Abstract
Fragile X syndrome (FXS) is one of the major causes for autism and mental retardation in humans. The etiology of FXS is linked to the expansion of the CGG trinucleotide repeats, r(CGG), suppressing the fragile X mental retardation 1 (FMR1) gene on the X chromosome, resulting in a loss of fragile X mental retardation protein (FMRP) expression, which is required for regulating normal neuronal connectivity and plasticity. Recent studies have further identified that microRNAs are involved in the mechanisms underlying FXS pathogenesis at three different developmental stages. During early embryogenesis before the blastocyst stage, an embryonic stem cell (ESC)-specific microRNA, miR-302, interferes with FMR1 mRNA translation to maintain the stem cell status and inhibit neural development. After blastocyst, the downregulation of miR-302 releases FMRP synthesis and subsequently leads to neuronal development; yet, in FXS, certain r(CGG)-derived microRNAs, such as miR-fmr1s, are expressed and accumulated and then induce DNA hypermethylation on the FMR1 gene promoter regions, resulting in transcriptional inactivation of the FMR1 gene and the loss of FMRP. In normal neuronal development, FMRP is an RNA-binding protein responsible for interacting with miR-125 and miR-132 to regulate the signaling of Group 1 metabotropic glutamate receptor (mGluR1) and N-methyl-d-aspartate receptor (NMDAR), respectively, and consequently affecting synaptic plasticity. As a result, the loss of FMRP impairs these signaling controls and eventually causes FXS-associated disorders, such as autism and mental retardation. Based on these current findings, this chapter will summarize the etiological causes of FXS and further provides significant insights into the molecular mechanisms underlying microRNA-mediated FXS pathogenesis and the related therapy development.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
A large portion of the genome is noncoding DNA, which often contains microsatellite-like short nucleotide repeats with unknown function. Recent studies have shown that the transcripts of certain trinucleotide repeats can fold into hairpin-like RNAs, which are then further processed by RNaseIII-associated Dicer s into microRNAs (miRNAs) or miRNA-like gene silencing molecules that are sized about 18–27 nucleotides and capable of interfering with specific target genes through either posttranscriptional mRNA degradation or translational suppression, or both [1–5].
These miRNAs play crucial roles in several triplet repeat expansion diseases (TREDs), including fragile X syndrome (FXS), Huntington’s disease (HD), myotonic dystrophy (DM), and a number of spinocerebellar ataxias (SCAs). Yet, most of the pathogenic mechanisms underlying these diseases remain largely unclear. To this, we will use FXS as a model to demonstrate how miRNAs are involved in the pathogenesis of FXS-associated disorders, such as autism and mental retardation.
FXS is one of the most common forms of inherited mental retardation, taking up approximately 30 % of total human mental retardation disorders [6]. This disease is originated from the transcriptional inactivation of fragile X mental retardation 1 (FMR1) gene and thus losing its encoded protein, FMRP. FMRP is associated with polyribosome assembly to form ribonucleoprotein (RNP) complexes that regulate certain protein translation involved in neuronal development and plasticity [7].
FMRP also contains a nuclear localization signal (NLS) and a nuclear export signal (NES) for shuttling certain mRNAs between the nucleus and cytoplasm [8, 9], albeit the function remains to be determined. As previous studies have shown that FMR1 gene is inactivated by the over-expansion of CGG trinucleotide repeats (i.e., r(CGG) ≥200 copies) and the hypermethylation of the FMR1 r(CGG) and promoter regions in 99 % of FXS patients [10], the pathological cause of FXS likely resides in certain interactions between r(CGG) over-expansion and FMR1 hypermethylation. Conceivably, r(CGG)-derived miRNAs may play a key role in this gene silencing mechanism.
Since RNA is a highly degradable material, research of the miRNA-associated FXS mechanisms requires the use of fresh samples isolated from either patient’s brain biopsies or comparable animal models. Due to lack of fresh human samples, almost all current studies adopt animal models instead. Zebrafish (Danio rerio) has been served as an excellent model for studying human mental disorders, including FXS and autism [11].
They possess three FMR1-related familial genes, fmr1, fxr1, and fxr2, which are orthologous to the human FMR1, FXR1, and FXR2 genes, respectively [12]. The tissue expression patterns of these familial genes in zebrafish are broadly consistent with those of humans as well [12, 13]. To investigate the effects of r(CGG)-derived miRNA over-expression on brain development, we had found and sequenced a 450-nucleotide r(CGG) expansion motif located in and beyond the zebrafish fmr1 5′-untranslational region (5′-UTR), which contains approximately 50 copies of r(CGG) similar to the numbers in normal human FMR1 gene [3, 4]. In this r(CGG)-rich region, we further identified and isolated several r(CGG)-derived miRNAs, providing the first in vivo evidence of miRNA involvement in FXS pathogenesis [3, 4]. In view of all these genetic similarities between human and zebrafish, studies using this zebrafish FXS model not only have shed significant light on the pathological mechanisms of human FXS but also are useful for developing potential therapies for treating FXS-associated disorders.
