella genus forms distinct clades largely based on level of psychrophilicity and Na + dependence (Kato and Nogi 2001; Zhao et al. 2010; Satomi 2014). Genomic comparisons of 17 Shewanella species and other members of the Gammaproteobacteria indicated that cold-adapted species have similar amino acid compositions, with enrichments in isoleucine, lysine, and asparagine, and low alanine, proline, and arginine content, in part due to differences in GC content (Zhao et al. 2010). Reciprocal best blasts of cold-adapted strains of Shewanella also showed similarity to the cold-adapted P. profundum SS9 and C. psychrerythraea 34H (Zhao et al. 2010).
While similar analyses are yet to be performed for all piezophilic and nonpiezophilic Shewanella species, studies comparing individual piezophiles to similar piezosensitive members have been completed. One study compared the genome of the psychropiezophilic microbe S. piezotolerans WP3 (Wang et al. 2008), isolated from sediment at a depth of 1914 m (Wang et al. 2004) and showing optimum growth at 20 MPa and 15–20 °C (Xiao et al. 2007), to that of the mesophile Shewanella oneidensis MR-1 (Heidelberg et al. 2002). WP3 showed moderately higher abundances of genes involved in cell wall/membrane/envelope biogenesis (M), energy production and conversion (C), intracellular trafficking, secretion, and vesicular transport (U), and inorganic ion transport and metabolism (P). The WP3 genome had higher numbers of duplicated genes, mostly involved in transport, secretion, energy metabolism, and transcriptional regulation. Consistent with the psychrophiles S. halifaxensis and S. sediminis, WP3 also showed higher abundances of cytochrome c oxidases when compared to MR-1 (Zhao et al. 2010). Like P. profundum SS9, WP3 has genes for both a polar and lateral flagellum (Wang et al. 2008), which are for swimming and swarming (i.e. movement along surfaces) respectively. The two sets of WP3 flagella were inversely regulated by low temperature and high pressure; the lateral flagellum was upregulated at low temperature but slightly repressed by high pressure (20 MPa) while the polar flagellum was repressed at low temperature but upregulated at high pressure. Further analysis showed that a mutant lacking a functional lateral flagellum showed no motility at 4 °C, indicating that this flagellum is essential for movement at low temperatures.
The genome sequence of the psychropiezophile S. violacea DSS12, isolated from sediment at a depth of 5110 m in the Ryukyu Trench and with optimum temperature and pressure of 8 °C and 30 MPa (Nogi et al. 1998a), has also been compared against that of the mesophile Shewanella oneidensis MR-1 (Aono et al. 2010). While most members of the Shewanella are capable of anaerobic growth using a number of electron acceptors, DSS12 contains only nitrate and trimethylamine N-oxide (TMAO) reductases and instead has more terminal oxidases for aerobic respiration. More proteases, polysaccharidases, chitinases, and cellulose hydrolases were identified in DSS12 than in MR-1, which may be useful for breaking down sinking POM-containing chitin exoskeletons. DSS12 also has genes for phosphatidylserine decarboxylase, an enzyme which catalyzes the conversion of phosphatidylserine to phosphatidylethanolamine (PE) and may play a role in cell division at low temperatures, and cardiolipin synthetase, which is thought to be important for maintaining the structural stability of the cytochrome c oxidase complex under high hydrostatic pressures. Both strain DSS12 and another Shewanella piezophile, S. benthica strain KT99, lack DNA photolyase genes. KT99 was isolated from amphipods collected at a depth of 9856 m in the Kermadec Trench and is obligately piezophilic, showing optimal growth at 90 MPa and no growth at 40 MPa (Lauro et al. 2013a). When COG abundances were compared against that of Shewanella frigidmarina NCIMB400, KT99 was enriched in genes for DNA replication, recombination, and repair, and transposases, all attributes of other deep-sea bacterial genomes.
2.3.5 Gammaproteobacteria: Mixed Orders (Oceanospirillales and Alteromonadales): Oceanospirillales and Colwellia
Two microbial taxa that have members adapted to high pressure and show interesting physiological plasticity include the order Oceanospirillales and the genus Colwellia within the Alteromonadales. Members of the Colwellia are some of the most common deep-sea isolates under high hydrostatic pressure conditions and have been isolated from at least four different trenches, including Colwellia sp. MT41 (Yayanos et al. 1981), C. piezophila (Nogi et al. 2004), and C. hadaliensis (Deming et al. 1988) (Fig. 2.1). Recently the first piezophile of the Oceanospirillales, Profundimonas piezophila YC-1, was obtained (Cao et al. 2014). YC-1 is a slow-growing, facultative anaerobic heterotroph most closely related to the uncultured symbiont of the deep-sea whale bone-eating Osedax worms. YC-1 has a doubling time of 41 h at its optimum pressure of 50 MPa, much slower than most piezophilic Gammaproteobacteria but similar to the 36 h seen for the piezophilic Roseobacter PRT1 also isolated from the Puerto Rico Trench (Eloe et al. 2011b), suggesting it might be adapted to an oligotrophic environment. Like other members of the Oceanospirillales YC-1 is capable of hydrocarbon utilization. Shortly after the Deepwater Horizon oil spill the associated microbial community was dominated by the Oceanospirillales (Mason et al. 2012). Metagenomics, single-cell genomics, and metatranscriptomics revealed that members of the Oceanospirillales were actively degrading alkanes (Mason et al. 2012). After the spill was capped, Colwellia spp. began to dominate the plume and sediment communities in concomitance with changes in hydrocarbon composition and abundance, leading to the hypothesis that non-gaseous n-alkanes and cycloalkanes were degraded by Oceanospirillales followed by gaseous and aromatic hydrocarbon degradation by Colwellia (Valentine et al. 2010; Mason et al. 2014a, b). Stable isotope probing experiments have shown that Colwellia spp. are capable of incorporating 13C from ethane, propane, and benzene and are therefore able to use a wide range of hydrocarbons (Redmond and Valentine 2012). However, a Colwellia SAG recovered from the Deepwater Horizon plume did not have complete pathways involved in hydrocarbon degradation (Mason et al. 2014a) and no isolate has been reported that degrades oil as the sole carbon source, although Colwellia sp. RC25 was shown to degrade hydrocarbons in the presence of Corexit (Bælum et al. 2012; Chakraborty et al. 2012). Corexit, a dispersant thought to make oil more bioavailable and which was used in the Deepwater Horizon oil spill, may allow some organisms to outcompete natural hydrocarbon degraders. Members of the Colwellia were enriched in dispersant-only and oil-Corexit mixtures but not in oil-only microcosms, which were dominated by the genus Marinobacter (Kleindienst et al. 2015). Rates of hydrocarbon oxidation were highest in oil-only microcosms and therefore Corexit may not support stimulation of oil biodegradation (Kleindienst et al. 2015). However, whether Colwellia outcompetes other hydrocarbon degraders in the presence of Corexit or if Corexit inhibits Marinobacter is unknown.
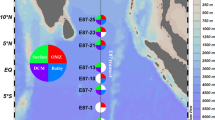
Phylogenetic tree of piezophilic bacterial strains and their isolation location, depth, and date. Many piezophiles have been isolated from many different oceanic basins, suggesting they have a widespread and relatively consistent distribution, over time, in the ocean. Samples without a species name (-) are recent, unpublished isolates. PR Puerto Rico Trench, JT Japan Trench, TT Tonga Trench, KT Kermadec Trench, MT Mariana Trench, PO Pacific Ocean, AO Atlantic Ocean, RT Ryukyu Trench, ST Sulu Trough, PE Patton Escarpment, JK Japan/Kurile-Kamchatcka Trench, PT Phillipine Trench, IB Izu-Bonin Trench, SB Suruga Bay
2.3.6 Gammaproteobacteria: Vibrionales: Photobacterium
The genus Photobacterium, composed of more than 20 species (Amaral et al. 2015), is a member of the Vibrionales order within the Gammaproteobacteria. Members of the genus Photobacterium are best known for their symbiotic relationships with marine eukaryotes (Amaral et al. 2015). One of the most studied piezophilic microbes is P. profundum SS9. SS9 was isolated at a depth of 2551 m from an amphipod in the Sulu Trough (DeLong 1986) and can grow over broad pressure (0.1–90 MPa) and temperature (2–20 °C) ranges, with optimum growth at 28 MPa and 15 °C (El-Hajj et al. 2010). At least three other strains of P. profundum have been isolated, including the piezophile DSJ4 from deep-ocean sediment (Nogi et al. 1998b) and the piezosensitive strains 3TCK and 1230sf1 (Campanaro et al. 2005; Lauro et al. 2014). Genomic comparisons of P. profundum bathytypes have revealed a number of attributes for a deep-sea adapted lifestyle (Campanaro et al. 2005; Lauro et al. 2014). SS9 has 15 copies of the rRNA operon that show variability in specific helices, which may confer ribosome stability under high-pressure conditions (Lauro et al. 2007). COG comparisons showed higher abundances of genes for motility and chemotaxis (N) and DNA replication, recombination and repair (L) but fewer genes involved in energy production (C) when SS9 was compared to 3TCK (Lauro et al. 2014). The overrepresentation of COG L was due to large numbers of transposable elements in SS9, which had 206 compared to 3 in 3TCK, while higher abundances of genes associated with motility were the result of a second cluster of genes for flagellum biosynthesis. These genes appear to encode for the production and function of a lateral flagella which also exist in related microbes, including piezophilic members of the Vibrionaceae (McCarter 2004) such as P. phosphoreum ANT-2200 (Zhang et al. 2014), Moritella sp. PE36 (Nagata et al. 2010), and S. piezoterolans WP3 as described in section IIID. Genetic experiments indicate that the SS9 surface motility system is primarily functional at high pressure, conditions that also favor the expression of the genes present within the lateral flagella gene cluster (Eloe et al. 2008). SS9 therefore regulates its lateral flagella genes differently than WP3. Regardless, in the surface ocean genes for chemotaxis and motility have been observed to be a characteristic of copiotrophic microbes, which possess the sensory and physiological capabilities to swim along the concentration plumes of organic matter, utilize it in high concentrations, and rapidly increase in cell numbers (Yooseph et al. 2010). These types of adaptations may also be important for deep-ocean microbes colonizing POM descending through the water column.
Pressure-related changes in regulation have also been found in genes affiliated with TMAO. SS9 has two torS genes which regulate the response to TMAO (Bordi et al. 2003). One of these genes, which is absent from 3TCK, is upregulated under high hydrostatic pressure conditions (Campanaro et al. 2005). SS9 also has multiple torA genes encoding TMAO reductase, one of which is upregulated at high pressure (Vezzi et al. 2005). The piezophile P. phosphoreum ANT-2200 has four copies of torA, including one homologous to the upregulated gene in SS9 (Zhang et al. 2014). These results suggest that TMAO reduction in deep-sea Photobacterium strains is an important respiratory adaptation at greater depths (Vezzi et al. 2005), perhaps within the anaerobic zones of particles or inside animal hosts. This is consistent with concentration measurements that show TMAO increasing in the tissues of some marine animals with depth of capture. This is thought to be an adaptation to high pressure (Yancey et al. 2014).
Unlike shallow Photobacterium spp., SS9 lacks a DNA photolyase (Vezzi et al. 2005; Lauro et al. 2014) and is extremely UV sensitive, a phenotype shared by many deep-sea microbes (Yayanos 1995). When the phr gene cluster was cloned from the shallow-water strain 3TCK into SS9 it dramatically enhanced UV-resistance. Because the phr gene cluster has a distinct codon usage in strain 3TCK it appears likely to have been acquired through horizontal gene transfer. P. profundum strains may be undergoing adaptive radiation driven by gene acquisition and loss and the horizontal gene transfer of the phr gene cluster into 3TCK could have been one of the evolutionary changes required for its adaptation from the deep-sea to shallower, sunlight-exposed waters (Lauro et al. 2014). This presents the intriguing possibility that the evolutionary path between shallow-water and deep-sea bacteria is not one-way, and shallow-water derivatives of piezophilic deep-sea bacteria can arise just as the reverse case. Horizontal gene transfer is also reflected in the genomes of deep-sea bacteria, with the nar gene cluster in the deep-sea isolate Pseudomonas sp. MT-1 being one example (Tamegai et al. 2004; Ikeda et al. 2009; Oikawa et al. 2015).
2.3.7 Gammaproteobacteria: Vibrionales: Vibrio
Despite being most well known for its pathogenic members, the genus Vibrio is composed of diverse species with representatives found in the deep sea (Ohwada et al. 1980; Tabor et al. 1981b; Reen et al. 2006). Recently the genome of Vibrio antiquarius, which was isolated from the East Pacific Rise hydrothermal vent system at 2520 m, was compared against other Vibrio spp. (Hasan et al. 2015). The genome of V. antiquarius suggests a unique ability to scavenge hydrogen peroxide and the potential for manganese oxidation and heavy metal resistance. V. antiquarius also encodes a fatty acid desaturase which may help maintain membrane fluidity under high pressures and low temperatures. Many psychrophilic and piezophilic bacteria produce unsaturated fatty acids or even polyunsaturated fatty acids (PUFAs) to help counter the effects of increasing pressure and decreasing temperature on their membranes (DeLong and Yayanos 1985; Allen et al. 1999; Bartlett 2002). Indeed, the benefits of polyunsatured fatty acid consumption on human health (Wall et al. 2010; Joffre et al. 2014) have led to interest in piezophile sources of PUFAs for use by humans or other animals. Hasan et al. note that the genome contains many homologs of virulence genes found in other Vibrio species, which they suggest may provide ecological functions outside of virulence, such as the establishment of host/cell relationships, provide a means of attachment to surfaces, signaling, or other interactions among aquatic communities. When recruited against metagenomic data V. antiquarius shows a ubiquitous distribution, identified in saltern, marine, coral, and human gut metagenomes. This distribution, along with the presence of a DNA photolyase, suggests that V. antiquarius may not be obligately adapted to the deep ocean.
2.3.8 Deltaproteobacteria: Desulfovibrio
Sulfate-reducing bacteria (SRB) contribute to the breakdown of organic matter and in some cases the coupling of anaerobic oxidation of methane to sulfate reduction in marine sediments (Hu et al. 2010; Stokke et al. 2012; Ruff et al. 2015). Members of the SRB within the genus Desulfovibrio, including D. profundus (Bale et al. 1997), D. hydrothermalis (Alazard et al. 2003), and D. piezophilus (Khelaifia et al. 2011), have been isolated from the deep sea. D. piezophilus C1TLV30, which was isolated from a wood fall at a depth of 1700 m and shows optimum growth at 30 °C and 10 MPa, was compared against other Desulfovibrio spp. and had its transcriptome analyzed at 0.1 and 10 MPa (Pradel et al. 2013). D. piezophilus has a cytochrome c gene that is overexpressed under high hydrostatic pressure conditions and is closely related to one found in P. profundum SS9. Amino acid composition, transport, and metabolism may be an important adaptive strategy for D. piezophilus as this bathytype showed an amino acid composition distinct from other Desulfovibrio strains and elevated expression of genes involved in alanine, histidine, and arginine biosynthesis and glutamine/glutamate metabolism and transport at high pressure. Similarly, transcriptomes from D. hydrothermalis, which was isolated from an East Pacific Rise hydrothermal vent at 2600 m, at 0.1, 10, and 26 MPa also showed differential expression of genes involved in glutamate metabolism (Amrani et al. 2014). Glutamate was shown to accumulate in D. hydrothermalis cells under high hydrostatic pressure, suggesting it may act as a piezolyte, an osmolyte that accumulates at high pressure (Martin et al. 2002). Genes involved in glutamate metabolism in P. profundum SS9 and S. violacea DSS12 also show differential expression with pressure (Campanaro et al. 2005; Vezzi et al. 2005; Ikegami et al. 2000). Thus, glutamate could serve an important global role as an extrinsic factor modulating protein structure or activity as a function of pressure in multiple piezophiles.