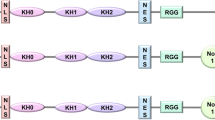
Based on the current findings, miRNAs are involved in FXS during three developmental stages, including pre-blastocyst, post-blastocyst, and neural development, as illustrated in Fig. 7.1. Embryonic cells prior to the blastocyst (32–64-cell) stage are undifferentiated. This is because embryonic stem cell (ESC)-specific miRNAs, in particular miR-302, induce the expression of ESC-specific transcription factors, such as Oct4, Sox2, and Nanog, to maintain the undifferentiated state of stem cells and inhibit developmental signals [14, 15]. In addition, miR-302 also targets the FMR1 mRNA 3′-UTR nucleotide 2774–2789 region (accession number NM_002024) for translationally suppressing FMRP synthesis. Such miR-302-mediated FMRP suppression is a normal developmental event during the pre-blastocyst stage in order to maintain the pluripotency of early ESCs.
Functional roles of FMR1/FMRP-associated miRNAs in neural development. Three groups of miRNAs were identified to affect FMR1 and FMRP expressions at different developmental stages: (1) Before the embryonic blastocyst stage, miR-302 suppresses FMR1 mRNA translation to maintain the ESC status and inhibit neural development. (2) After blastocyst, the expression of FMR1 mRNA generates r(CGG)-derived miR-fmr1 miRNAs. In FXS, the over-expression and over-accumulation of miR-fmr1s in the neuron nuclei trigger DNA hypermethylation and hence inactivate FMR1 transcription, the most prevalent event in FXS. (3) During neural development, miR-125a, miR-125b, and miR-132 may interact with FMRP to regulate the translation of certain neural proteins required for maintaining normal synaptic connection and plasticity. In FXS, the loss of FMRP may abolish these miRNA-mediated regulations and thus leads to the pathologies of FXS-related disorders, such as mental retardation and autism
After blastocyst, most of the ESC-specific miRNAs are downregulated and then ectodermic differentiation begins. At this post-blastocyst stage, FMR1 expression is escalated and its transcripts are quickly accumulated. As a result, the processes of FMR1 mRNA maturation generate r(CGG)-derived miRNAs. In normal individuals, the level of r(CGG)-derived miRNAs is not sufficient to affect FMR1 gene expression; yet, the high expansion of r(CGG) in FXS causes over-expression and over-accumulation of these r(CGG)-derived miRNAs and hence leads to the inhibition of FMR1 and its protein FMRP expression. Since FMRP is responsible for regulating miR-125- and miR-132-mediated protein translation required for maintaining neuronal connectivity and synaptic plasticity [16, 17], the deficiency of FMRP disrupts these miRNA-mediated regulations and consequently results in premature neuronal development. In view of this established disease model, we will further discuss the functional roles of r(CGG)-derived miRNA, miR-125 and miR-132 in FXS pathogenesis.
Identification of r(CGG)-Derived miRNAs in a Zebrafish FXS Model
Human and zebrafish brains share many genetic and structural similarities [11–13]. As it is impossible to directly experiment on living human brain tissues, the current studies of r(CGG)-derived miRNAs are mainly based on a transgenic zebrafish FXS model. To mimic human FXS, an r(CGG)-rich DNA motif isolated from the zebrafish fmr1 5′-UTR r(CGG) expansion region (accession number NW001511047 from the 124001st to 124121st nucleotide) is transgenically overexpressed with high titer retroviral delivery into zebrafish brains [3, 4].
The over-expression of this r(CGG)-rich motif is mediated by an isolated gamma-aminobutyric acid receptor ßZ2 (GABAR2) promoter and hence only occurs in the GABAergic neurons of cortex, hippocampus, and cerebellum [4, 5]. Since previous studies have established that GABAR2 and FMR1 genes are co-expressed in the GABAergic neurons [18, 19], the zebrafish FXS model so obtained can specifically reflect the damages of r(CGG)-derived miRNAs in the FMR1-positive neurons and subsequently prevents any potential off-target effect in other kinds of neuronal cells. Furthermore, by adjusting the multiplicity of retroviral infection (MOI), we can control the expression levels of r(CGG)-derived miRNAs to a certain amount sufficient to trigger the transcriptional suppression of FMR1, resembling the same etiological mechanism of human FXS [4, 5].
As depicted in Fig. 7.2a, wild-type zebrafish contains approximately 35–50 copies of CGG repeats in the fmr1 5′-UTR, whereas the r(CGG) number in the FMR1 of human FXS is over 200, which conceivably may generate more than four times of r(CGG)-derived miRNAs.
Model of the transgenic r(CGG)-overexpressing zebrafish resembling human FXS. Normal zebrafish fmr1 5′-UTR region contains 35–50 r(CGG) copies, whereas the estimated r(CGG) number in human FXS FMR1 is ≥200. In order to mimic the expression levels of human miR-fmr1s in FXS, the fmr1 r(CGG) region was overexpressed ≥6-folds, leading to pathological results similar to human FXS. (a) Dose-dependent correlation between r(CGG)-derived miR-fmr1 expression and transcriptional fmr1 suppression, determined by northern blotting (p < 0.01). The levels of miR-fmr1 expression and fmr1 knockdown were proportional to the expression rates of r(CGG) expression. (b) Sequence and structural features of the r(CGG)-derived miR-fmr1 precursor. From the 5′ to 3′ end, it contains several key motifs, such as a poly-pyrimidine tract (PPT), an r(CGG)-rich hairpin-like pre-miRNA structure, a NIS, a short poly(A) tail, and sometimes a G/C-rich tract. Two miRNA isoforms had been identified as miR-fmr1-27 and miR-fmr1-42. (c) In situ hybridization with an anti-miR-fmr1-27 probe showed the different miR-fmr1 expression patterns between normal (wild-type) and FXS pallium–neocortical neurons
To create a condition similar to the r(CGG) over-expression in human FXS, we have used retroviral delivery to introduce a GABAR2-mediated r(CGG)-expressing red-shifted green fluorescent protein (RGFP) transgene into zebrafish [4, 5]. An r(CGG)-rich DNA motif isolated from the zebrafish fmr1 5′-UTR is manually placed in the 5′-UTR of the RGFP gene. After increasing the r(CGG) expression over sixfolds of the normal level in zebrafish, FXS-like disorders start to be observed in the targeted GABAergic neurons, indicating the minimal threshold of the r(CGG) over-expression levels required for inducing fmr1 gene suppression. Accordingly, since the diseased human FMR1 possesses over 200 copies of r(CGG), this threshold may be actually lower than sixfolds in human FXS. Following the elevation of r(CGG) over-expression, we have further identified the presence of two r(CGG)-derived miRNAs, namely miR-fmr1s, both of which are increased corresponding to the levels of r(CGG) expression. Meanwhile, the fmr1 gene transcription is decreased in response to the increase of miR-fmr1 expression, suggesting that the expressed r(CGG) RNA transcripts can be processed into miR-fmr1s and then result in suppressing the fmr1 gene transcription.
The sequences of these two r(CGG)-derived miR-fmr1s have been confirmed to be miR-fmr1-27 and miR-fmr1-42, respectively, as shown in Fig. 7.2b. They share a 26-nucleotide overlapping sequence in their 5′-end and are derived from the fmr1 5′-UTR r(CGG) expansion region approximately 65-nucleotide upstream of the translational start codon (accession number NM_152963). Both miR-fmr1s contain the same CCG-rich seed sequence for targeting the r(CGG) expansion region of the fmr1 gene.
Notably, the miR-fmr1-42 further possesses three unique structures in its precursor microRNA (pre-miRNA) sequence, including (1) multiple matched CGG-CCG base pairs in the stem arm of the hairpin-like pre-miRNA, (2) a nuclear import signal (NIS) motif located in the 3-end of the hairpin structure, and (3) multiple CCG-rich DNA binding motifs in the mature miR-fmr1-42 sequence. Based on these structures, it is conceivable that the NIS motif may allow the entry of mature miR-fmr1-42 into the cell nucleus via a certain unidentified mechanism, while the multiple CCG-rich DNA binding motifs are involved in transcriptional suppression of the fmr1 gene. In addition, the NIS motif is often flanked with a short poly-A tail, which may facilitate the decay of miR-fmr1-42 and hence prevents the miRNA accumulation in the normal neurons.
The expression patterns of both miR-fmr1 isoforms have been identified in the wild-type zebrafish brain, particularly located in the lateral pallium-neocortical and cerebellar neurons, as determined by fluorescent in situ hybridization (FISH) staining with a specific locked nucleic acid (LNA) probe directed against the miR-fmr1-27 sequence (Fig. 7.3). As shown in Fig. 7.2c, the FISH staining also indicated that wild-type pallium neurons (equivalent of human hippocampal stratum radiatum) present a limited amount of miR-fmr1s in the cytoplasmic region surrounding the cell nucleus, whereas the r(CGG)-overexpressing FXS neurons exhibit abundant miR-fmr1 expression and accumulation in all over the dendrite, soma, and nucleus areas. Further analyses using northern blotting of the two miR-fmr1s isolated from either cytoplasm or nuclei of the pallium neurons revealed that miR-fmr1-42 is the only miRNA accumulated in the nuclei of the FXS neurons [4, 5].
After deleting the NIS motif, the accumulation of miR-fmr1-42 is dramatically reduced in the nucleus portion but found more in the cytoplasm, suggesting that NIS may be required for the transporting of miR-fmr1-42 from the cytoplasm to the nucleus [4].