2.3.9 Firmicutes: Carnobacterium
Members of the genus Carnobacterium, which belongs to the Lactobacillales within the Firmicutes, are lactic acid bacteria of wide interest in the food and aquaculture industries. Carnobacteria have been isolated from processed meat, fish, and dairy products and can produce antimicrobial peptides to stunt the growth of other microbes, leading to speculation that members of this genus may cause or inhibit food spoilage (Hammes and Hertel 2006; Leisner et al. 2007). Similarly, carnobacteria have been identified within fish and may have both probiotic and pathogenetic effects on their host (Hammes and Hertel 2006; Leisner et al. 2007, 2012). Isolates have also been obtained from many different environments, including permafrost (Pikuta et al. 2005), and some strains even show growth under the low temperature, low pressure, and anoxic conditions similar to those found on Mars (Nicholson et al. 2013). Two isolates from 2500 m in the Aleutian Trench, AT7 and AT12, show optimum growth at 15 MPa (Lauro et al. 2007; Yayanos and DeLong 1987) and are the only gram-positive piezophilic bacteria currently isolated. Comparative genomics of Carnobacterium sp. AT7 with other Carnobacterium spp. and Enterococcus faecalis have shown this microbe is distinct (Stratton 2008; Leisner et al. 2012; Voget et al. 2011). While AT7 does not contain a photolyase, it does contain two endonucleases that may be involved in UV damage repair (Stratton 2008). When compared to C. maltaromaticum ATCC 35586, a host-associated species, AT7 lacks genes affiliated with host colonization, invasion, and metabolism (Leisner et al. 2012). A unique characteristic of AT7 is the presence of two plasmids that contain genes putatively affiliated with cadmium, tellurium, and copper efflux which are likely to contribute to heavy-metal resistance (Stratton 2008). The identification of Carnobacterium spp. showing growth under high pressure may have important repercussions for foods that are sterilized using pascalization.
2.3.10 Archaea: Marine Group I Thaumarchaea
Marine Group I Thaumarchaea (MGI) are ammonia-oxidizing archaea recognized as important drivers of nitrification in marine environments. MGI can be found at all depths (Nunoura et al. 2015) and in some cases compose the majority of deep-ocean communities (Karner et al. 2001). The presence of their ammonia monooxygenase gene (amoA), which helps catalyze a key intermediate step in nitrification by converting ammonia to nitrite, can be identified throughout the water column (Sintes et al. 2013). Genes involved in the 3-hydroxypropionate/4-hydroxybutyrate (3H/4H) pathway for carbon fixation and ammonia oxidation seem to be conserved among members of the Thaumarchaea (Ngugi et al. 2015), highlighting the importance of these functions within this lineage. However, mixotrophy has also been suggested in these archaea based on substrake uptake (Ouverney and Fuhrman 2000; Teira et al. 2006; Seyler et al. 2014; Qin et al. 2014) and genomic data (Hallam et al. 2006; Agogué et al. 2008; Swan et al. 2014). Despite their abundance and important role in the global nitrogen cycle cultivation and isolation of Thaumarchaea has proven difficult (Könneke et al. 2005; Tourna et al. 2011; Qin et al. 2014, 2015). Current isolates are physiologically distinct, displaying different tolerances to light, pH, and salinity, and varying capacities for the use of certain carbon compounds (Qin et al. 2014). While no deep-ocean Thaumarchaea are currently in culture, phylogenetic analyses using 16S rRNA, amoA, and concatenated single-copy marker genes have shown that MGI fall into shallow- and deep-water clades (Hallam et al. 2006; Beman et al. 2008; Nicol et al. 2011; Ngugi et al. 2015). These clades likely represent distinct ecotypes as niche separation has been identified according to ammonia concentrations, among other factors (Sintes et al. 2013; Nunoura et al. 2015). Analysis of single-amplified genomes have also shown that deep-ocean MGI represent distinct bathytypes. While thaumarchaeal photolyase genes have been identified in surface waters, they were absent in mesopelagic SAGs, suggesting they are not exposed to light-induced damage (Luo et al. 2014). It has been proposed that sensitivity to photoinhibition may be partially responsible for depth distributions (Mincer et al. 2007; Church et al. 2010) as some Thaumarchaea are inhibited by light (Merbt et al. 2012; Qin et al. 2014). Analysis of MGI SAGs collected from mesopelagic depths at Station ALOHA and the South Atlantic identified nine phylotypes within the deep clade, with six containing SAGs from both regions, suggesting these members have a cosmopolitan deep-ocean distribution (Swan et al. 2014). COG distributions associated with signal transduction and urea utilization were also found to differ between bathytypes, with the former more abundant in the shallow ecotype and the latter in the deep one (Luo et al. 2014). A SAG related to Nitrosopumilus from the Puerto Rico Trench also has genes for urea degradation, along with genes involved in fatty acid and lipoic acid synthesis and glycine cleavage (Léon-Zayas et al. 2015). This SAG also has an aquaporin, which function in osmotic pressure adaptation by effluxing water from cells exposed to hypotonic environments or help in the retention of small-molecule compatible solutes. This aquaporin may therefore play a role in high-pressure adaptation.
2.4 What Do These Deep-Sea Microbial Communities Actually Look like?
Deep-ocean bathytypes can provide insight into adaptations in specific microbial lineages but are these results ecologically important? While SAR11 and Thaumarchaea are widespread, dominant members of the deep-ocean community, many deep bathytypes are present in low abundance in deep ocean samples (Eloe et al. 2011c; Vandieken et al. 2012; Smedile et al. 2013). Many bathytypes are opportunistic heterotrophs, capable of rapid growth in nutrient-rich environments, and show increases in abundance when associated with enrichments, such as Colwellia and the Oceanospirillales at oil spills (Valentine et al. 2010; Redmond and Valentine 2012; Mason et al. 2014b) and Pseudoalteromonas, Colwellia, and Shewanella with polyaromatic hydrocarbons (Dong et al. 2015). Perhaps unsurprisingly, therefore, the same species are consistently isolated in rich media under high hydrostatic pressure conditions. These species have been isolated over long time scales and geographic distances, suggesting a widespread distribution in the deep ocean. Even strains within specific bathytypes, like those of S. benthica, can display distinct pressure optima and roughly cluster based on depth, suggesting they may colonize distinct depth-zones (Fig. 2.1). Comparisons of these strains may reveal small but significant differences required for adaptation to increasing depth. However, no studies thus far have assessed the in situ depth restrictions of piezophiles.
In contrast to culture-based studies, community analysis using deep 16S rRNA gene sequencing suggests deep-ocean microbial communities are composed of abundant, widely distributed species and a number of rare members with a more limited distribution (Sogin et al. 2006; Salazar et al. 2015a, b). Deep-ocean communities tend to be more similar to one another when compared to shallower samples as deep-sea datasets cluster together despite disparate environmental locations (DeLong et al. 2006; Eloe et al. 2011c; Thureborn et al. 2013; Jacob et al. 2013; Nunoura et al. 2015), a finding that also seems to be true with viral communities at depth (Yoshida et al. 2013; Winter et al. 2014; Hurwitz et al. 2015). Within these deep-sea communities composition has been shown to differ depending upon their size-fraction (DeLong et al. 1993; Moeseneder et al. 2001; Simon et al. 2002, 2014; Wilkins et al. 2013; Ganesh et al. 2014; Salazar et al. 2015a, b), which is thought to separate free-living from particle-associated taxa. Comparisons of bathypelagic communities have shown that free-living communities are dependent on temperature and depth and are similar between different sites, whereas particle-attached communities are more basin-specific and dependent on dispersal limitation, water age, and mixing (Salazar et al. 2015a). Deep-ocean particles may be produced at depth and thus lack a surface ocean connection, leading to their distinctive basin-specific microbial community compositions (Herndl and Reinthaler 2013; Salazar et al. 2015a). A high proportion of taxa, including 7 of 15 identified phyla, were consistently found to be associated with either the free-living or particle-attached size fraction in the bathypelagic. The archaea, including the Thaumarchaea, SAR86, SAR324, SAR406, and SAR202 clades were all associated with the free-living fraction, while the Bacteroidetes, Firmicutes, and Planctomycetes were associated with the particle-attached fraction. These findings suggest that these size-fractions have distinct biogeochemical roles associated with differing lifestyles and are consistent with our understanding of the metabolism of many of these taxa (Salazar et al. 2015b). Future studies should focus on how mesoscale events, seasonal cycles, and decadal shifts may alter community composition due to the tight coupling between the deep ocean and the surface by sampling at the same sites over longer time scales.
The most abundant deep-ocean prokaryotic groups are the Gammaproteobacteria (López-García et al. 2001; Delong et al. 2006; Konstantinidis et al. 2009; Smedile et al. 2013; Wilkins et al. 2013; Nunoura et al. 2015; Salazar et al. 2015a, b), Alphaprotebacteria (Martin-Cuadrado et al. 2007; Eloe et al. 2011c), Deltaproteobacteria, (Salazar et al. 2015a, b), Actinobacteria (Salazar et al. 2015a, b), and Thaumarchaea (Martin-Cuadrado et al. 2009; Eloe et al. 2011c; Yakimov et al. 2011). Archaeal abundances have been seen to range from 2% (Salazar et al. 2015a, b) to 39% (Karner et al. 2001) of deep-sea communities. In a variety of bathypelagic settings Salazar and colleagues found that the only abundant and cosmopolitan OTUs belonged to Alteromonas, MGI Thamaurchaea, and SAR324, despite the presence of similar microbial community compositions when assessed at broader phylogenetic classifications (Salazar et al. 2015a, b). The Deltaproteobacteria SAR324 clade can be divided into distinct clusters (Brown and Donachie 2007) and is specifically enriched within oxygen minimum zones (Wright et al. 2012) and in meso- and bathy-pelagic dark-ocean locales (Wright et al. 1997; Treusch et al. 2009; Wright et al. 2012; Nunoura et al. 2015). Other groups have been seen to increase with depth along depth profiles, including the Planctomycetes, Gemmatimonadetes, Acidobacteria, Alteromonadaceae, Nitrospina, SAR11 (Delong et al. 2006; Konstantinidis et al. 2009), SAR406 (Nunoura et al. 2015), and SAR202 of the Chloroflexi (Morris et al. 2004; Varela et al. 2008). The candidate phylum SAR406 (known as the ‘Marinimicrobia’ or Marine Group A) is one of the most consistent groups of microbes found preferentially at depth (Nunoura et al. 2015; Salazar et al. 2015a, b). More than 10 subgroups have been identified (Allers et al. 2013; Wright et al. 2014) and many members are prevalent in low oxygen environments. This is consistent with fosmid library gene studies suggesting that Marinimicrobia have adaptations to O2 deficient conditions and may be capable of sulfur-based energy metabolism (Wright et al. 2014). Single-amplified genomes have been published (Rinke et al. 2013) but while SAR406 is consistently identified in deep-ocean samples (López-García et al. 2001; Wilkins et al. 2013; Salazar et al. 2015a, b), including trenches (Eloe et al. 2011c; Nunoura et al. 2015), little is known about the physiological properties of its deeper dwelling members.
Much more is known about community composition at bathyal and abyssal depths than of the hadal zone. Recently a community depth profile down to 10,257 m in the Challenger Deep portion of the Mariana Trench identified microbial assemblage stratification through the water column (Nunoura et al. 2015). An increase in heterotrophic microbial groups was present at depths exceeding 7000 m at the expense of chemoautotrophs, potentially as a result of higher availability of POC. Communities deeper than 9000 m were dominated by Pseudomonas spp. In addition, ammonia oxidizers and nitrifiers also showed distinct depth trends, with ammonia oxidizing bacteria (AOB) of the genus Nitrospira abundant in the surface and upper water column and in the hadal zone, while ammonia oxidizing archaea (AOA) and Nitrospina were abundant in the intermediate depths. These results are consistent with niche separation due to the increased availability of ammonia and nitrite resulting from the decomposition of organic matter in the upper water column and within the trench, as AOB prefers higher ammonia concentrations than AOA. These results add to a growing body of evidence that the hadal ocean may display increased rates of heterotrophy because of increased carbon deposition due to the funneling and concentration of POC into the deeper trench regions (Glud et al. 2013; Ichino et al. 2015; Nunoura et al. 2015). Interestingly, in contrast to the Challenger Deep pelagic waters most bacterial sequences obtained from the water column at 6000 m in the Puerto Rico Trench were members of the SAR11 clade (Eloe et al. 2011c). The difference in community composition between these two hadal settings, even when considering depth and distance from the seafloor, suggests that trench communities can differ markedly from one another.
2.5 What Is the Effect of Decompression?
Sample handling is an especially important consideration when working with deep-ocean samples. One normal consequence following the collection of materials from within the deep ocean is sample decompression during recovery. Because sample collection can take upwards of four hours to return to the surface from the hadal zone, followed by time for sample processing, it is likely that community composition changes occur as a result of physiological stress. All piezophiles currently in culture are able to withstand moderate periods of decompression. This is evident by the decompression that is routinely employed during most sample recoveries, as well as during the preparation of piezophile subcultures and frozen stocks. While incubation at 0.1 MPa for <10 h did not lead to large decreases in CFUs for the obligate piezophile Colwellia sp. MT41 or the moderate piezophile Psychromonas sp. CNPT3, decompression of MT41 for 50 h led to CFU decreases by over five orders of magnitude and cell ultrastructural alterations (Yayanos and Dietz 1983; Chastain and Yayanos 1991). This suggests that given enough time at atmospheric pressure many obligately piezophilic deep-ocean microbes will lose viability.
While isolate viability may be maintained during short-term decompression the effects of decompression or decompression-recompression cycles on community composition and their activity is relatively unexplored. Some research groups have developed pressure-retaining samplers or equipment for in situ filtration and preservation (Yayanos 1977, Tabor et al. 1981a; Jannasch and Wirsen 1982; Bianchi et al. 1999; Kato 2006; Tamburini et al. 2009). One study developed and deployed a sampler to ~2200 m water depth and compared transcriptomes of seawater filtered in situ and after collection at the surface (Edgcomb et al. 2014). Almost two times more total and classifiable reads were recovered from the in situ filtered and fixed samples as compared with those filtered post-recovery, and changes in gene expression were also noted between the two sample collection methods. Members of the Thaumarchaea may be some of the microbes most affected by sampling methods that include decompression (La Cono et al. 2015). La Cono and colleagues collected samples from 500 m and 2222 m in the Mediterranean Sea and filtered immediately after collection or after incubation for 24 or 72 h at in situ temperatures. In samples left for three days archaeal populations decreased three-fold while their bacterial populations doubled. Thaumarchaea composition shifted over time, perhaps reflecting the existence of autochthonous and allochthonous ecotypes. Regardless of how communities and activities shift as a function of pressure, maintaining in situ pressure conditions should be an important consideration when sampling the deep ocean to obtain ecologically relevant results.
2.6 How Do Bathytype Features Compare with Results from Culture-Independent Metagenome Analyses?
Comparisons of deep-ocean bathytypes with shallow-water ecotypes have indicated the presence of a variety of characteristics that correlate with growth and survival at depth. These include larger genome sizes, increased abundances of transposable elements, the ability to colonize POM and break down recalcitrant DOC, heavy metal resistance, and the absence of genes for repairing UV-induced DNA damage (Table 2.1). A valuable complement to the genome comparisons of cultured deep and shallow ecotypes is the comparison of deep and shallow metagenomic sequence data. Metagenomic data can provide insight into broad community genetic and physiological characteristics, and when compared with specific bathytypes, can shed light on genomic changes required for deep-ocean lifestyles. Below these two approaches are compared with regard to (1) particle attachment, (2) mobile elements and genome size, (3) heavy metal resistance, (4) recalcitrant organic carbon utilization and (5) autotrophy and lithotrophy.
Particle attachment. Deep-ocean microbes are thought to be largely dependent on particulate organic matter (POM) sinking from the surface. Sinking POM are microbial “hot spots,” harboring dense aggregations of bacteria that can be orders of magnitude higher than the surrounding seawater thanks to higher nutrient levels (Azam and Long 2001; Simon et al. 2002; Lyons and Dobbs 2012; Turner 2015). Despite the notion that the deep ocean is carbon-limited, large fluxes of organic matter into the deep ocean have been seen, showing that there are pulses of detritus to the deep-sea floor (Billett et al. 1983; Rice et al. 1986; Lochte and Turley 1988; Thiel et al. 1989; Danovaro et al. 2002; Wu et al. 2013; Agusti et al. 2014), and trenches may be enriched in organic matter when compared to shallower sites thanks to topography (Boetius et al. 1996; Danovaro et al. 2003; Gooday et al. 2010; Glud et al. 2013; Ichino et al. 2015). It is likely that deep-ocean communities are adapted to generally low and sporadic fluxes of organic matter and have methods to cope with resource scarcity, such as increased numbers of signal transduction pathways in the Puerto Rico Trench (Eloe et al. 2011a). They may also require movement for quick colonization when POM becomes available. Deep bathytypes are typically enriched in methyl-accepting chemotaxis proteins which may be involved in finding POM and DOM (Lauro and Bartlett 2008). For example, the piezophilic bathytype Psychromonas sp. CNPT3 and a Psychromonas SAG from the Puerto Rico Trench have genes involved in motility and chemotaxis not present in the psychrophile Psychromonas ingrahamii (Stratton 2008; Léon-Zayas et al. 2015). Genes for pilus synthesis, type II secretion systems, polysaccharide synthesis, and antibiotic synthesis, which are associated with microbes who live particle-attached lifestyles or are part of a biofilm, were enriched at 4000 m (DeLong et al. 2006), consistent with other deep-sea sites (Tringe et al. 2005; Martin-Cuadrado et al. 2007). In contrast, flagellar biosynthesis and bacterial chemotaxis proteins were more highly represented in photic zone samples than those in the deep water at Station ALOHA (Delong et al. 2006). Deep-ocean microbes may also colonize particles, reach high densities, and produce bioluminescence in order to be engulfed by larger predators and colonize their gastrointestinal tracts (Zarubin et al. 2012; Martini et al. 2013). The moderate piezophile Photobacterium phosphoreum ANT-2200 showed three times more bioluminescence when grown at 22 MPa than at 0.1 MPa, forming aggregates under these high pressure conditions (Martini et al. 2013). Threefold more luciferase oxygenase homologs for bioluminescence production were found at 4000 m than in surface datasets (Konstantinidis et al. 2009). However, release of these microbes from larger organisms as fecal pellets in shallower waters, which then sink into the deep ocean, may also be responsible for increased numbers of genes involved in bioluminescence present at depth.
Genome size and mobile elements. Genome size estimates in both cultured deep-ocean microbes and metagenomic analyses show increases versus their shallow-water counterparts. Estimates of genome size at Station ALOHA showed a ~1.35 fold increase in the bathypelagic versus the surface ocean (Konstantinidis et al. 2009; Beszteri et al. 2010). One potential reason for increased genome size is an enrichment of transposases within deep-ocean microbes, such as in bathytypes of Photobacterium, Shewanella, and Pseudoalteromonas. A Marinosulfonomonas SAG from the Puerto Rico Trench also has a large number of transposases (Léon-Zayas et al. 2015). One of the most striking findings in the Station ALOHA metagenomes was increased abundances of transposases, integrases, and ratios 2–3 times higher of nonsynonymous to synonymous (Dn/Ds) mutations at depth (Delong et al. 2006; Konstantinidis et al. 2009). Transposase sequences were from different families and present in diverse microbial taxa suggesting this is not attributable to one specific group in high abundance. Similar abundances have been noted in the deep Mediterranean at 4908 m where transposases, phage integrases, and plasmids were ~10 fold more abundant than in the surface ocean (Smedile et al. 2013). Furthermore, deep-ocean bathytypes and metagenomes are enriched in genes involved in phage interaction, such as CRISPR regions (Ivars-Martínez et al. 2008b; Smedile et al. 2013; Thrash et al. 2014; Mason et al. 2014a). The ratio of viruses to prokaryotes typically increases with depth within the water column (Parada et al. 2007; De Corte et al. 2010; Nunoura et al. 2015) but not in the sediment (Corinaldesi 2015). Still, in one study almost half of deep-ocean sediment isolates harbored prophages (Engelhardt et al. 2011) and viral lysis may be responsible for most prokaryotic mortality in deep-sea sediments (Danovaro et al. 2008), highlighting the importance of viruses in these ecosystems. Protein clusters found exclusively in viruses in the aphotic deep Pacific paralleled those that function in adaptation to high hydrostatic pressure, including those involved in DNA replication, DNA repair, and motility, potentially boosting their hosts’ fitness (Hurwitz et al. 2015). Taken together with increases in intergenic spacer regions, these findings suggest more relaxed purifying selection in deep-ocean microbes. Microbes with large genomes may also be more successful in environments with low and diverse resources and where there is no drawback for slow growth (Konstantinidis and Tiedje 2004), such as in SAR11 (Thrash et al. 2014). Both the ability to quickly colonize energy sources when available or grow slowly in an oligotrophic environment are niches found in the deep-ocean.
Heavy-metal toxicity. Another adaptation that may reflect particle attachment is the abundance of genes involved in heavy-metal efflux. Like other bathytypes the deep-ocean isolate Halomonas zincidurans contains heavy-metal resistance genes involved in copper homeostasis and tolerance, cobalt–zinc–cadmium resistance, mercuric reduction, and arsenic resistance, and was experimentally shown to tolerate elevated levels of zinc (Xu et al. 2013). Similarly, metagenomic data has shown enrichment in heavy-metal resistance genes at depth. Both the deep Puerto Rico Trench and Mediterranean metagenomes were enriched in genes for heavy metal efflux and detoxification (Eloe et al. 2011a; Smedile et al. 2013). Live sediment trap communities collected from 500 m at Station ALOHA, which were essentially enrichments of actively growing microbes associated with sinking particulate matter, had higher abundances of genes for heavy-metal resistance (czcABCD), copper efflux (cusAB) and copper two-component sensory systems (cusRS), and mercury resistance (merABR) and transport (merTP) (Fontanez et al. 2015). These genes were affiliated with Alteromonas, Marinobacter, and Glaciecola, genera typically associated with POM and which have been isolated from deep-sea sites. Further analysis of size-fractionated metagenomes, coupled with culture-based studies, will provide insight into the microbial groups and their underlying physiology that are responsible for enrichments in heavy-metal efflux genes and mobile elements at depth.
Recalcitrant organic carbon utilization. DOC export into the mesopelagic and bathypelagic is thought to represent between 10 and 20% of total primary productivity at the surface (Carlson et al. 2010, Giering et al. 2014), leading to concentrations of approximately 40 umol L−1 in the deep ocean (Arrieta et al. 2015). Refractory DOC is the dominant form of DOC present in the deep ocean because of its resistance to rapid microbial degradation and subsequent accumulation (Hansell 2013). However, a recent study showed that deep bathypelagic communities are capable of using in situ DOC but low concentrations make its use not energetically feasible (Arrieta et al. 2015), a finding that has been previously suggested (Kujawinski 2011; Hansell 2013). Surface water communities amended with high molecular weight DOM showed that Alteromonas spp. and Idiomarina spp. were the most highly represented taxa soon after addition (McCarren et al. 2010), consistent with the hypothesis that A. macleodii AltDE is capable of breaking down DOM (Ivars-Martínez et al. 2008b). The class Dehalococcoidia of the Chloroflexi, another microbial group identified in marine sediment communities at depth, may also be able to perform oxidation of complex organic compounds (Wilms et al. 2006; Fry et al. 2008) or carbon fixation via the Wood-Ljungdahl pathway (Wasmund et al. 2014). Metabolic genes related to the degradation of refractory DOC have also been identified in deep-ocean metagenomes. Genes for glyoxylate and dicarboxylate metabolism for degradation of oxidized and degraded DOM have been identified in deep samples (Delong et al. 2006; Eloe et al. 2011a) and the 3010 m Mediterranean metagenome was enriched in pathways for the breakdown of a number of recalcitrant forms of carbon (Martin-Cuadrado et al. 2007). Interestingly, whole-body extracts of the hadal amphipod Hirondellea gigas showed the ability to degrade plant-derived polysaccharides using amylase, cellulose, mannanase, xylanase, and α-glucosidase (Kobayashi et al. 2012). Bacteria were not able to be isolated from these samples and bacterial or archaeal DNA was unable to be amplified, leading the authors to believe this activity is performed by the amphipod itself. However, dockerin type I repeats, which are involved in cellulose degradation, were more abundant in deep Station ALOHA samples (Konstantinidis et al. 2009) and P. profundum SS9 upregulates pathways to degrade chitin and cellulose under high pressure (Vezzi et al. 2005). Furthermore, piezophilic bacteria have been isolated from H. gigas previously (Yayanos et al. 1981), so microbes may be responsible for the breakdown of these hard-to-process recalcitrant materials in amphipods.
Autotrophy and lithotrophy. Because of the assumed reliance of deep-ocean communities on POC, estimates of POC flux should theoretically be balanced with community metabolism. However, the metabolic activity of deep-sea microbial communities can be up to 2 orders of magnitude higher than that which is sustainable through measured estimates of sinking POC (Reinthaler et al. 2006; Baltar et al. 2009; Giering et al. 2014). This mismatch highlights potential problems estimating microbial activity or POC flux or could reflect the presence of as yet unidentified sources of organic carbon, such as neutrally buoyant macroscopic particles (Bochdansky et al. 2010) or virus decomposition (Dell’Anno et al. 2015).
One alternative source of organic carbon could be the fixation of dissolved inorganic carbon in the dark ocean. The abundance of genes involved in autotrophy were comparable at 4000 m to those at the surface at Station ALOHA (Konstantinidis et al. 2009) and genes for autotrophy in the 6000 m Puerto Rico Trench metagenome showed comparable abundances to those at the surface, although some key enzymes were missing (Eloe et al. 2011a). These findings are consistent with activity measurements that have shown that carbon fixation in the deep sea is an important source of organic carbon, with estimates of dark primary production being similar to rates of heterotrophic production and equal to 15–53% of the carbon that is exported from the surface (Reinthaler et al. 2010). It is thought that Thaumarchaea may be predominantly responsible for autotrophy in many deep-sea settings (Herndl et al. 2005; Hallam et al. 2006; Ingalls et al. 2006; Wuchter et al. 2006; Yakimov et al. 2011; Smedile et al. 2013; Swan et al. 2014). However, bacteria may also perform DIC fixation in the dark ocean and in some situations could incorporate DIC at higher rates than archaea (Varela et al. 2011). Screening of 502 bacterial single-amplified genomes from 770 and 800 m showed that 12% were positive for RuBisCO, with 25% of the Gammaproteobacteria and 47% of SAR324 encoding the RuBisCO large subunit (Swan et al. 2014). In the case of the deep-sea clade SAR324, the consistent co-occurrence of RuBisCO and sulfur oxidation genes may mean that dissimilatory sulfur oxidation is used for energetic support of autotrophic carbon fixation, although other studies have concluded that members of SAR324 could also be heterotrophic (Chitsaz et al. 2011; Sheik et al. 2014). Sulfur oxidation and autotrophy have also been identified in the SUP05 clade, a subgroup within the Gammaproteobacteria that predominates at suboxic and anoxic sites (Walsh et al. 2009; Wright et al. 2012; Anantharaman et al. 2013). Interestingly, genomes of viruses associated with SUP05 contained genes for subunits of reverse dissimilatory sulfite reductase and may represent a genetic reservoir responsible for conferring this ability to SUP05 (Anantharaman et al. 2014; Roux et al. 2014).
In addition to chemoautotrophy it has been noted that heterotrophic microbes could be responsible for some of the measured dark primary productivity by incorporating CO2 during anaplerotic reactions, which provide no net gain of carbon. Heterotrophs, including Alteromonas macleodii AltDE, may actively take up CO2 in nutrient replete conditions to replace necessary metabolic precursors or intermediates (Alonso-Sáez et al. 2010; Yakimov et al. 2014). Deep-ocean microbial communities are important drivers of carbon remineralization and sequestration and a greater understanding of their function, both in using and fixing organic carbon, will be fundamental to evaluating the role of the deep ocean as a carbon sink.
Electron donors in the deep ocean are scarce (Reinthaler et al. 2010) and some deep-ocean microbes may use unique sources. One such example is the prevalence of genes for aerobic carbon monoxide oxidation in the deep sea. The coxL, coxM, and coxS genes, which encode for carbon monoxide dehydrogenase, have been identified in many deep-ocean metagenomes (Martin-Cuadrado et al. 2007, 2009; Eloe et al. 2011b; Quaiser et al. 2011; Smedile et al. 2013). Carbon monoxide oxidation may provide an alternative or supplementary energy source (Martin-Cuadrado et al. 2007) and while the possible role of CO in the deep ocean has been questioned because the source is unclear (Quaiser et al. 2011), it is possible that it is produced during organic matter degradation (Eloe et al. 2011b). Most Thaumarchaea in the deep ocean are thought to be capable of oxidizing ammonia based on abundances of amoA genes (Konstantinidis et al. 2009; Yakimov et al. 2011) and their recovery from the majority of mesopelagic SAGs from the South Atlantic (60%) and North Pacific (81%) (Swan et al. 2014). However, low ammonia levels in the deep ocean may result in archaeal nitrification that is too low to support the measured rates of carbon fixation (Herndl et al. 2005; Reinthaler et al. 2010). The identification of urease genes within MG1 SAGs suggests they may also oxidize urea, providing an alternative energy production pathway (Alonso-Sáez et al. 2012; Swan et al. 2014).
2.7 Conclusions and Future Directions
Deep-ocean communities contain genotypic and phenotypic properties that confer adaptation to the deep ocean. Many attributes associated with deep-ocean bathytypes, such as genes for particle-association, heavy-metal resistance, and increased numbers of mobile elements are also found in metagenomes, suggesting they are relatively conserved across major groups of deep-sea taxa. Most of the bathytypes discussed here show growth at atmospheric as well as high pressure and more extreme comparisons with obligate piezophiles may allow for a better understanding of the adaptations to the deep ocean. One of the major challenges moving forward will be to determine the taxonomic and functional traits that distinguish not just “deep” versus “shallow” pelagic prokaryotes, but “ultra-deep” or hadal prokaryotes from everything else, and to uncover the environmental conditions that lead to these differences. To accomplish these tasks it will be important to apply genomics technologies to a far larger number of piezophiles and hadal locations than the handful examined thus far, along with the collection of robust suites of associated biological, chemical, and physical data. Large sampling expeditions comparable to the Global Ocean Sampling Expedition or the TARA Expedition (Rusch et al. 2007; Karsenti et al. 2011) should be considered for global analyses of microbial communities and their genomic adaptations within hadal environments.
Another major emphasis must be to distinguish autochthonous from allochthonous microorganisms present at great depths. Because pressure is clearly a major driver of the vertical distribution of marine life, one way to address this question will be to determine the impact of retaining or changing the in situ pressure on the identities and activities of the microbes present. This objective becomes progressively more complicated, technically challenging and expensive with increasing depth and pressure. Another approach is to consider ways in which the microbiology to be performed can be accomplished in the deep sea prior to sample retrieval. This could involve the fixation of samples at depth for later processing topside or through analyses done remotely like that conducted with the Environmental Sample Processor (Scholin et al. 2009).
Another major research direction must include more mechanistic studies of the basis of life at great depth. In most cases this will involve pure cultures, but innovative interrogations of microbial communities present in microcosms might also yield new insights. These efforts would benefit from the collection of a much more diverse and representative collection of piezophiles than now exist. Enrichments with more complex organic sources over longer time scales may provide new species of microbes for study, and transitioning from batch to recirculating culturing approaches to maintain carbon sources, electron donors, and electron acceptors (notably dissolved oxygen) at constant concentrations under high-hydrostatic pressure should be pursued (Parkes et al. 2009; Deusner et al. 2010; Zhang et al. 2011; Sauer et al. 2012; Ohtomo et al. 2013; Foustoukos and Perez-Rodriquez 2015).