Following the nuclear accumulation of miR-fmr1-42, a marked increase of epigenetic DNA methylation in the fmr1 promoter and its upstream region has been detected by bisulfite PCR and sequencing assays [4]. It is also noted that these DNA hypermethylation events occur mostly in the CpG-rich binding sites of several fmr1-associated transcriptional cofactors, such as NRF1 (GCGCGC), SP1 (GC box), and USF1/USF2 (E box). As previous studies have reported that blocking the binding of these transcriptional cofactors may cause transcriptional inactivation of FMR1 in human FXS [20], the currently established r(CGG)-overexpressing zebrafish model not only successfully reveals this pathological process but also provides significant insights into the molecular mechanism underlying miRNA-mediated FMR1 inactivation.
Role of miR-fmr1 in the Etiological Mechanism of FXS
In view all of the above studies, Fig. 7.4 summarizes the etiological mechanism of FXS, in which excessive expression of r(CGG)-derived miRNAs, such as miR-fmr1s, causes miRNA accumulation in the neuron nuclei and then forms RNA-induced transcriptional silencing (RITS) complexes to trigger massive DNA methylation in the FMR1 gene promoter, consequently leading to the inactivation of FMR1 transcription. As in this zebrafish FXS model we only overexpressed one-third of the ~50 r(CGG) copies in wild-type fmr1 and had already identified two miR-fmr1 isoforms, it is estimated that the ≥200 r(CGG) expansion in the diseased FMR1 gene of human FXS may generate over 12 different kinds of r(CGG)-derived miRNA isoforms to inactivate FMR1 transcription.
Mechanism of miRNA-mediated FXS pathogenesis. Over-expansion of r(CGG) in the FMR1 gene that encodes FMRP underlies FXS-associated disorders, such as mental retardation and autism. Repeats expanded over 200 copies (full mutation) lead to a complete loss of FMR1 and FMRP expression. The pathological progression causing such FMRP deficiency includes eight steps: (1) Transcription of the FMR1 gene starts after the embryonic blastocyst stage (i.e., day 7–10 human embryo or 10–12 h post-fertilization zebrafish). (2) RNA splicing and further processing excise the 5′-UTR r(CGG) expansion of FMR1 mRNA into fragments. (3) The excised r(CGG) fragments are further processed into various repeat-associated miRNA precursors by certain ribonucleases. (4) These miRNA precursors are exported to cytoplasm and then cleaved into mature miR-fmr1s by RNaseIII Dicers. (5) Excessive miR-fmr1s are accumulated in cytoplasm around the cell nucleus. (6) miR-fmr1s with a NIS signal enter the cell nucleus. (7) Nuclear accumulation of miR-fmr1s induces the formation of RNA-induced transcriptional silencing (RITS) complexes with Rad54l and MeCP2 near the FMR1 gene promoter. (8) RITS complexes trigger DNA hypermethylation and hence lead to transcriptional FMR1 inactivation, the most prevalent event in over 99 % of the FXS patients
As a result, it will be very difficult to target these various miR-fmr1s for treatment. Also, how to demethylate the methylated FMR1 gene promoter in FXS neurons is another problem. To overcome these problems, we need to understand the detail RITS compositions in order to prevent the assembly between RITS and miR-fmr1s; yet, till now the RITS components remain largely unknown.
To unveil the compositions of RITS, our studies have found that Dicer1 rather than Drosha is required for the r(CGG)-derived miRNA biogenesis, while Rad54l and MeCP2 may play crucial roles in the RITS assembly with miR-fmr1s [4, 5]. Using antisense morpholino oligonucleotides directed against certain key RNAi- and CpG methylation-associated effector genes [4], we discovered that the mechanism of miR-fmr1-mediated gene silencing requires activities of Dicer 1 RNase, Rad54-like protein (Rad54l) and methyl-CpG binding protein 2 (MeCP2), but not Drosha RNase. Since the processing of intronic miRNAs has been known to bypass Drosha [21–23], the miR-fmr1 biogenesis may function through a similar process.
Moreover, our results also showed that both miR-fmr1 biogenesis and fmr1 inactivation can be prevented by the morpholino-mediated knockdown of Dicer 1, while only the fmr1 inactivation but not miR-fmr1 biogenesis is affected by the knockdown of either Rad54l or MeCP2. To this, further studies revealed that the fmr1 gene transcription in FXS neurons is increasingly re-activated in response to the decrease of either Rad54l or MeCP2 expression, suggesting that both Rad54l and MeCP2 activities are likely required for the miRNA-mediated fmr1 inactivation. As previous reports have manifested that both Rad54l and MeCP2 are involved in the CpG methylation of repetitive chromatin sequences in autism spectrum disorders [24], a similar mechanism may be reiterated to cause the miRNA-induced hypermethylation of the FMR1 r(CGG) expansion and promoter regions in human FXS.
Pathologies of miR-fmr1-Induced FXS Disorders
The pathological outcomes of the currently established miR-fmr1-mediated FXS animal model are completely reminiscent of the neurodegenerative and cognitive impairments in human FXS disorders, including neuronal deformity, immature synapse formation, long dendritic spine shaping, diminishment of long-term potentiation (LTP), and augmentation of group 1 metabotropic glutamate receptor-dependent long-term depression (mGluR1-LTD).
Alterations of neurite growth and synaptic connectivity have been examined in the FXS zebrafish brains, resembling the exact pathological features of human FXS neurons (Fig. 7.5). Formation of long stripe neuronal dendrites is an important mark of human FXS. In fish lateral pallium (equivalent of human hippocampal stratum radiatum), wild-type neurons present normal neurite growth and branching dendrification, whereas miR-fmr1-affected FXS neurons exhibit long stripe dendrites and disconnected synapses similar to those found in the human FXS hippocampal–neocortical junction [3–5]. In particular, high density of long, immature dendritic spines is markedly increased, indicating failures in forming normal synaptic connections between these FXS-affected neurons (Fig. 7.5a).
Pathological alterations of FXS neurons. (a) Morphological changes of lateral pallium neurons in FXS (middle row) and Nurr1-knockdown (Nurr1 KO; bottom row) zebrafish as compared to the wild-types (top row). Using fluorescent 3D-micrograph examination, three different kinds of neuronal connectivity were observed, including normal (wild-type), disrupted (FXS), tangled (Nurr1 KO) neurite growth and synaptic circuit formation. White arrows indicated the formation of dendritic spines (most right panels). The Nurr1 KO transgenics were generated by retroviral transfection of miR-739 into zebrafish, showing the distinctions between different miRNA functions. Abbreviations indicated: Pa pallium, sP subpallium, Te tectum, H hypothalamus. (b) Standard curves of synaptic LTP responses in pallium slices isolated from FXS zebrafish. Synaptic input–output fEPSP curves were evoked by varying bipolar current intensities, from 5.0, 10.0, 15.0, 25.0, 45.0, 65.0, 100.0 to 155.0 μA (pulse duration 0.1 ms). Calibration: 1 mV, 10 ms. (c) Pulse-induced LTD curves were measured at 40 μA for 125 ms. Calibration: 50 pA, 50 ms
In FXS patients, changes in spine shape are often linked to the absence of FMRP function [25]. FMRP is a translational inhibitor associated with local protein synthesis of certain mRNA species involved in neurite growth and synaptic connection, providing a crucial regulatory process for eliminating immature synapses and enhancing synaptic strength during normal brain development [26–28]. As a result, the miR-fmr1-mediated FMR1 suppression causes FMRP deficiency and hence hinders synaptic strengthening through the formation of protein synthesis-dependent synaptic connections, consequently leading to a cascade of events that FXS is strongly implicated.
Impairment of synaptic plasticity is another major symptom of human FXS. Two types of synaptic plasticity are dependent on protein synthesis: LTP and long-term depression (LTD). LTP is a long-term increase in synaptic strength in response to high-frequency stimulation, whereas LTD is a long-lasting decrease in synaptic strength to below the normal baseline level after prolonged, low frequency stimulation. Both LTP and LTD underline the encoding activities of new declarative memories in brain [29]. LTP in hippocampus is a learning-associated form of synaptic plasticity that is highly involved in the shape change of dendritic spines [30].
It is also known that theta activity (3–8 Hz) is the major late-phase (protein synthesis-dependent) stimulation during the hippocampal encoding of long-term memory [29]. As shown in FXS zebrafish neurons (Fig. 7.5b), theta burst-stimulated synapses of the pallium–neocortical junction (equivalent of human hippocampal CA1–CA3) exhibit diminished LTP compared to that of the wild-type controls [4, 5]. This decreased LTP remains after the blockade of metabotropic GABAalpha receptor (GABAAR)-dependent synaptic inhibition by picotoxin treatment, similar to the reported response of human FXS neurons [4]. In addition, a decrease of the input–output currents, measured at the peak amplitude of dendritic field excitatory postsynaptic potentials (fEPSPs), occurs corresponding to the diminished LTP, indicating a lower excitatory membrane response in the FXS neurons [4]. Given that the excitability of LTP of GABAergic neurons is mediated by activation of mGluR1 [31], further studies of the connection between miRNA-mediated FMRP deficiency and mGluR1 activity may provided answers to the pathological mechanism underlying LTP diminishment in FXS.
Post-synaptic stimulation of mGluR1 has been reported to increase the synthesis of certain neural proteins that trigger internalization of α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptors (AMPAR), leading to a process crucial for LTD expression [32, 33]. FMRP, one of mGluR1-stimulated proteins, is responsible for quenching this LTD process [33]. In FXS, the loss of FMRP in hippocampal neurons fails to quench LTD and hence results in a marked decrease of the LTP excitability, as shown in Fig. 7.5c. Yet, this prolonged LTD remains after co-treatment of an mGluR agonist, 3,5-dihydroxyphenyglycine (DHPG), and an N-methyl-D-aspartate receptor (NMDAR) antagonist, D-2-amino-5-phosphonovalerate (D-APV), suggesting that it is not mediated by NMDAR [4, 5].