Important information already exists on the cellular changes needed for life at elevated pressure, most clearly in the case of the requirement for elevated unsaturated fatty acids in the membranes of psychrophilic or psychrotolerant piezophiles. However, despite the close phylogenetic relationship that exists between piezophilic and nonpiezophilic microbes, fundamental gaps remain in the understanding of the basis of high-pressure life. Thus, high-pressure growth appears to arise from a modest number of subtle changes. The application of systems biology approaches (transcriptomics, proteomics, lipidomics, metabolomics) in concert with metabolic flux analyses, osmolyte measurements, directed evolution, genetics, biochemistry, and modeling will all be needed for further progress. Genetic studies are currently possible for only a small number of piezophiles (Chen et al. 2011; El-Hajj et al. 2010; Thiel et al. 2014; Li et al. 2015).
There are many reasons to study deep-sea life in general and ultra-deep microbes in particular. Deep-sea microbes are considered important targets for the search for bioactive secondary metabolites and drugs (Hopwood 2007) or as sources of enzymes and metabolic pathways for the breakdown of complex hydrocarbons, plastics or other organics (Sekiguchi et al. 2011). The study of deep-sea microbes also makes it possible to examine life under different boundaries of pressure and temperature than is the case for virtually all other habitats, excluding the deep subsurface. In this regard they are useful as proxies for possible life forms existing in extraterrestrial habitats (Schrenk and Brazelton 2013). But, they should primarily be appreciated as the members of a major portion of the biosphere, responsible for much of the carbon mineralization and sequestration that takes place on Earth.
References
Agogué H, Brink M, Dinasquet J, Herndl GJ (2008) Major gradients in putatively nitrifying and non-nitrifying Archaea in the deep North Atlantic. Nature 456:788–792. doi:10.1038/nature07535
Agusti S, González-Gordillo JI, Vaqué D, Estrada M, Cerezo MI, Salazar G, Gasol JM, Duarte CM (2014) Ubiquitous healthy diatoms in the deep sea confirm deep carbon injection by the biological pump. Nat Commun 6:1–8. doi:10.1038/ncomms8608
Alazard D, Dukan S, Verhé F, Bouabida N, Morel F, Thomas P, Garcia JL, Ollivier B (2003) Desulfovibrio hydrothermalis sp. nov., a novel sulfate-reducing bacterium isolated from hydrothermal vents. Int J Syst Evol Microbiol 53:173–178. doi:10.1099/ijs.0.02323-0
Allen EE, Bartlett DH (2000) FabF is required for piezoregulation of cis-Vaccenic acid levels and piezophilic growth of the deep-sea bacterium Photobacterium profundum strain SS9. J Bacteriol 182(5):1264–1271. doi:10.1128/JB.182.5.1264-1271.2000
Allen EE, Facciotti D, Bartlett DH (1999) Monounsaturated but not polyunsaturated fatty acids are required for growth of the deep-sea bacterium Photobacterium profundum SS9 at high pressure and low temperature. Appl Environ Microbiol 65(4):1710–1720
Allers E, Wright JJ, Konwar KM, Howes CG, Beneze E, Hallam SJ, Sullivan MB (2013) Diversity and population structure of Marine Group A bacteria in the Northeast subarctic Pacific Ocean. ISME J 7:256–268. doi:10.1038/ismej.2012.108
Alonso-Sáez L, Galand PE, Casamayor EO, Pedrós-Alió C, Bertilsson S (2010) High bicarbonate assimilation in the dark by Arctic bacteria. ISME J 4:1581–1590. doi:10.1038/ismej.2010.69
Alonso-Sáez L, Sánchez O, Gasol JM (2012) Bacterial uptake of low molecular weight organics in the subtropical Atlantic: are major phylogenetic groups functionally different? Limnol Oceanogr 57(3):798–808
Amaral GRS, Campeao ME, Swings J, Thompson FL, Thompson CC (2015) Finding diagnostic phenotypic features of Photobacterium in the genome sequences. Antonie Van Leeuwenhoek 107:1351–1358
Amrani A, Bergon A, Holota H, Tamburini C, Garel M, Ollivier B, Imbert J, Dolla A, Pradel N (2014) Transcriptomics reveal several gene expression patterns in the piezophile Desulfovibrio hydrothermalis in response to hydrostatic pressure. PLoS one 9(9):1–10. doi:10.1371/journal.pone.0106831
Anantharaman K, Breier JA, Sheik CS, Dick GJ (2013) Evidence for hydrogen oxidation and metabolic plasticity in widespread deep-sea sulfur-oxidizing bacteria. Proc Natl Acad Sci 110(1):330–335. doi:10.1073/pnas.1215340110
Anantharaman K, Duhaime MB, Breier JA, Wendt KA, Toner BM, Dick GJ (2014) Sulfur oxidation genes in diverse deep-sea viruses. Science 344:757–760. doi:10.1126/science.1252229
Aono E, Baba T, Ara T, Nishi T, Nakamichi T, Inamoto E, Toyonaga H, Hasegawa M, Takai Y, Okumura Y, Baba M, Tomita M, Kato C, Oshima T, Nakasone K, Mori H (2010) Complete genome sequence and comparative analysis of Shewanella violacea, a psychrophilic and piezophilic bacterium from deep sea floor sediments. Mol BioSyst 6:1216–1226. doi:10.1039/C000396D
Arrieta JM, Mayol E, Hansman RL, Herndl GJ, Dittmar T, Duarte CM (2015) Dilution limits dissolved organic carbon utilization in the deep ocean. Science 348:331–333. doi:10.1126/science.1258955
Azam F, Long RA (2001) Oceanography: sea snow microcosms. Nature 414:495–498. doi:10.1038/35107174
Bælum J, Borglin S, Chakraborty R, Fortney JL, Lamendella R, Mason OU, Auer M, Zemla M, Bill M, Conrad ME, Malfatti SA, Tringe SG, Holman H, Hazen TC, Jansson JK (2012) Deep-sea bacteria enriched by oil and dispersant from the Deepwater Horizon spill. Environ Microbiol 14(9):2405–2416. doi:10.1111/j.1462-2920.2012.02780.x
Bale SJ, Goodman K, Rochelle PA, Marchesi JR, Fry JC, Weightman AJ, Parkes RJ (1997) Desulfovibrio profundus sp. nov., a novel barophilic sulfate-reducing bacterium from deep sediment layers in the Japan Sea. Int J Syst Evol Microbiol 47(2):515–521. doi:10.1099/00207713-47-2-515
Baltar F, Arístegui J, Gasol J, Sintes E, Herndl GJ (2009) Evidence of prokaryotic metabolism on suspended particulate organic matter in the dark waters of the subtropical North Atlantic. Limnol Oceanogr 54(1):182–193. doi:10.4319/lo.2009.54.1.0182
Bartlett DH (2002) Pressure effects on in vivo microbial processes. Biochim Biophys Acta 1595:367–381. doi:10.1016/S0167-4838(01)00357-0
Batzke A, Engelen B, Sass H, Cypionka H (2007) Phylogenetic and physiological diversity of cultured deep-biosphere bacteria from equatorial Pacific Ocean and Peru margin sediments. Geomicrobiol J 24:261–273. doi:10.1080/01490450701456453
Baumann L, Baumann P, Mandel M, Allen RD (1972) Taxonomy of aerobic marine Eubacteria. J Bacteriol 110(1):402–429
Beman JM, Popp BN, Francis CA (2008) Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J 2:429–441. doi:10.1038/ismej.2007.118
Beszteri B, Temperton B, Frickenhaus S, Giovannoni SJ (2010) Average genome size: a potential source of bias in comparative metagenomics. ISME Journal 4:1075–1077. doi:10.1038/ismej.2010.29
Bianchi A, Garcin J, Tholosan O (1999) A high-pressure serial sampler to measure microbial activity in the deep sea. Deep-Sea Res I 46:2129–2142
Biddle JF, House CH, Brenchley JE (2005) Enrichment and cultivation of microorganisms from sediment from the slope of the Peru Trench (ODP Site 1230). Proc ODP Sci Results 201:1–19
Billett DSM, Lampitt RS, Rice AL, Mantoura RFC (1983) Seasonal sedimentation of phytoplankton to the deep-sea benthos. Nature 302:520–522. doi:10.1038/302520a0
Bochdanksy AB, van Aken HM, Herndl GJ (2010) Role of macroscopic particles in deep-sea oxygen consumption. Proc Natl Acad Sci 107(18):8287–8291. doi:10.1073/pnas.0913744107
Boetius A, Scheibe S, Tselepides A, Thiel H (1996) Microbial biomass and activities in deep-sea sediments of the Eastern Mediterranean: trenches are benthic hotspots. Deep-Sea Res I 43(9):1439–1460. doi:10.1016/S0967-0637(96)00053-2
Bordi C, Iobbi-Nivol I, Méjean V, Patte J (2003) Effects of ISSo2 insertions in structural and regulatory genes of the trimethylamine oxide reductase of Shewanella oneidensis. J Bacteriol 185(6):2042–2045. doi:10.1128/JB.185.6.2042-2045.2003
Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P (2008) Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nature Rev Microbiol 6:245–252. doi:10.1038/nrmicro1852
Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, Wilkins MJ, Wrighton KC, Williams KH, Banfield JF (2015) Unusual biology across a group comprising more than 15% of domain bacteria. Nature 523(7559):208–211. doi:10.1038/nature14486
Brown MV, Donachie SP (2007) Evidence for tropical endemicity in the Deltaproteobacteria Marine Group B/SAR324 bacterioplankton clade. Aquat Microb Ecol 46:107–115
Brown MV, Fuhrman JA (2005) Marine bacterial microdiversity as revealed by internal transcribed spacer analysis. Aquat Microb Ecol 41(1):15–23
Brown MV, Lauro FM, DeMaere MZ, Muir L, Wilkins D, Thomas T, Riddle MJ, Fuhrman JA, Andrews-Pfannkoch C, Hoffman JM, McQuaid JB, Allen A, Rintoul SR, Cavicchioli R (2012) Global biogeography of SAR11 marine bacteria. Mol Syst Biol 8(595):1–13. doi:10.1038/msb.2012.28
Burke C, Steinberg P, Rusch D, Kjelleberg S, Thomas T (2011) Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci 108(34):14288–14293. doi:10.1073/pnas.1101591108
Campanaro S, Vezzi A, Vitulo N, Lauro F, D’Angelo M, Simonato F, Cestaro A, Malacrida G, Bertoloni G, Valle G, Bartlett DH (2005) Laterally transferred elements and high pressure adaptation in Photobacterium profundum strains. BMC Genom 6(122):1–15. doi:10.1186/1471-2164-6-122
Cao Y, Chastain RA, Eloe EA, Nogi Y, Kato C, Bartlett DH (2014) Novel psychropiezophilic Oceanospirillales species Profundimonas piezophila gen. nov., sp. nov., isolated from the deep-sea environment of the Puerto Rico Trench. Appl Environ Microbiol 80(1):54–60. doi:10.1128/AEM.02288-13
Carlson CA, Hansell DA, Nelson NB, Siegel DA, Smethie WM, Khatiwala S, Meyers MM, Halewood E (2010) Dissolved organic carbon export and subsequent remineralization in the mesopelagic and bathypelagic realms of the North Atlantic basin. Deep-Sea Res II 57(16):1433–1445. doi:10.1016/j.dsr2.2010.02.013
Carlson CA, Morris R, Parsons R, Treusch AH, Giovannoni SJ, Vergin K (2009) Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J 3:283–295. doi:10.1038/ismej.2008.117
Chakraborty R, Borglin SE, Dubinsky EA, Andersen GL, Hazen TC (2012) Microbial response to the MC-252 oil and Corexit 9500 in the Gulf of Mexico. Front Microbiol 3(357):1–6. doi:10.3389/fmicb.2012.00357
Chastain RA, Yayanos AA (1991) Ultrastructural changes in an obligately barophilic marine bacterium after decompression. Appl Environ Microbiol 57(5):1489–1497
Chen X, Zhang Y, Gao P, Laun X (2003) Two different proteases produced by a deep-sea psychrotrophic bacterial strain, Pseudoalteromonas sp. SM9913. Mar Biol 143(5):989–993
Chen Y, Wang F, Xu J, Mehmood M, Xiao X (2011) Physiological and evolutionary studies of NAP systems in Shewanella piezotolerans WP3. ISME J 5:843–855. doi:10.1038/ismej.2010.182
Chikuma S, Kasahara R, Kato C, Tamegai H (2007) Bacterial adaptation to high pressure: a respiratory system in the deep-sea bacterium Shewanella violacea DSS12. FEMS Microbiol Lett 267:108–112. doi:10.1111/j.1574-6968.2006.00555.x
Chitsaz H, Yee-Greenbaum JL, Tesler G, Lombardo MJ, Dupont CL, Badger JH, Novotny M, Rusch DB, Fraser LJ, Gormley NA, Schulz-Trieglaff O, Smith GP, Evers DJ, Pevzner PA, Lasken RS (2011) Efficient de novo assembly of single-cell bacterial genomes from short-read data sets. Nat Biotechnol 29(10):915–922. doi:10.1038/nbt.1966
Church MJ, Wai B, Karl DM, Delong EF (2010) Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environ Microbiol 12(3):679–688. doi:10.1111/j.1462-2920.2009.02108.x
Corinaldesi C (2015) New perspectives in benthic deep-sea microbial ecology. Front Mar Sci 2(17):1–12. doi:10.3389/fmars.2015.00017
Danovaro R, Croce ND, Dell’Anno A, Pusceddu A (2003) A depocenter of organic matter at 7800 m depth in the SE Pacific Ocean. Deep-Sea Res I 50(12):1411–1420. doi:10.1016/j.dsr.2003.07.001
Danovaro R, Dell’Anno A, Corinaldesi C, Magagnini M, Noble R, Tamburini C, Weinbauer M (2008) Major viral impact on the functioning of benthic deep-sea ecosystems. Nature 454:1084–1088. doi:10.1038/nature07268
Danovaro R, Gambi C, Croce ND (2002) Meiofauna hotspot in the Atacama Trench, eastern South Pacific Ocean. Deep-Sea Res I 49(5):843–857. doi:10.1016/S0967-0637(01)00084-X
De Corte D, Sintes E, Winter C, Yokokawa T, Reinthaler T, Herndl GJ (2010) Links between viral and prokaryotic communities throughout the water column in the (sub)tropical Atlantic Ocean. ISME J 4:1431–1442. doi:10.1038/ismej.2010.65
Dell’Anno A, Corinaldesi C, Danovaro R (2015) Virus decomposition provides an important contribution to benthic deep-sea ecosystem functioning. Proc Natl Acad Sci 112(16):E2014–E2019. doi:10.1073/pnas.1422234112
DeLong EF (1986) Adaptations of deep-sea bacteria to the abyssal environment. PhD thesis, University of California, San Diego
DeLong EF, Franks DG, Alldredge AL (1993) Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr 38(5):924–934. doi:10.4319/lo.1993.38.5.0924
DeLong EF, Franks DG, Yayanos AA (1997) Evolutionary relationships of cultivated psychrophilic and barophilic deep-sea bacteria. Appl Environ Microbiol 63(5):2105–2108
DeLong EF, Karl DM (2005) Genomic perspectives in microbial oceanography. Nature 437:336–342. doi:10.1038/nature04157
DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard N, Martinez A, Sullivan MB, Edwards R, Brito BR, Chisholm SW, Karl DM (2006) Community genomics among stratified microbial assemblages in the ocean’s interior. Science 311:496–503. doi:10.1126/science.1120250
DeLong EF, Yayanos AA (1985) Adaptation of the membrane lipids of a deep-sea bacterium to changes in hydrostatic pressure. Science 228:1101–1103. doi:10.1126/science.3992247
Deming JW, Somers LK, Straube WL, Swartz DG, Macdonell MT (1988) Isolation of an obligately barophilic bacterium and description of a new genus, Colwellia gen. nov. Syst Appl Microbiol 10(2):152–160. doi:10.1016/S0723-2020(88)80030-4
Deusner C, Meyer V, Ferdelman TG (2010) High-pressure systems for gas-phase free continuous incubation of enriched marine microbial communities performing anaerobic oxidation of methane. Biotechnol Bioeng 105(3):524–533. doi:10.1002/bit.22553
Dong C, Bai X, Sheng H, Jiao L, Zhou H, Shao Z (2015) Distribution of PAHs and the PAH-degrading bacteria in the deep-sea sediments of the high-latitude Arctic Ocean. Biogeosciences 12:2163–2177. doi:10.5194/bg-12-2163-2015
El-Hajj ZW, Allcock D, Tryfona T, Lauro FM, Sawyer L, Bartlett DH, Ferguson GP (2010) Insights into piezophily from genetic studies on the deep-sea bacterium, Photobacterium profundum SS9. Ann. N.Y. Acad Sci 1189:143–148. doi:10.1111/j.1749-6632.2009.05178.x
El-Hajj ZW, Tryfona T, Allcock DJ, Hasan F, Lauro FM, Sawyer L, Bartlett DH, Ferguson GP (2009) Importance of proteins controlling initiation of DNA replication in the growth of the high-pressure-loving bacterium Photobacterium profundum SS9. J Bacteriol 191(20):6383–6393. doi:10.1128/JB.00576-09
Edgcomb VP, Taylor C, Pachiadaki MG, Honjo S, Engstrom I, Yakimov M (2014) Comparison of Niskin vs. in situ approaches for analysis of gene expression in deep Mediterranean Sea water samples. Deep-Sea Res 2:1–10. doi:10.1016/j.dsr2.2014.10.020
Eloe EA, Lauro FM, Vogel RF, Bartlett DH (2008) The deep-sea bacterium Photobacterium profundum SS9 utilizes separate flagellar systems for swimming and swarming under high-pressure conditions. Appl Environ Microbiol 74(20):6298–6305. doi:10.1128/AEM.01316-08
Eloe EA, Fadrosh DW, Novotny M, Allen LZ, Kim M, Lombardo MJ, Yee-Greenbaum J, Yooseph S, Allen EE, Lasken R, Williamson SJ, Bartlett DH (2011a) Going deeper: metagenome of a hadopelagic microbial community. PLoS one 6(5):1–15. doi:10.1371/journal.pone.0020388
Eloe EA, Malfatti F, Gutierrez J, Hardy K, Schmidt WE, Pogliano K, Pogliano J, Azam F, Bartlett DH (2011b) Isolation and characterization of a psychropiezophilic Alphaproteobacterium. Appl Environ Microbiol 77(22):8145–8153. doi:10.1128/AEM.05204-11
Eloe EA, Shulse CN, Fadrosh DW, Williamson SJ, Allen EE, Bartlett DH (2011c) Compositional differences in particle-associated and free-living microbial assemblages from an extreme deep-ocean environment. Environ Microbiol Rep 3(4):449–458. doi:10.1111/j.1758-2229.2010.00223.x
Engelhardt T, Sahlberg M, Cypionka H, Engelen B (2011) Induction of prophages from deep-subseafloor bacteria. Environ Microbiol 3(4):459–465. doi:10.1111/j.1758-2229.2010.00232.x
Field KG, Gordon D, Wright T, Rappe M, Urbach E, Vergin K, Giovannoni SJ (1997) Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. App Environ Microbiol 63(1):63–70
Fontanez KM, Eppley JM, Samo TJ, Karl DM, DeLong EF (2015) Microbial community structure and function on sinking particles in the North Pacific Subtropical Gyre. Front Microbiol 6(469):1–14. doi:10.3389/fmicb.2015.00469
Foustoukos DI, Perez-Rodriguez I (2015) A continuous culture system for assessing microbial activities in the piezosphere. Appl Environ Microbiol 81(19):6850–6856. doi:10.1128/AEM.01215-15
Fry JC, Parks RJ, Cragg BA, Weightman AJ, Webster G (2008) Prokaryotic biodiversity and activity in the deep subseafloor biosphere. FEMS Microbiol Ecol 66:181–196. doi:10.1111/j.1574-6941.2008.00566.x
Ganesh S, Parris DJ, Delong EF, Stewart FJ (2014) Metagenomic analysis of size-fractionated picoplankton in a marine oxygen minimum zone. ISME 8:187–211. doi:10.1038/ismej.2013.144
García-Martínez J, Rodríguez-Valera F (2000) Microdiversity of uncultured marine prokaryotes: the SAR11 cluster and the marine Archaea of Group 1. Mol Ecol 9(7):935–948. doi:10.1046/j.1365-294x.2000.00953.x
Giering SLC, Sanders R, Lampitt RS, Anderson TR, Tamburini C, Boutrif M, Zubkov MV, Marsay CM, Henson SA, Saw K, Cook K, Mayor DJ (2014) Reconciliation of the carbon budget in the ocean’s twilight zone. Nature 507:480–483. doi:10.1038/nature13123
Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, Bibbs L, Eads J, Richardson TH, Noordewier M, Rappé MS, Short JM, Carrington JC, Mathur EJ (2005) Genome streamlining in a cosmopolitan oceanic bacterium. Science 309(5738):1242–1245. doi:10.1126/science.1114057
Glud RN, Wenzhöfer F, Middelboe M, Oguri K, Turnewitsch R, Canfield DE, Kitazato H (2013) High rates of microbial carbon turnover in sediments in the deepest oceanic trench on Earth. Nat Geosci 6:284–288. doi:10.1038/ngeo1773
Gooday AJ, Uematsu K, Kitazato H, Toyofuku T, Young JR (2010) Traces of dissolved particles, including coccoliths, in the tests of agglutinated foraminifera from the Challenger Deep (10,890 m) water depth, western equatorial Pacific). Deep-Sea Res I 57:239–247. doi:10.1016/j.dsr.2009.11.003
Grote J, Thrash JC, Huggett MJ, Landry ZC, Carini P, Giovannoni SJ, Rappé MS (2012) Streamlining and core genome conservation among highly divergent members of the SAR11 clade. Mbio 3(5):1–13. doi:10.1128/mBio.00252-12
Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, Delong EF (2006) Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol 4(4):0520–0536. doi:10.1371/journal.pbio.0040095
Hammes WP, Hertel C (2006) The genera Lactobacillus and Carnobacterium. In: Dworkin et al (eds) The Prokaryotes 3rd ed, vol 4.1.2.10. Springer, US, pp 320–403. doi:10.1007/0-387-30744-3_10
Hansell DA (2013) Recalcitrant dissolved organic carbon fractions. Annu Rev Mar Sci. 5:421–445. doi:10.1146/annurev-marine-120710-100757
Hasan NA, Grim CJ, Lipp EK, Rivera ING, Chun J, Haley BJ, Taviana E, Choi SY, Hoq M, Munk AC, Brettin TS, Bruce D, Challacombe JF, Detter JC, Han CS, Eisen JA, Huq A, Colwell RR (2015) Deep-sea hydrothermal vent bacteria related to human pathogenic Vibrio species. Proc Natl Acad Sci 112(21):E2813–E2819. doi:10.1073/pnas.1503928112
Hedlund BP, Dodsworth JA, Murugapiran SK, Rinke C, Woyke T (2014) Impact of single-cell genomics and metagenomics on the emerging view of extremophile “microbial dark matter”. Extremophiles 18(5):865–875. doi:10.1007/s00792-014-0664-7
Heidelberg JF, Paulsen IT, Nelson KE, Gaidos EJ, Nelson WC, Read TD, Eisen JA, Seshadri R, Ward N, Methe B, Clayton RA, Meyer T, Tsapin A, Scott J, Beanan M, Brinkac L, Daugherty S, DeBoy RT, Dodson RJ, Durkin AS, Haft DH, Kolonay JF, Madupu R, Peterson JD, Umayam LA, White O, Wolf AM, Vamathevan J, Weidman J, Impraim M, Lee K, Berry K, Lee C, Mueller J, Khouri H, Gill J, Utterback TR, McDonald LA, Feldblyum TV, Smith HO, Venter JC, Nealson KH, Fraser CM (2002) Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat Biotechnol 20:1118–1123. doi:10.1038/nbt749
Herndl GJ, Reinthaler T (2013) Microbial control of the dark end of the biological pump. Nat Geosci 6:718–724. doi:10.1038/ngeo1921
Herndl GJ, Reinthaler T, Teira E, van Aken H, Veth C, Pernthaler A, Pernthaler J (2005) Contribution of archaea to total prokaryotic production in the deep Atlantic Ocean. Appl Environ Microbiol 71(5):2303–2309. doi:10.1128/AEM.71.5.2303-2309.2005
Hopwood DA (2007) Therapeutic treasures from the deep. Nat Chem Biol 3:457–458. doi:10.1038/nchembio0807-457
Hu A, Jiao N, Zhang R, Yang Z (2011) Niche partitioning of Marine Group I Crenarchaeota in the euphotic and upper mesopelagic zones of the East China Sea. Appl Environ Microbiol 77(21):7469–7478. doi:10.1128/AEM.00294-11
Hu Y, Fu C, Huang Y, Yin Y, Cheng G, Lei F, Lu N, Li J, Ashforth EJ, Zhang L, Zhu B (2010) Novel lipolytic genes from the microbial metagenomic library of the South China Sea marine sediment. FEMS Microbiol Ecol 72:228–237. doi:10.1111/j.1574-6941.2010.00851.x
Hurwitz BL, Brum JR, Sullivan MB (2015) Depth-stratified functional and taxonomic niche specialization in the ‘core’ and ‘flexible’ Pacific Ocean Virome. ISME J 9:472–484. doi:10.1038/ismej.2014.143
Ichino MC, Clark MR, Drazen JC, Jamieson A, Jones DOB, Martin AP, Rowden AA, Shank TM, Yancey PH, Ruhl HA (2015) The distribution of benthic biomass in hadal trenches: a modeling approach to investigate the effect of vertical and lateral organic matter transport to the seafloor. Deep Sea Res Part I 100:21–33. doi:10.1016/j.dsr.2015.01.010
Ikeda E, Andou S, Iwama U, Kato C, Horikoshi K, Tamegai H (2009) Physiological roles of two dissimilatory nitrate reductases in the deep-sea denitrifier Pseudomonas sp. strain MT-1. Biosci Biotechnol Biochem 73(4):896–2009
Ikegami A, Nakasone K, Kato C, Nakamura Y, Koshikawa I, Usami R, Horikoshi K (2000) Glutamine synthetase gene expression at elevated hydrostatic pressure in a deep-sea piezophilic Shewanella violacea. FEMS Microbiol Lett 192:91–95. doi:10.1111/j.1574-6968.2000.tb09364.x
Ingalls AE, Shah SR, Hansman RL, Aluwihare LI, Santos GM, Druffel ERM, Pearson A (2006) Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc Natl Acad Sci 103(17):6442–6447. doi:10.1073/pnas.0510157103
Ishii A, Nakason K, Sato T, Wachi M, Sugai M, Nagai K, Kato C (2002) Isolation and characterization of the dcw cluster from the piezophilic deep-sea bacterium Shewanella violacea. J Biochem 132(2):183–188
Ishii A, Sato T, Wachi M, Nagai K, Kato C (2004) Effects of high hydrostatic pressure on bacterial cytoskeleton FtsZ polymers in vivo and in vitro. Microbiology 150:1955–1972. doi:10.1099/mic.0.26962-0
Ivanova EP, Ng HJ, Webb HK (2014) The family Pseudoalteromonadaceae. In: Rosenberg E et al (eds) The Prokaryotes—Gammaproteobacteria. Springer, Berlin, pp 575–582. doi:10.1007/978-3-642-38922-1_229
Ivars-Martínez E, D’Auria G, Rodríguez-Valera F, Sánchez-Porro C, Ventosa A, Joint I, Mühling M (2008a) Biogeography of the ubiquitous marine bacterium Alteromonas macleodii determined by multilocus sequence analysis. Mol Ecol 17:4092–4106. doi:10.1111/j.1365-294X.2008.03883.x
Ivars-Martínez E, Martin-Cuadrado AB, D’Auria G, Mira A, Ferriera S, Johnson J, Friedman R, Rodríguez-Valera F (2008b) Comparative genomics of two ecotypes of the marine planktonic copiotroph Alteromonas macleodii suggests alternative lifestyles associated with different kinds of particulate organic matter. ISME J 2:1194–1212. doi:10.1038/ismej.2008.74
Jacob M, Soltwedel T, Boetius A, Ramette A (2013) Biogeography of deep-sea benthic bacteria at regional scale (LTER HAUSGARTEN, Fram Strait, Arctic). PLoS one 8(9):1–10. doi:10.1371/journal.pone.0072779
Jannasch HW, Wirsen CO (1982) Microbial activities in undecompressed and decompressed deep sea water samples. Appl Env Microbiol 43(5):1116–1124
Jebbar M, Franzetti B, Girard E, Oger P (2015) Microbial diversity and adaptation to high hydrostatic pressure in deep-sea hydrothermal vents prokaryotes. Extremophiles 19(4):721–740. doi:10.1007/s00792-015-0760-3
Ji B, Gimenez G, Barbe V, Vacherie B, Rouy Z, Amrani A, Fardeau ML, Bertin P, Alazard D, Leroy S, Talla E, Ollivier B, Dolla A, Pradel N (2013) Complete genome sequence of the piezophilic, mesophilic, sulfate-reducing bacterium Desulfovibrio hydrothermalis AM13. Genome Announc 1(1):1–2. doi:10.1128/genomeA.00226-12
Joffre C, Nadjar A, Lebbadi M, Calon F, Layea S (2014) n-3 LCPUFA improves cognition: the young, the old and the sick. Prostaglandins Leukot Essent Fatty Acids 91:1–20. doi:10.1016/j.plefa.2014.05.001
Jun X, Lupeng L, Minjuan X, Oger P, Fengping W, Jebbar M, Xiang X (2011) Complete genome sequence of the obligate piezophilic hyperthermophilic archaeon Pyrococcus yayanosii CH1. J Bacteriol 193(16):4297–4298. doi:10.1128/JB.05345-11
Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D’Hondt S (2012) Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci 109(40):16213–16216. doi:10.1073/pnas.1203849109
Kaneko H, Takami H, Inoue A, Horikoshi K (2000) Effects of hydrostatic pressure and temperature on growth and lipid composition of the inner membrane of barotolerant Pseudomonas sp. BT1 isolated from the deep-sea. Biosci Biotechnol Biochem 64(1):72–79. doi:10.1271/bbb.64.72
Karner MB, Delong EF, Karl DM (2001) Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507–510. doi:10.1038/35054051
Karsenti E, Acinas SG, Bork P, Bowler C, De Vargas C, Raes J, Sullivan M, Arendt D, Benzoni F, Claverie JM, Follows M, Gorsky G, Hingamp P, Iudicone D, Jaillon O, Kandels-Lewis S, Krzic U, Not F, Ogata H, Pesant S, Reynaud EG, Sardet C, Sieracki ME, Speich S, Velayoudon D, Weissenbach J, Wincker P, the Tara Oceans Consortium (2011) A holistic approach to marine eco-systems biology. PloS Biol 9(10):1–5
Kashtan N, Roggensack SE, Roddrigue S, Thompson JW, Biller SJ, Coe A, Ding H, Marttinen P, Malmstrom RR, Stocker R, Follows MJ, Stepanauskas R, Chisholm SW (2014) Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science 344:416–420. doi:10.1126/science.1248575