Further treatment of DHPG with anisomycin, a protein synthesis inhibitor, completely prevents the mGluR1-LTD augmentation in normal rather than FXS neurons [5], indicating that FMRP may regulate mGluR1-LTD through both protein synthesis-dependent and protein synthesis-independent mechanisms. Since FMRP functions to suppress the translation of certain proteins involved in neural development and synaptic plasticity [8, 9], its deficiency in FXS may lead to excessive accumulation of these FMRP-suppressed proteins which in turn augment the protein synthesis-dependent mGluR1-LTD. On the other hand, emerging studies also showed that FMRP is associated with certain neuron-specific miRNAs, which may contribute to the protein synthesis-independent LTD inhibition.
Roles of FMRP-Associated miRNAs in Neural Development
Recent studies have found several miRNA species associated with FMRP in maintenance of normal neuronal development and synaptic plasticity. Using microarray analyses of miRNAs co-precipitated with FMRP isolated from wild-type and FMR1-knockout mouse brains, 12 candidate miRNAs were identified to specifically interact with wild-type FMRP, including let-7c, miR-9, 100, 124, 125a, 125b, 127, 128, 132, 138, 143, and 219 [17]. Among them, only miR-125a, 125b, and 132 have known functions in relation to FXS [16, 17], while others remain under investigation.
Current studies showed that FMRP does not directly contact with miR-125a; yet, FMRP phosphorylation promotes the assembly of AGO2 and miR-125a into an RNA-induced silencing complex (RISC), which in turn suppresses post-synaptic density protein 95 (PSD-95) mRNA translation [16]. PSD-95 is a protein member of the membrane-associated guanylate kinase (MAGUK) family, which is almost exclusively located in the post-synaptic density of neurons and is involved in anchoring synaptic proteins [34, 35]. It directly or indirectly binds with neuroligin, NMDAR, AMPA receptors, and potassium channels to form a multimeric scaffold for their clustering and hence plays an important role in synaptic plasticity and the stabilization of synaptic changes during LTP [35, 36].
Activation of mGluR1 stimulates the translation of PSD-95 mRNA and leads to AMPAR internalization [37], a process important for LTD expression. Interestingly, studies also found that mGluR1 signaling further triggers the dephosphorylation of FMRP to dissociate the AGO2–miR-125a RISC complex and thus prevents the inhibitory effect of FMRP on PSD-95 translation [16]. Based on this finding, it is conceivable that the loss of FMRP in FXS may cause PSD-95 over-expression and ultimately results in mGluR1-LTD augmentation.
FMRP was also recently reported to affect NMDAR signaling via a negatively regulatory mechanism involving miR-125b and miR-132 [17], albeit no alteration of NMDAR-mediated LTD found in human FXS. Studies showed that both miR-125b and miR-132 have inhibitory effects on dendritic spine morphology and synaptic physiology in mouse hippocampal neurons, while FMRP knockdown can alleviate these effects [17].
Further target screening analyses revealed that miR-125b and AGO1 form certain RISC assembly to specifically suppress NMDAR subunit NR2A mRNA translation [17]. This silencing effect of miR-125b on NR2A may be one of many reasons causing dendritic spine changes; nevertheless, how does it relate to the known FXS pathologies such as diminished LTP and prolonged mGluR1-LTD remains unclear. Since there is no prior report of such an NMDAR-LTD alteration in the established mouse or zebrafish FXS model, research in this new direction may need to depend on the development of a novel animal model, in which NMDAR-LTD is affected by the loss of FMRP. Alternatively, it is possible that this NMDAR-LTD alteration may be covered or compensated by some unknown mechanisms in the current FXS animal models. To solve these questions, further investigation is needed.
Research of miRNA involvement in FXS is still at its very early stage. Although there are many miRNAs associated with FMRP, the majority of them currently do not have a clear function in relation to FXS. Yet, in view of this vast number of FMRP-associated miRNAs, it is conceivable that studies in this direction may discover fruitful knowledge to understand the detail processes of FXS pathogenesis and, in the long run, may lead to the development of novel medical interventions for treating FXS-related disorders, such as mental retardation and autism. The future application is very promising.
Conclusion
In sum, FXS is a genetic disease caused by dysregulation of multiple miRNA functions, including r(CGG)-derived miR-fmr1s and neuron-specific miR-125a, 125b, 132, and maybe many more waiting to be discovered. Based on the current understanding, the onset of FXS occurs after the embryonic blastocyst stage when ectodermic differentiation starts to form primordial neurons. At this critical moment for neural development, the expression of FMR1 and its protein FMRP functions to regulate the translation of certain neural proteins, so as to enhance the establishment of normal synaptic connection and plasticity. However, due to the mutated over-expansion of r(CGG) in FXS patients’ FMR1 5′-UTR, the expression of such mutated FMR1 also generates an excessive amount of r(CGG)-rich fragments, which are further processed into various miRNA isoforms, namely miR-fmr1s, by RNaseIII Dicers and are gradually accumulated in the cytoplasm and nuclei of neuronal cells. Since many of these miR-fmr1s carry NIS motifs for entering the cell nuclei, their nuclear over-accumulation results in the formation of RITS complexes with Rad54l and MeCP2, which then cause DNA hypermethylation of the r(CGG) expansion and its adjacent promoter regions of the FMR1 gene, leading to completely shutdown of FMR1 and FMRP expression. Without FMRP regulation, mGluR1 signaling will trigger the synthesis of excessive neural proteins that cause AMPAR internalization, pre-mature synaptic plasticity, LTP diminishment, and prolonged LTD expression, a cascade of events strongly implicated in FXS. In addition, since FMRP interacts with miR-125a and miR-125b/132 to regulate the signaling pathways of mGluR1 and NMDAR, respectively, its deficiency may abolish these miRNA-mediated regulations and further aggravate the FXS pathologies. To clarify the involvement of these miRNAs in FXS, their functional roles are depicted in Fig. 7.1.
The development of possible therapy for FXS is still under investigation. There are three major difficulties in its therapeutic design: First, how to demethylate the highly methylated r(CGG) expansion and promoter regions in FXS patients’ FMR1 gene? Second, how to deliver the drug through the blood–brain barrier (BBB)? Last, how to limit the drug effect only in GABAergic neurons? Currently, there are a few chemical drugs capable of causing DNA demethylation; yet, none of them are safe for treating neurons. Alternatively, recent findings of reprogramming-related miRNAs, such as miR-302, may offer a new approach to solve these problems.
Previous studies have shown that miR-302 enhances somatic cell DNA demethylation and hence reprograms the somatic cells into an ESC-like state [14, 15]. Using retroviral deliver of an inducible miR-302 transgene into zebrafish FXS neurons, we have found that there is a narrow window of miR-302 concentrations capable of silencing MeCP2 expression to demethylate the fmr1 promoter region but not strong enough to suppress fmr1 mRNA translation. Due to the similarity between human FMR1 and zebrafish fmr1 genes, this miR-302-mediated partial reprogramming approach may be useful for developing a novel therapy treating FXS. Nevertheless, how to maintain the miR-302 expression within such a narrow dosage window in a specific group of brain neurons will be challenging.
The findings in zebrafish and mouse FXS models have signified a vast similarity between these animal models and human FXS, which may shed light on new therapeutic interventions. Furthermore, these animal models may provide significant insights into the mechanisms of microsatellite-like nucleotide repeats in brain development for understanding their functional effects on human intelligence quotient (IQ) and autism.
Given that there are many different microsatellite-like nucleotide repeats in the human genome, which may encode a variety of repeat-associated miRNAs (ramRNAs), the availability of these animal models is surely useful for studying the functional roles of these various ramRNAs in vivo as a forthcoming challenge.
References
Handa V, Saha T, Usdin K. The fragile X syndrome repeats form RNA hairpins that do not activate the interferon-inducible protein kinase, PKR, but are cut by Dicer. Nucleic Acids Res. 2003;31:6243–8.
Krol J, Fiszer A, Mykowska A, Sobczak K, de Mezer M, Krzyzosiak WJ. Ribonuclease dicer cleaves triplet repeat hairpins into shorter repeats that silence specific targets. Mol Cell. 2007;25:575–86.
Lin SL, Chang SJE, Ying SY. First in vivo evidence of microRNA-induced fragile X mental retardation syndrome. Mol Psychiatry. 2006;11:616–7.
Chang SJE, Chang-Lin S, Chang D, Ying SY, Lin SL. Repeat-associated microRNA triggers fragile X syndrome in zebrafish. Open Neuropsychopharmacol J. 2008;1:6–18.
Lin SL, Ying SY. Role of repeat-associated microRNAs (ramRNA) in fragile X syndrome (FXS). In: Ying SY, editor. Current perspectives in microRNAs. New York: Springer; 2008. p. 245–66.
Hagerman RJ, Staley LW, O’Conner R, Lugenbeel K, Nelson D, McLean SD, Taylor A. Learning-disabled males with a fragile X CGG expansion in the upper premutation size range. Pediatrics. 1996;97:122–6.
Jin P, Alisch RS, Warren ST. RNA and microRNAs in fragile X mental retardation. Nat Cell Biol. 2004;6:1048–53.
Eberhart DE, Malter HE, Feng Y, Warren ST. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum Mol Genet. 1996;5:1083–91.
Tamanini F, Van Unen L, Bakker C, Sacchi N, Galjaard H, Oostra BA, Hoogeveen AT. Oligomerization properties of fragile-X mental-retardation protein (FMRP) and the fragile-X-related proteins FXR1P and FXR2P. Biochem J. 1999;343(Pt 3):517–23.
Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, Warren ST. DNA methylation represses FMR1 transcription in fragile X syndrome. Hum Mol Genet. 1992;1:397–400.