Kato C (2006) Handling of piezophilic microorganisms. Methods Microbiol 35:733–741
Kato C, Li L, Tamaoka J, Horikoshi K (1997) Molecular analyses of the sediment of the 11000-m deep Mariana Trench. Extremophiles 1(3):117–123. doi:10.1007/s007920050024
Kato C, Nogi Y (2001) Correlation between phylogenetic structure and function: examples from deep-sea Shewanella. FEMS Microbiol Ecol 35:223–230. doi:10.1111/j.1574-6941.2001.tb00807.x
Kawai M, Uchiyama I, Takami H, Inagaki F (2015) Low frequency of endospore-specific genes in subseafloor sedimentary metagenomes. Environ Microbiol Rep 7(2):341–350. doi:10.1111/1758-2229.12254
Kawano H, Nakasone K, Matsumoto M, Yoshida Y, Usami R, Kato C, Abe F (2004) Differential pressure resistance in the activity of RNA polymerase isolated from Shewanella violacea and Escherichia coli. Extremophiles 8:367–375. doi:10.1007/s00792-004-0397-0
Kaye JZ, Sylvan JB, Edwards KJ, Baross JA (2011) Halomonas and Marinobacter ecotypes from hydrothermal vent, subseafloor and deep-sea environments. FEMS Microbiol Ecol 75:123–133. doi:10.1111/j.1574-6941.2010.00984.x
Khelaifia S, Fardeau M, Pradel N, Aussignargues C, Garel M, Tamburini C, Cayol J, Gaudron S, Gaill F, Ollivier B (2011) Desulfovibrio piezophilus sp. nov., a piezophilic, sulfate-reducing bacterium isolated from wood falls in the Mediterranean Sea. Int J Syst Evol Microbiol 61:2706–2711. doi:10.1099/ijs.0.028670-0
Kleindienst S, Seidel M, Ziervogel K, Grim S, Loftis K, Harrison S, Malkin SY, Perkins MJ, Field J, Sogin ML, Dittmar T, Passow U, Medeiros PM, Joye SB (2015) Chemical dispersants can suppress the activity of natural oil-degrading microorganisms. Proc Natl Acad Sci 112(48):14900–14905. doi:10.1073/pnas.1507380112
Kobayashi H, Hatada Y, Tsubouchi T, Nagahama T, Takami H (2012) The hadal amphipod Hirondellea gigas possessing a unique cellulose for digesting wooden debris buried in the deepest seafloor. PLoS one 7(8):1–8. doi:10.1371/journal.pone.0042727
Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA (2005) Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546. doi:10.1038/nature03911
Konstantinidis KT, Braff J, Karl DM, Delong EF (2009) Comparative metagenomic analysis of a microbial community residing at a depth of 4,000 meters at Station ALOHA in the North Pacific Subtropical Gyre. Appl Environ Microbiol 75(16):5345–5355. doi:10.1128/AEM.00473-09
Konstantinidis KT, Tiedje JM (2004) Trends between gene content and genome size in prokaryotic species with larger genomes. Proc Natl Acad Sci 101(9):3160–3165. doi:10.1073/pnas.0308653100
Kujawinski EB (2011) The impact of microbial metabolism on marine dissolved organic matter. Annu Rev Mar Sci 3:567–599. doi:10.1146/annurev-marine-120308-081003
La Cono V, Smedile F, La Spada G, Arcadi E, Genovese M, Ruggeri G, Genovese L, Giuliano L, Yakimov MM (2015) Shifts in the meso- and bathypelagic archaea communities composition during recovery and short-term handling of decompressed deep-sea samples. Environ Microbiol Rep 7(3):450–459. doi:10.1111/1758-2229.12272
Langerhuus AT, Røy H, Lever MA, Morono Y, Inagaki F, Jørgensen BB, Lomstein BA (2012) Endospore abundance and D:L-amino acid modeling of bacterial turnover in Holocene marine sediment (Aarhus Bay). Geochim Cosmochim Acta 99:87–99. doi:10.1016/j.gca.2012.09.023
Lauro FM, Bartlett DH (2008) Prokaryotic lifestyles in deep sea habitats. Extremophiles 12(1):15–25. doi:10.1007/s00792-006-0059-5
Lauro FM, Chastain RA, Blankenship LE, Yayanos AA, Bartlett DH (2007) The unique 16S rRNA genes of piezophiles reflect both phylogeny and adaptation. Appl Environ Microbiol 73(3):838–845. doi:10.1128/AEM.01726-06
Lauro FM, Chastain RA, Ferriera S, Johnson J, Yayanos AA, Bartlett DH (2013a) Draft genome sequence of the deep-sea bacterium Shewanella benthica KT99. Genome Announc 1(3):1–2. doi:10.1128/genomeA.00210-13
Lauro FM, Eloe-Fadrosh EA, Richter TKS, Vitulo N, Ferriera S, Johnson JH, Bartlett DH (2014) Ecotype diversity and conversion in Photobacterium profundum strains. PLoS one 9(5):1–10. doi:10.1371/journal.pone.0096953
Lauro FM, Stratton TK, Chastain RA, Ferriera S, Johnson J, Goldberg SMD, Yayanos AA, Bartlett DH (2013b) Complete genome sequence of the deep-sea bacterium Psychromonas strain CNPT3. Genome Announc 1(3):1–2. doi:10.1128/genomeA.00304-13
Leisner JJ, Hansen MA, Larsen MH, Hansen L, Ingmer H, Sørensen SJ (2012) The genome sequence of the lactic acid bacterium, Carnobacterium maltaromaticum ATCC 35586 encodes potential virulence factors. Int J Food Microbiol 152:107–115. doi:10.1016/j.ijfoodmicro.2011.05.012
Leisner JJ, Laursen BG, Prévost H, Drider D, Dalgaard P (2007) Carnobacterium: positive and negative effects in the environment and in foods. FEMS Microbiol Rev 31:592–613. doi:10.1111/j.1574-6976.2007.00080.x592-613
Lennon JT, Jones SE (2011) Microbial seed banks: the ecological and evolutionary implications of dormancy. Nature Rev Microbiol 9:119–130. doi:10.1038/nrmicro2504
Léon-Zayas R, Novotny M, Podell S, Shepard CM, Berkenpas E, Nikolenko S, Pevzner P, Lasken RS, Bartlett DH (2015) Single cells within the Puerto Rico Trench suggest hadal adaptation of microbial lineages. Appl Environ Microbiol 81(24):8265–8276. doi:10.1128/AEM.01659-15
Li L, Kato C, Horikoshi K (1999) Microbial diversity in sediments collected from the deepest cold-seep area, the Japan Trench. Mar Biotechnol 1(4):391–400. doi:10.1007/PL00011793
Li X, Fu L, Li Z, Ma X, Xiao X, Xu J (2015) Genetic tools for the piezophilic hyperthermophilic archaeon Pyrococcus yayanosii. Extremophiles 19(1):59–67. doi:10.1007/s00792-014-0705-2
Liu SB, Chen XL, He HL, Zhang XY, Xie BB, Yu Y, Chen B, Zhou BC, Zhang YZ (2013) Structure and ecological roles of a novel exopolysaccharide from the Arctic sea ice bacterium Pseudoalteromonas sp. strain SM20310. Appl Environ Microbiol 79(1):224–230. doi:10.1128/AEM.01801-12
Lochte K, Turley CM (1988) Bacteria and cyanobacteria associated with phytodetritus in the deep sea. Nature 333:67–69. doi:10.1038/333067a0
Lomstein BA, Langerhuus AT, D’Hondt S, Jørgensen BB, Spivack AJ (2012) Endospore abundance, microbial growth and necromass turnover in deep sub-seafloor sediment. Nature 484(7392):101–104. doi:10.1038/nature10905
López-García P, López-López A, Moreira D, Rodríguez-Valera F (2001) Diversity of free-living prokaryotes from a deep-sea site at the Antarctic Polar Front. FEMS Microb Ecol 36:193–202. doi:10.1111/j.1574-6941.2001.tb00840.x
López-López A, Bartual SG, Stal L, Onyshchenko O, Rodríguez-Valera F (2005) Genetic analysis of housekeeping genes reveals a deep-sea ecotype of Alteromonas macleodii in the Mediterranean Sea. Environ Microbiol 7(5):649–659. doi:10.1111/j.1462-2920.2005.00733.x
Lucas S, Han J, Lapidus A, Cheng JF, Goodwin LA, Pitluck S, Petgers L, Milkhailova N, Teshima H, Detter JC, Han C, Tapia R, Land M, Hauser L, Kyrpides NC, Ivanova N, Pagani I, Vannier P, Oger P, Bartlett DH, Noll KM, Woyke T, Jebbar M (2012) Complete genome sequence of the thermophilic, piezophilic, heterotrophic bacterium Marinitoga piezophila KA3. Genome Announc 194(21):5974–5975. doi:10.1128/JB.01430-12
Luo H, Tolar BB, Swan BK, Zhang CL, Stepanauskas R, Moran MA, Hollibaugh JT (2014) Single-cell genomics shedding light on marine Thaumarchaeota diversification. ISME J 8:732–736. doi:10.1038/ismej.2013.202
Lyons MM, Dobbs FC (2012) Differential utilization of carbon substrates by aggregate-associated and water-associated heterotrophic bacterial communities. Hydrobiologia 686:181–193. doi:10.1007/s10750-012-1010-7
Martin D, Bartlett DH, Roberts MF (2002) Solute accumulation in the deep-sea bacterium Photobacterium profundum. Extremophiles 6:507–514. doi:10.1007/s00792-002-0288-1
Martin-Cuadrado AB, Ghai R, Gonzaga A, Rodriguez-Valera F (2009) CO dehydrogenase genes found in metagenomic fosmid clones from the deep Mediterranean Sea. Appl Environ Microbiol 75(32):7436–7444. doi:10.1128/AEM.01283-09
Martin-Cuadrado AB, López-García P, Alba JC, Moreira D, Monticelli L, Strittmatter A, Gottschalk G, Rodríguez-Valera F (2007) Metagenomics of the deep Mediterranean, a warm bathypelagic habitat. PLoS one 9:1–15. doi:10.1371/journal.pone.0000914
Martini S, Al Ali B, Garel M, Nerini D, Grossi V, Pacton M, Casalot L, Cuny P, Tamburini C (2013) Effects of hydrostatic pressure on growth and luminescence of a moderately-piezophilic luminous bacteria Photobacterium phosphoreum ANT-2200. PLoS one 8(6):1–9. doi:10.1371/journal.pone.0066580
Martiny AC, Tai APK, Veneziano D, Primeau F, Chisholm SW (2009) Taxonomic resolution, ecotypes and the biogeography of Prochlorococcus. Env Microbiol 11(4):823–832. doi:10.1111/j.1462-2920.2008.01803.x
Mason OU, Han J, Woyke T, Jansson JK (2014a) Single-cell genomics reveals features of a Colwellia species that was dominant during the Deepwater Horizon oil spill. Front Microbiol 5(332):1–8. doi:10.3389/fmicb.2014.00332
Mason OU, Hazen TC, Borglin S, Chain PSG, Dubinsky EA, Fortney JL, Han J, Holman HYN, Hultman J, Lamendella R, Mackelprang R, Malfatti S, Tom LM, Tringe SG, Woyke T, Zhou J, Rubin EM, Jansson JK (2012) Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J 6:1715–1727. doi:10.1038/ismej.2012.59
Mason OU, Scott NM, Gonzalez A, Robbins-Pianka A, Bælum J, Kimbrel J, Bouskill NJ, Prestat E, Borglin S, Joyner DC, Fortney JL, Jurelevicius D, Stringfellow WT, Alvarez-Cohen L, Hazen TC, Knight R, Gilbert JA, Jansson JK (2014b) Metagenomics reveals sediment microbial community response to Deepwater Horizon oil spill. ISME J 8:1464–1475. doi:10.1038/ismej.2013.254
McCarren J, Becker JW, Repeta DJ, Shi Y, Young CR, Malmstrom RR, Chisholm SW, DeLong EF (2010) Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc Natl Acad Sci 107(38):16420–16427. doi:10.1073/pnas.1010732107
McCarter LL (2004) Dual flagellar systems enable motility under different circumstances. J Mol Microbiol Biotechnol 7:18–29. doi:10.1159/000077866
Médigue C, Krin E, Pascal G, Barbe V, Bernsel A, Bertin PN, Cheung F, Cruveiller S, D’Amico S, Duilio A, Fang G, Feller G, Ho C, Mangenot S, Marino G, Nilsson J, Parrilli E, Rocha EPC, Rouy Z, Sekowska A, Tutino ML, Vallenet D, von Heijne G, Danchin A (2005) Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res 15:1325–1335. doi:10.1101/gr.4126905
Moeseneder MM, Winter C, Herndl GJ (2001) Horizontal and vertical complexity of attached and free-living bacteria of the eastern Mediterranean Sea, determined by 16S rDNA and 16S rRNA fingerprints. doi:10.4319/lo.2001.46.1.0095
Merbt SN, Stahl DA, Casamayor EO, Martí E, Nicol GW, Prosser JI (2012) Differential photoinhibition of bacterial and archaeal ammonia oxidation. FEMS Microbiol Lett 327:41–46. doi:10.1111/j.1574-6968.2011.02457.x
Mincer TJ, Church MJ, Taylor LT, Preston C, Karl DM, Delong EF (2007) Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol 9(5):1162–1175. doi:10.1111/j.1462-2920.2007.01239.x
Miyazaki M, Nogi Y (2014) The family Psychromonadaceae. In: Rosenberg E et al (eds) The Prokaryotes—Gammaproteobacteria. Springer, Berlin, pp 583–590. doi:10.1007/978-3-642-38922-1_228
Morris RM, Rappé M, Connon SA, Vergin KL, Siebold WA, Carlson CA, Giovannoni SJ (2002) SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806–810. doi:10.1038/nature01240
Morris RM, Rappé MS, Urbach E, Connon SA, Giovannoni SJ (2004) Prevalence of the Chloroflexi-related SAR202 bacterioplankton cluster throughout the mesopelagic zone and deep ocean. Appl Environ Microbiol 70(5):2836–2842. doi:10.1128/AEM.70.5.2836-2842.2004
Morris RM, Vergin KL, Cho JC, Rappé MS, Carlson CA, Giovannoni SJ (2005) Temporal and spatial response of bacterioplankton lineages to annual convective overturn at the Bermuda Atlantic Time-series Study site. Limnol Oceanogr 50(5):1687–1696. doi:10.4319/lo.2005.50.5.1687
Nagata T, Tamburini C, Arístegui J, Baltar F, Bochdansky AB, Fonda-Umani S, Fukuda H, Gogou A, Hansell DA, Hansman RL, Herndl GJ, Panagiotopoulos C, Reinthaler T, Sohrin R, Verdugo P, Yamada N, Yamashita Y, Yokokawa T, Bartlett DH (2010) Emergicing concepts on microbial processes in the bathypelagic ocean—ecology, biogeochemistry, and genomics. Deep-Sea Res II 57:1519–1536. doi:10.1016/j.dsr2.2010.02.019
Ngugi DK, Blom J, Alam I, Rashid M, Ba-Alawi W, Zhang G, Hikmawan T, Guan Y, Antunes A, Siam R, El Dorry H, Bajic V, Stingl U (2015) Comparative genomics reveals adaptations of a halotolerant Thaumarchaeon in the interfaces of brine pools in the Red Sea. ISME J 9:396–411. doi:10.1038/ismej.2014.137
Nichols CM, Lardiér SG, Bowman JP, Nichols PD, Gibson JAE, Guézennec J (2005) Chemical characterization of exopolysaccharides from Antarctic marine bacteria. Microb Ecol 49(4):578–589. doi:10.1007/s00248-004-0093-8
Nicholson WL, Krivushin K, Gilichinsky D, Schuerger AC (2013) Growth of Carnobacterium spp. from permafrost under low pressure, temperature, and anoxic atmosphere has implications for Earth microbes on Mars. Proc Natl Acad Sci 110(2):666–671. doi:10.1073/pnas.1209793110
Nicol GW, Leininger S, Schleper C (2011) Distribution and activity of ammonia-oxidizing archaea in natural environments. Nitrification. ASM P. Press., Washington, DC. ISBN-13 7, pp 157–178