Tropepe V, Sive HL. Can zebrafish be used as a model to study the neurodevelopmental causes of autism? Genes Brain Behav. 2003;2:268–81.
Tucker B, Richards R, Lardelli M. Expression of three zebrafish orthologs of human FMR1-related genes and their phylogenetic relationships. Dev Genes Evol. 2004;214:567–74.
van ’t Padje S, Engels B, Blonden L, Severijnen LA, Verheijen F, Oostra BA, Willemsen R. Characterization of FMRP in zebrafish: evolutionary dynamics of the fmr1 gene. Dev Genes Evol. 2005;215:198–206.
Lin SL, Chang D, Lin CH, Ying SY, Leu D, Wu DTS. Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res. 2011;39:1054–65.
Lin SL. Deciphering the mechanism behind induced pluripotent stem cell generation. Stem Cells. 2011;29:1645–9.
Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell. 2011;42:673–88.
Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–84.
Lopez-Bendito G, Shigemoto R, Kulik A, Vida I, Fairen A, Lujan R. Distribution of metabotropic GABA receptor subunits GABAB1a/b and GABAB2 in the rat hippocampus during prenatal and postnatal development. Hippocampus. 2004;14:836–48.
Selby L, Zhang C, Sun QQ. Major defects in neocortical GABAergic inhibitory circuits in mice lacking the fragile X mental retardation protein. Neurosci Lett. 2007;412:227–32.
Kumari D, Gabrielian A, Wheeler D, Usdin K. The roles of Sp1, Sp3, USF1/USF2 and NRF-1 in the regulation and three-dimensional structure of the fragile X mental retardation gene promoter. Biochem J. 2005;386:297–303.
Lin SL, Chang D, Wu DY, Ying SY. A novel RNA splicing-mediated gene silencing mechanism potential for genome evolution. Biochem Biophys Res Commun. 2003;310:754–60.
Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–6.
Lin SL, Kim H, Ying SY. Intron-mediated RNA interference and microRNA (miRNA). Front Biosci. 2008;13:2216–30.
Lopez-Rangel E, Lewis ME. Loud and clear evidence for gene silencing by epigenetic mechanisms in autism spectrum and related neurodevelopmental disorders. Clin Genet. 2006;69:21–2.
Irwin SA, Patel B, Idupulapati M, Harris JB, Crisostomo RA, Larsen BP, Kooy F, Willems PJ, Cras P, Kozlowski PB, Swain RA, Weiler IJ, Greenough WT. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am J Med Genet. 2001;98:161–7.
Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–87.
Li Z, Zhang Y, Ku L, Wilkinson KD, Warren ST, Feng Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001;29:2276–83.
Jin P, Zarnescu DC, Zhang F, Pearson CE, Lucchesi JC, Moses K, Warren ST. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron. 2003;39:739–47.
Axmacher N, Mormann F, Fernandez G, Flger CE, Fell J. Memory formation by neuronal synchronization. Brain Res Rev. 2006;52:170–82.
Patenaude C, Chapman CA, Bertrand S, Congar P, Lacaille JC. GABAB receptor- and metabotropic glutamate receptor-dependent cooperative long-term potentiation of rat hippocampal GABAA synaptic transmission. J Physiol. 2003;553:155–67.
Godfraind JM, Reyniers E, De Boulle K, D’Hooge R, De Deyn PP, Bakker CE, Oostra BA, Kooy RF, Willems PJ. Long-term potentiation in the hippocampus of fragile X knockout mice. Am J Med Genet. 1996;64:246–51.
Snyder EM, Philpot BD, Huber KM, Dong X, Fallon JR, Bear MF. Internalization of ionotropic glutamate receptors in response to mGluR activation. Nat Neurosci. 2001;4:1079–85.
Nosyreva ED, Huber KM. Metabotropic receptor-dependent long-term depression persists in the absence of protein synthesis in the mouse model of fragile X syndrome. J Neurophysiol. 2006;95:3291–5.
Hunt CA, Schenker LJ, Kennedy MB. PSD-95 is associated with the postsynaptic density and not with the presynaptic membrane at forebrain synapses. J Neurosci. 1996;16:1380–8.
Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1–29.
Meyer D, Bonhoeffer T, Scheuss V. Balance and stability of synaptic structures during synaptic plasticity. Neuron. 2014;82:430–43.
Bhattacharyya S, Biou V, Xu W, Schluter O, Malenka RC. A critical role for PSD-95/AKAP interactions in endocytosis of synaptic AMPA receptors. Nat Neurosci. 2009;12:172–81.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Lin, SL. (2015). microRNAs and Fragile X Syndrome. In: Santulli, G. (eds) microRNA: Medical Evidence. Advances in Experimental Medicine and Biology, vol 888. Springer, Cham. https://doi.org/10.1007/978-3-319-22671-2_7
Download citation
DOI: https://doi.org/10.1007/978-3-319-22671-2_7
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-22670-5
Online ISBN: 978-3-319-22671-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)