Nogi Y, Hosoya S, Kato C, Horikoshi K (2004) Colwellia piezophila sp. nov., a novel piezophilic species from deep-sea sediments of the Japan Trench. Int J Syst Evol Microbiol 54:1627–1631. doi:10.1099/ijs.0.03049-0
Nogi Y, Kato C, Horikoshi K (1998a) Taxonomic studies of deep-sea barophilic Shewanella strains and description of Shewanella violacea sp. nov. Arch Microbiol 170(5):331–338. doi:10.1007/s002030050650
Nogi Y, Masui N, Kato C (1998b) Photobabacterium profundum sp. nov., a new, moderately barophilic bacterial species isolated from a deep-sea sediment. Extremophiles 2(1):1–8. doi:10.1007/s007920050036
Nunoura T, Takaki Y, Hirai M, Shimamura S, Makabe A, Koide O, Kikuchi T, Miyazaki J, Koba K, Yoshida N, Sunamura M, Takai K (2015) Hadal biosphere: insight into the microbial ecosystem in the deepest ocean on Earth. Proc Natl Acad Sci 112(11):E1230–E1236. doi:10.1073/pnas.1421816112
Ohtomo Y, Ijiri A, Ikegawa Y, Tsutsumi M, Imachi H, Uramoto GI, Hoshino T, Morono Y, Sakai S, Saito Y, Tanikawa W, Hirose T, Inagaki F (2013) Biological CO2 conversion to acetate in subsurface coal-sand formation using a high-pressure reactor system. Front Microbiol 4(361):1–17. doi:10.3389/fmicb.2013.00361
Ohwada K, Tabor PS, Colwell RR (1980) Species composition and barotolerance of gut microflora of deep-sea benthic macrofauna collected at various depths in the Atlantic Ocean. Appl Environ Microbiol 40(4):746–755
Oikawa Y, Sinmura Y, Ishizaka H, Midorikawa R, Kawamoto J, Kurihara T, Kato C, Horikoshi K, Tamegai H (2015) Nar is the dominant dissimilatory nitrate reductase under high pressure conditions in the deep-sea denitrifier Pseudomonas sp. MT-1. J Gen Appl Microbiol 61:10–14. doi:10.2323/jgam.61.10
Orcutt BN, Sylvan JB, Knab NJ, Edwards KJ (2011) Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol Mol Biol Rev 75(2):361–422. doi:10.1128/MMBR.00039-10
Ouverney CC, Fuhrman JA (2000) Marine planktonic archaea take up amino acids. Appl Environ Microbiol 66(11):4829–4833. doi:10.1128/AEM.66.11.4829-4833.2000
Parada V, Sintes E, van Aken HM, Weinbauer MG, Herndl GJ (2007) Viral abundance, decay, and diversity in the meso- and bathypelagic waters of the North Atlantic. 73(14):4429-4438. doi: 10.1128/AEM.00029-07
Parkes RJ, Sellek G, Webster G, Martin D, Anders E, Weightman AJ, Sass H (2009) Culturable prokaryotic diversity of deep, gas hydrate sediments: first use of a continuous high-pressure, anaerobic, enrichment and isolation system for subseafloor sediments (DeepIsoBUG). Environ Microbiol 11(12):3140–3153. doi:10.1111/j.1462-2920.2009.02018.x
Pathom-aree W, Stach JEM, Ward AC, Horikoshi K, Bull AT, Goodfellow M (2006) Diversity of actinomycetes isolated from Challenger Deep sediment (10,898 m) from the Mariana Trench. Extremophiles 10(3):181–189. doi:10.1007/s00792-005-0482-z
Pikuta EV, Marsic D, Bej A, Tang J, Krader P, Hoover RB (2005) Carnobacterium pleistocenium sp. nov., a novel psychrotolerant, facultative anaerobe isolated from permafrost of the Fox Tunnel in Alaska. Int J Syst Evol Microbiol 55:473–478. doi:10.1099/ijs.0.63384-0
Pradel N, Ji B, Gimenez G, Talla E, Lenoble P, Garel M, Tamburini C, Fourquet P, Lebrn R, Bertin P, Denis Y, Pophillat M, Barbe V, Ollivier B, Dolla A (2013) The first genomic and proteomic characterization of a deep-sea sulfate reducer: insights into the piezophilic lifestyle of Desulfovibrio piezophilus. PLoS one 8(1):1–11. doi:10.1371/journal.pone.0055130
Puig P, Palanques A, Sanchez-Cabeza JA, Masqué P (1999) Heavy metals in particulate matter and sediments in the southern Barcelona sedimentation system. Mar Chem 63:311–329. doi:10.1016/S0304-4203(98)00069-3
Qin G, Zhu L, Chen X, Wang PG, Zhang Y (2007) Structural characterization and ecological roles of a novel exopolysaccharide from the deep-sea psychrotolerant bacterium Pseudoalteromonas sp. SM9913. Microbiology 153:1566–1572. doi:10.1099/mic.0.2006/003327-0
Qin QL, Li Y, Zhang YJ, Zhou ZM, Zhang WX, Chen XL, Zhang XY, Zhou BC, Wang L, Zhang YZ (2011) Comparative genomics reveals a deep-sea sediment-adapted life style of Pseudoalteromonas sp. SM9913. ISME J 5:274–284. doi:10.1038/ismej.2010.103
Qin W, Amin SA, Martens Habbena W, Walker CB, Urakawa H, Devol AH, Ingalls AE, Moffett JW, Armbrust EV, Stahl DA (2014) Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc Natl Acad Sci 111(34):12504–12509. doi:10.1073/pnas.1324115111
Qin W, Carlson LT, Armbrust EV, Devol AH, Moffett JW, Stahl DA, Ingalls AE (2015) Confounding effects of oxygen and temperature on the TEX86 signature of marine Thaumarchaeota. Proc Natl Acad Sci 112(35):10979–10984. doi:10.1073/pnas.1501568112
Quaiser A, Zivanovic Y, Moreira D, López-García P (2011) Comparative metagenomics of bathypelagic plankton and bottom sediment from the Sea of Marmara. ISME J 5:285–304. doi:10.1038/ismej.2010.113
Raguenes G, Pignet P, Gauthier G, Peres A, Christen R, Rougeaux H, Barbier G, Guezennec J (1996) Description of a new polymer-secreting bacterium from a deep-sea hydrothermal vent, Alteromonas macleodii subsp. Fijiensis, and preliminary characterization of the polymer. Appl Environ Microbiol 62(1):67–73
Redmond MC, Valentine DL (2012) Natural gas and temperature structured a microbial community response to the Deepwater Horizon oil spill. Proc Natl Acad Sci 109(50):20292–20297. doi:10.1073/pnas.1108756108
Reen FJ, Almagro-Moreno S, Ussery D, Boyd EF (2006) The genomic code: inferring Vibrionaceae niche specialization. Nature Rev Microbiol 4:697–704. doi:10.1038/nrmicro1476
Reinthaler T, van Aken HM, Herndl GJ (2010) Major contribution of autotrophy to microbial carbon cycling in the deep North Atlantic’s interior. Deep-Sea Res II 57(16):1572–1580. doi:10.1016/j.dsr2.2010.02.023
Reinthaler T, van Aken H, Veth C, Arístegui J, Robinson C, Williams PJIB, Lebaron P, Herndl GJ (2006) Prokaryotic respiration and production in the meso- and bathypelagic realm of the eastern and western North Atlantic basin. Limnol Oceanogr 51(3):1262–1273. doi:10.4319/lo.2006.51.3.1262
Rice AL, Billett DSM, Fry J, John AWG, Lampitt RS, Mantoura RFC, Morris RJ (1986) Seasonal deposition of phytodetritus to the deep-sea floor. Proc Roy Soc Edinb B 88:265–279. doi:10.1017/S0269727000004590
Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T (2013) Insights into the phylogeny and coding potential of microbial dark matter. Nature 499(7459):431–437. doi:10.1038/nature12352
Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, Arellano A, Coleman M, Hauser L, Hess WR, Johnson ZI, Land M, Lindell D, Post AF, Regala W, Shah M, Shaw SL, Steglich C, Sullivan MB, Ting CS, Tolenen A, Webb EA, Zinser ER, Chisholm SW (2003) Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424:1042. doi:10.1038/nature01947
Roux S, Hawley AK, Beltran MT, Scofield M, Schwientek P, Stepanauskas R, Woyke T, Hallam SJ, Sullivan MB (2014) Ecology and evolution of viruses infecting uncultivated SUP05 bacteria as revealed by single-cell- and meta-genomics. eLife 3:1–20. doi:10.7554/eLife.03125
Ruff SE, Biddle JF, Teske AP, Knittel K, Boetius A, Ramette A (2015) Global dispersion and local diversification of the methane seep microbiome. Proc Natl Acad Sci 112(13):4015–4020. doi:10.1073/pnas.1421865112
Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers YH, Falcon LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, Platt T, Bermingham E, Gallardo V, Tamayo-Castillo G, Ferrari MR, Strausberg RL, Nealson K, Friedman R, Frazier M, Venter JC (2007) The Sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol 5(3):0398–0431. doi:10.1371/journal.pbio.0050077
Salazar G, Cornejo-Castillo FM, Benítez-Barrios V, Fraile-Nuez E, Alvarez-Salgado XA, Duarte CM, Gasol JM, Acinas SG (2015a) Global diversity and biogeography of deep-sea pelagic prokaryotes. ISME J,1–13. doi:10.1038/ismej.2015.137
Salazar G, Cornejo-Castillo FM, Borrull E, Díez-Vives C, Lara E, Vaqué D, Arrieta JM, Duarte CM, Gasol JM, Acinas SG (2015b) Particle-association lifestyle is a phylogenetically conserved trait in bathypelagic prokaryotes. Mol Ecol 24(22):5692–5706. doi:10.1111/mec.13419
Satomi M (2014) The family Shewanellaceae. In: Rosenberg E et al (eds) The Prokaryotes—Gammaproteobacteria. Springer, Berlin, pp 597–625. doi:10.1007/978-3-642-38922-1_226
Satomi M, Fujii T (2014) The family Oceanospirillaceae. In: Rosenberg E et al (eds) The Prokaryotes—Gammaproteobacteria. Springer, Berlin, pp 491–527. doi:10.1007/978-3-642-38922-1_286
Sauer P, Glombitza C, Kallmeyer J (2012) A system for incubations at high gas partial pressure. Front Microbiol 3(25):1–9. doi:10.3389/fmicb.2012.00025
Scholin C, Doucette G, Jensen S, Roman B, Pargett D, Marin IIIIR, Preston C, Jones W, Feldman J, Everlove C, Harris A, Alvarado N, Massion E, Birch J, Greenfield D, Vrijenhoek R, Mikulski C, Jones K (2009) Remote detection of marine microbes, small invertebrates, harmful algae, and biotoxins using the environmental sample processor (ESP). Oceanography 22(2):158–167. doi:10.5670/oceanog.2009.46
Schrenk MO, Brazelton WJ (2013) Serpentinization, carbon, and deep life. Rev Mineral Geochem 75:575–606. doi:10.2138/rmg.2013.75.18
Sekiguchi T, Saika A, Nomura K, Watanabe T, Watanabe T, Fujimoto Y, Enoki M, Sato T, Kato C, Kanehiro H (2011) Biodegradation of aliphatic polyesters soaked in deep seawaters and isolation of poly(ε-caprolactone)-degrading bacteria. Polym Degrad Stabil 96:1397–1403. doi:10.1016/j.polymdegradstab.2011.03.004
Seyler LM, McGuinness LM, Kerkhof LJ (2014) Crenarchaeal heterotrophy in salt marsh sediments. ISME J 8:1534–1543. doi:10.1038/ismej.2014.15
Sheik CS, Jain S, Dick GJ (2014) Metabolic flexibility of enigmatic SAR324 revealed through metagenomics and metatranscriptomics. Environ Microbiol 16(1):304–317. doi:10.1111/1462-2920.12165
Shi Y, McCarren J, DeLong EF (2012) Transcriptional responses of surface water marine microbial assemblages to deep-sea water amendment. Environ Microbiol 14(1):191–206. doi:10.1111/j.1462-2920.2011.02598.x
Sikorski J, Möhle M, Wackernagel W (2002) Identification of complex composition, strong strain diversity and directional selection in local Pseudomonas stutzeri populations from marine sediment and soils. Environ Microbiol 4(8):465–476. doi:10.1046/j.1462-2920.2002.00325.x
Simon HM, Smith MW, Herfort L (2014) Metagenomic insights into particles and their associated microbiota in a coastal margin ecosystem. Front Microbiol 5(466):1–10. doi:10.3389/fmicb.2014.00466
Simon M, Grossart HP, Schweitzer B, Ploug H (2002) Microbial ecology of organic aggregates in aquatic ecosystems. Aquat Microb Ecol 28:175–211
Sintes E, Bergauer K, De Corte D, Yokokawa T, Herndl GJ (2013) Archaeal amoA gene diversity points to distinct biogeography of ammonia-oxidizing Crenarchaeota in the ocean. Environ Microbiol 15(5):1647–1658. doi:10.1111/j.1462-2920.2012.02801.x
Smedile F, Messina E, la Cono V, Tsoy O, Monticelli LS, Borghini M, Giuliano L, Golyshin PN, Mushegian A, Yakimov MM (2013) Metagenomic analysis of hadopelagic microbial assemblages thriving at the deepest part of Mediterranean Sea. Matapan-Vavilov Deep Environ Microbiol 15(1):167–182. doi:10.1111/j.1462-2920.2012.02827.x
Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ (2006) Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci 103(32):12115–12120. doi:10.1073/pnas.0605127103
Stokke R, Roalkvam I, Lanzen A, Haflidason H, Steen IH (2012) Integrated metagenomic and metaproteomic analyses of an ANME-1-dominated community in marine cold seep sediments. Environ Microbiol 14(5):1333–1346. doi:10.1111/j.1462-2920.2012.02716.x
Stratton TK (2008) Genomic analysis of high pressure adaptation in deep sea bacteria. Proquest UC San Diego Electronic Theses and Dissertations, pp 1–164
Swan BK, Chaffin MD, Martinez-Garcia M, Morrison HG, Field EK, Poulton NJ, Masland EDP, Harris CC, Sczyrba A, Chain PSG, Koren S, Woyke T, Stepanauskas R (2014) Genomic and metabolic diversity of Marine Group I Thaumarchaeota in the mesopelagic of two subtropical gyres. PLoS one 9(4):1–9. doi:10.1371/journal.pone.0095380
Tabor PS, Deming JW, Ohwada K, Davis H, Waxman M, Colwell RR (1981a) A pressure-retaining deep ocean sampler and transfer system for measurement of microbial activity in the deep sea. Microb Ecol 7(1):51–65. doi:10.1007/BF02010478
Tabor PS, Ohwada K, Colwell RR (1981b) Filterable marine bacteria found in the deep sea: distribution, taxonomy, and response to starvation. Microb Ecol 7(1):67–83. doi:10.1007/BF02010479
Takami H, Inoue A, Fuji F, Horikoshi K (1997) Microbial flora in the deepest sea mud of the Mariana Trench. FEMS Microbiol Lett, 279–285. doi:10.1111/j.1574-6968.1997.tb10440.x
Takami H, Kobata K, Nagahama T, Kobayashi H, Inoue A, Horikoshi K (1999) Biodiversity in deep-sea sites located near the south part of Japan. Extremophiles 3(2):97–102. doi:10.1007/s007920050104
Tamburini C, Boutrif M, Garel M, Colwell RR, Deming JW (2013) Prokaryotic responses to hydrostatic pressure in the ocean—a review. Environ Microbiol 15(5):1262–1274. doi:10.1111/1462-2920.12084
Tamburini C, Garel M, Al Ali B, Mérigot B, Kriwy P, Charriére B, Budillon G (2009) Distribution and activity of bacteria and archaea in the different water masses of the Tyrrhenian Sea. Deep-Sea Res II 56:700–712. doi:10.1016/j.dsr2.2008.07.021
Tamegai H, Kato C, Horikoshi K (2004) Lateral gene transfer in the deep sea of Mariana Trench: identification of nar gene cluster encoding membrane-bound nitrate reductase from Pseudomonas sp. strain MT-1. DNA Seq 15:338–343. doi:10.1080/10425170400009293
Tamegai H, Li L, Masui N, Kato C (1997) A denitrifying bacterium from the deep sea at 11000-m depth. Extremophiles 1(4):207–211. doi:10.1007/s007920050035
Teira E, van Aken H, Veth C, Herndl GJ (2006) Archaeal uptake of enantiomeric amino acids in the meso- and bathypelagic waters of the North Atlantic. Limnol Oceanogr 51(1):60–69. doi:10.4319/lo.2006.51.1.0060
Thiel A, Michoud G, Moalic Y, Flament D, Jebbar M (2014) Genetic manipulations of the hyperthermophilic piezophilic archaeon Thermococcus barophilus. Appl Environ Microbiol 80(7):2299–2306. doi:10.1128/AEM.00084-14
Thiel H, Pfannkuche O, Schriever G, Lochte K, Gooday AJ, Hemleben C, Mantoura RFG, Turley CM, Patching JW, Riemann F (1989) Phytodetritus on the deep-sea floor in a central oceanic region of the northeast Atlantic. Biol Oceanogr 6(2):203–239. doi:10.1080/01965581.1988.10749527
Thrash JC, Temperton B, Swan BK, Landry ZC, Woyke T, Delong EF, Stepanauskas R, Giovannoni SJ (2014) Single-cell enabled comparative genomics of a deep ocean SAR11 bathytype. ISME J 8:1440–1451. doi:10.1038/ismej.2013.243
Thureborn P, Lundin D, Plathan J, Poole AM, Sjöberg BM, Sjöling S (2013) A metagenomics transect into the deepest point of the Baltic Sea reveals clear stratification of microbial functional capacities. PLoS one 8(9):1–11. doi:10.1371/journal.pone.0074983
Ting CS, Rocap G, King J, Chisholm SW (2002) Cyanobacterial photosynthesis in the oceans: the origins and significance of divergent light-harvesting strategies. Trends Microbiol 10(3):134–142. doi:10.1016/S0966-842X(02)02319-3
Todo T (1999) Functional diversity of the DNA photolyase/blue light receptor family. Mutat Res/DNA Repair 434:89–97
Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Ulrich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C (2011) Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci 108(20):8420–8425. doi:10.1073/pnas.1013488108
Treusch AH, Vergin KL, Finaly LA, Donatz MG, Burton RM, Carlson CA, Giovannoni SJ (2009) Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J 3:1148–1163. doi:10.1038/ismej.2009.60
Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, Bork P, Hugenholtz P, Rubin EM (2005) Comparative metagenomics of microbial communities. Science 308:554–557. doi:10.1126/science.1107851
Turner JT (2015) Zooplankton fecal pellets, marine snow, phytodetritus and the ocean’s biological pump. Prog Oceanogr 130:205–248. doi:10.1016/j.pocean.2014.08.005
Urakawa H (2014) The family Moritellaceae. In: Rosenberg E et al (eds) The Prokaryotes—Gammaproteobacteria. Springer, Berlin, pp 477–489. doi:10.1007/978-3-642-38922-1_227
Valentine DL, Kessler JD, Redmond MC, Mendes SD, Heintz MB, Farwell C, Hu L, Kinnaman FS, Yvon-Lewis S, Du M, Chan EW, Tigreros FG, Villanueva CJ (2010) Propane respiration jump-starts microbial response to a deep oil spill. Science 330(6001):208–211. doi:10.1126/science.1196830
Vandieken V, Pester M, Finke N, Hyun JH, Friedrich MW, Loy A, Thamdrup B (2012) Three manganese oxide-rich marine sediments harbor similar communities of acetate-oxidizing manganese-reducing bacteria. ISME J 6:2078–2090. doi:10.1038/ismej.2012.41
Vannier P, Marteinsson VT, Fridjonsson OH, Oger P, Jebbar M (2011) Complete genome sequence of the hyperthermophilic, piezophilic, heterotrophic, and carboxydotrophic archaeon Thermococcus barophilus MP. J Bacteriol 193(6):1481–1482. doi:10.1128/JB.01490-10
Varela MM, van Aken HM, Herndl GJ (2008) Abundance and activity of Chloroflexi-type SAR202 bacterioplankton in the meso- and bathypelagic waters of the (sub)tropical Atlantic. Environ Microbiol 10(7):1903–1911. doi:10.1111/j.1462-2920.2008.01627.x
Varela MM, van Aken HM, Sintes E, Reinthaler T, Herndl GJ (2011) Contribution of Crenarchaeota and Bacteria to autotrophy in the North Atlantic interior. Environ Microbiol 13(6):1524–1533. doi:10.1111/j.1462-2920.2011.02457.x
Vezzi A, Campanaro S, D’Angelo M, Simonato F, Vitulo N, Lauro FM, Cestaro A, Malacrida G, Simionati B, Cannata N, Romualdi C, Bartlett DH, Valle G (2005) Life at depth: Photobacterium profundum genome sequence and expression analysis. Science 307(5714):1459–1461. doi:10.1126/science.1103341
Voget S, Klippel B, Daniel R, Antranikian G (2011) Complete genome sequence of Carnobacterium sp. 17-4. J Bacteriol 193(13):3403–3404. doi:10.1128/JB.05113-11
Wall R, Ross RP, Fitzgerald GF, Stanton C (2010) Fatty acids from fish: the anti-inflammatory potential of long-chain ω-3 fatty acids. Nutr Rev 68:280–289. doi:10.1111/j.1753-4887.2010.00287.x
Walsh DA, Zaikova E, Howes CG, Song YC, Wright JJ, Tringe SG, Tortell PD, Hallam SJ (2009) Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science 326(5952):578–582. doi:10.1126/science.1175309
Wang F, Wang P, Chen M, Xiao X (2004) Isolation of extremophiles with the detection and retrieval of Shewanella strains in deep-sea sediments from the west Pacific. Extremophiles 8(2):165–168. doi:10.1007/s00792-003-0365-0
Wang F, Wang J, Jian H, Zhang B, Li S, Wang F, Zeng X, Gao L, Bartlett DH, Yu J, Hu S, Xiao X (2008) Environmental adaptation: Genomic analysis of the piezotolerant and psychrotolerant deep-sea iron reducing bacterium Shewanella piezotolerans WP3. PLoS one 3(4):1–12. doi:10.1371/journal.pone.0001937
Wasmund K, Schreiber L, Lloyd KG, Petersen DG, Schramm A, Stepanauskas R, Jørgensen BB, Adrian L (2014) Genome sequencing of a single cell of the widely distributed marine subsurface Dehalococcoidia, phylum Chloroflexi. The ISME Journal 8:383–397. doi:10.1038/ismej.2013.143
Welch TJ, Farewell A, Neidhardt FC, Bartlett DH (1993) Stress response of Escherichia coli to elevated hydrostatic pressure. J Bacteriol 175(22):7170–7177
Whitman WB, Coleman DC, Wiebe WJ (1998) Prokaryotes: the unseen majority. Proc Natl Acad Sci 95(12):6578–6583
Wilkins D, van Sebille E, Rintoul SR, Lauro FM, Cavicchioli R (2013) Advection shapes Southern Ocean microbial assemblages independent of distance and environment effects. Nat Commun 4(2457):1–7. doi:10.1038/ncomms3457
Wilms R, Köpke B, Sass H, Chang TS, Cypionka H, Engelen B (2006) Deep biosphere-related bacteria within the subsurface of tidal flat sediments. Environ Microbiol 8(4):709–719. doi:10.1111/j.1462-2920.2005.00949.x
Winter C, Garcia JAL, Weinbauer MG, DuBow MS, Herndl GJ (2014) Comparison of deep-water viromes from the Atlantic Ocean and the Mediterranean Sea. PloS one 9(6):1–8. doi:10.1371/journal.pone.0100600
Wright JJ, Konwar KM, Hallam SJ (2012) Microbial ecology of expanding oxygen minimum zones. Nature Rev Microbiol 10:381–394. doi:10.1038/nrmicro2778
Wright JJ, Mewis K, Hanson NW, Konwar KM, Maas KR, Hallam SJ (2014) Genomic properties of Marine Group A bacteria indicate a role in the marine sulfur cycle. ISME J 8:455–468. doi:10.1038/ismej.2013.152
Wright TD, Vergin KL, Boyd PW, Giovannoni SJ (1997) A novel delta-subdivision proteobacterial lineage from the lower ocean surface layer. Appl Environ Microbiol 63(4):1441–1448
Wu J, Gao W, Johnson RH, Zhang W, Meldrum DR (2013) Integrated metagenomic and metatranscriptomic analyses of microbial communities in the meso- and bathypelagic realm of North Pacific Ocean. Mar Drugs 11(10):3777–3801. doi:10.3390/md11103777
Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk J, Timmers P, Strous M, Teira E, Herndl GJ, Middelburg JJ, Schouten S, Damsté JSS (2006) Archaeal nitrification in the ocean. Proc Natl Acad Sci 104(13):12317–12322. doi:10.1073/pnas.0600756103
Xiao X, Wang P, Zeng X, Bartlett DH, Wang F (2007) Shewanella psychrophila sp. nov. and Shewanella piezotolerans sp. nov., isolated from west Pacific deep-sea sediment. Int J Syst Evol Microbiol 57:60–65. doi:10.1099/ijs.0.64500-0
Xu M, Wang P, Wang F, Xiao X (2005) Microbial diversity at a deep-sea station of the Pacific nodule province. Biodivers Conserv 14(14):3363–3380. doi:10.1007/s10531-004-0544-z
Xu L, Xu XW, Meng FX, Huo YY, Oren A, Yang JY, Wang CS (2013) Halomonas zincidurans sp. nov., a heavy-metal-tolerant bacterium isolated from the deep-sea environment. Int J Syst Evol Microbiol 63:4230–4236. doi:10.1099/ijs.0.051656-0
Yakimov MM, La Cono V, Smedile F, DeLuca TH, Juárez S, Ciordia S, Fernández M, Albar JP, Ferrer M, Golyshin PN, Giuliano L (2011) Contribution of crenarchaeal autotrophic ammonia oxidizers to the dark primary production in Tyrrhenian deep waters (Central Mediterranean Sea). ISME J 5:945–961. doi:10.1038/ismej.2010.197
Yakimov MM, La Cono V, Smedile F, Crisafi F, Arcadi E, Leonardi M, Decembrini F, Catalfamo M, Bargiela R, Ferrer M, Golyshin PN, Giuliano L (2014) Heterotrophic bicarbonate assimilation is the main process of de novo organic carbon synthesis in hadal zone of the Hellenic Trench, the deepest part of Mediterranean Sea. Environ Microbiol Rep 6(6):709–722. doi:10.1111/1758-2229.12192
Yanagibayashi M, Nogi Y, Li L, Kato C (1999) Changes in the microbial community in Japan Trench sediment from a depth of 6292 m during cultivation without decompression. FEMS Microbiol Lett 170:271–279. doi:10.1111/j.1574-6968.1999.tb13384.x
Yancey PH, Gerringer ME, Drazen JC, Rowden AA, Jamieson A (2014) Marine fish may be biochemically constrained from inhabiting the deepest ocean depths. Proc Natl Acad Sci 111(12):4461–4465. doi:10.1073/pnas.1322003111
Yayanos AA (1977) Simply actuated closure for a pressure vessel: design for use to trap deep-sea animals. Rev Sci Instrum 48:786–789. doi:10.1063/1.1135150
Yayanos AA (1986) Evolutional and ecological implications of the properties of deep-sea barophilic bacteria. Proc Natl Acad Sci 83(24):9542–9546
Yayanos AA (1995) Microbiology to 10,500 meters in the deep sea. Annu Rev Microbiol 49:777–805. doi:10.1146/annurev.mi.49.100195.004021
Yayanos AA, Delong EF (1987) Deep-sea bacterial fitness to environmental temperatures and pressures. Curr perspect high pressure biol, 17–32
Yayanos AA, Dietz AS (1983) Death of a hadal deep-sea bacterium after decompression. Science 220(4596):497–498. doi:10.1126/science.220.4596.497
Yayanos AA, Dietz AS, Van Boxtel R (1981) Obligately barophilic bacterium from the Mariana Trench. Proc Natl Acad Sci 78(8):5212–5215
Yayanos AA, Pollard EC (1969) A study of the effects of hydrostatic pressure on macromolecular synthesis in Escherichia coli. Biophys J 9(12):1464–1482. doi:10.1016/S0006-3495(69)86466-0
Yayanos AA, Van Boxtel R, Dietz AS (1983) Reproduction of Bacillus stearothermophilus as a function of temperature and pressure. Appl Environ Microbiol 46(6):1357–1363
Yooseph S, Nealson KH, Rusch DB, McCrow JP, Dupont CL, Kim M, Johnson J, Montgomery R, Ferriera S, Beeson K, Williamson SJ, Tovchigrechko A, Allen AE, Zeigler LA, Sutton G, Eisenstadt E, Rogers YH, Friedman R, Frazier M, Venter JC (2010) Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature 468:60–67. doi:10.1038/nature09530
Yoshida M, Takaki Y, Eitoku M, Nunoura T, Takai K (2013) Metagenomic analysis of viral communities in (hado)pelagic sediments. PLoS one 8(2):1–14. doi:10.1371/journal.pone.0057271
Zarubin M, Belkin S, Ionescu M, Genin A (2012) Bacterial bioluminescence as a lure for marine zooplankton and fish. Proc Natl Acad Sci 109(3):853–857. doi:10.1073/pnas.1116683109
Zhang SD, Barbe V, Garel M, Zhang WJ, Chen H, Santini CL, Murat D, Jing H, Zhao Y, Lajus A, Martini S, Pradel N, Tamburini C, Wu LF (2014) Genome sequence of luminous piezophile Photobacterium phosphoreum ANT-2200. Genome Announc 2(2):1–2. doi:10.1128/genomeA.00096-14
Zhang Y, Maignien L, Zhao X, Wang F, Boon N (2011) Enrichment of a microbial community performing anaerobic oxidation of methane in a continuous high-pressure bioreactor. BMC Microbiol 11(137):1–8. doi:10.1186/1471-2180-11-137
Zhao JS, Deng Y, Manno D, Hawari J (2010) Shewanella spp. genomic evolution for a cold marine lifestyle and in-situ explosive biodegradation. PLoS one 5(2):1–22. doi:10.1371/journal.pone.0009109
Zhao Y, Temperton B, Thrash JC, Schwalbach MS, Vergin KL, Landry ZC, Ellisman M, Deerinck T, Sullivan MB, Giovannoni SJ (2013) Abundant SAR11 viruses in the ocean. Nature 494:357–360. doi:10.1038/nature11921
Zhou MY, Chen XL, Zhao HL, Dang HY, Luan XW, Zhang XY, He HL, Zhou BC, Zhang YZ (2009) Diversity of both the cultivable protease-producing bacteria and their extracellular proteases in the sediments of the South China Sea. Microb Ecol 58(3):582–590. doi:10.1007/s00248-009-9506-z
Acknowledgements
We are grateful for support from the National Science Foundation (grants 0801973, 0827051, and 1536776), the National Aeronautics and Space Administration (grant NNX11AG10G and PLANET14F-0048), and the Prince Albert II Foundation (Project 1265).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Peoples, L.M., Bartlett, D.H. (2017). Ecogenomics of Deep-Ocean Microbial Bathytypes. In: Chénard, C., Lauro, F. (eds) Microbial Ecology of Extreme Environments. Springer, Cham. https://doi.org/10.1007/978-3-319-51686-8_2
Download citation
DOI: https://doi.org/10.1007/978-3-319-51686-8_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-51684-4
Online ISBN: 978-3-319-51686-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)