Abstract
Phosphorus (P) is the second important key plant nutrient after nitrogen. An adequate supply of P is therefore required for proper functioning and various metabolisms of plants. Majority of P in soils is fixed, and hence, plant available P is scarcely available despite the abundance of both inorganic and organic P forms in soils. A group of soil microorganisms capable of transforming insoluble P into soluble and plant accessible forms across different genera, collectively called phosphate-solubilizing microorganisms (PSM), have been found as best eco-friendly option for providing inexpensive P to plants. These organisms in addition to supplying soluble P to plants also facilitate the growth of plants by several other mechanisms, for instance, improving the uptake of nutrients and stimulating the production of some phytohormones. Even though several bacterial, fungal and actinomycetal strains have been identified as PSM, the mechanism by which they make P available to plants is poorly understood. This chapter focuses on the mechanism of P-solubilization and physiological functions of phosphate solubilizers in order to better understand the ecophysiology of PSM and consequently to gather knowledge for managing a sustainable environmental system. Conclusively, PSM are likely to serve as an efficient bio-fertilizer especially in areas deficient in P to increase the overall performance of crops.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
2.1 Introduction
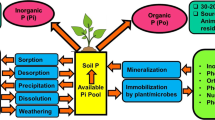
Phosphorus (P) is one of the most essential plant nutrients which profoundly affect the overall growth of plants (Wang et al. 2009) by influencing various key metabolic processes such as cell division and development, energy transport, signal transduction, macromolecular biosynthesis, photosynthesis and respiration of plants (Shenoy and Kalagudi 2005; Ahemad et al. 2009; Khan et al. 2009). On the contrary, unlike N, the atmosphere does not provide soluble P to plants. And hence, the source of P is largely the primary and secondary minerals and/or organic compounds. In comparison to other nutrients, P concentration in soil solution is much lower and ranges from 0.001 to 1 mg/l (Brady and Weil 2002). Broadly, P compounds in soil can be placed into three categories: (i) inorganic compounds, (ii) organic compounds of the soil humus and (iii) organic and inorganic P compounds associated with the cells of living matter. Mineral compounds of P usually contain aluminium (Al), iron (Fe), manganese (Mn) and calcium (Ca) and vary from soils to soils. For example, P forms a complex with Al, Fe and Mn in acidic soils, while in alkaline soils it reacts very strongly with Ca. However, under all conditions, the types of soil P compounds are determined mainly by soil pH and by the type and concentrations of soil minerals. Some of the most common P minerals are presented in Table 2.1. Of the total soil P pool, about 50 % of P is in the organic forms (Richardson 1994), which varies between 4 and 90 % in most soils (Yadav and Verma 2012). The organic P in plants includes (i) inositol phosphate (10–50 % in soil) which represents a series of phosphate esters ranging from monophosphates up to hexaphosphates. Phytic acid (inositol hexakisphosphate) is the main compound that plants use to store P in seeds to support early seedling growth following germination. Phytin (a Ca–Mg salt of phytic acid) is the most abundant of the known organophosphorus compounds in soils. Other organic P in soils occur as sugar phosphates, nucleotides (0.2–2.5 %), phosphoprotein (trace), phosphonates (Tate 1984) and phospholipids (1–5 %) (Yadav and Verma 2012). Of the various forms of P, plants take up only negatively charged primary and secondary orthophosphate ions (H2PO4 − and HPO4 2−) as nutrient. Indeed, the amount of plant available P is very low relative to the total soil P. Moreover, majority of the soil P is fixed, and only a small fraction of P is available for uptake by plants. Therefore, P deficiency results in stunted growth, dark leaves, and inhibition of flowering and root system development. In most plants, these symptoms will appear when P concentration in the leaves goes below 0.2 %. And, hence, in many cases, phosphatic fertilizers which are quite soluble and manures that also contain P (soluble P, organic P and inorganic P) are applied to overcome P deficiency in soils and to provide adequate P to plants. The P of the phosphatic fertilizers or the manure reacts very strongly with soil constituents and becomes unavailable to plants. The insoluble and inaccessible forms of P are hydrolysed to soluble and available forms through the process of solubilization (inorganic P)/mineralization (organic P). The immobilization in contrast is the reverse reaction of mineralization. During immobilization, microorganisms convert inorganic forms to organic phosphate, which are then incorporated into their living cells. Mineralization and immobilization of P occur simultaneously and are influenced by structure and compositions of microbes and physico-chemical characteristics of soils besides the exudates of various plant genotypes.
2.2 Phosphate Solubilization by Microbes: Current Perspective
The insoluble forms of P such as tricalcium phosphate (Ca3PO4)2, aluminium phosphate (Al3PO4), iron phosphate (Fe3PO4), etc. may be converted to soluble P by P-solubilizing organisms inhabiting different soil ecosystems (Gupta et al. 2007; Song et al. 2008; Khan et al. 2013; Sharma et al. 2013). Soil microorganisms in this regard have generally been found more effective in making P available to plants from both inorganic and organic sources by solubilizing (Toro 2007; Wani et al. 2007a) and mineralizing complex P compounds (Bishop et al. 1994; Ponmurugan and Gopi 2006), respectively. Several workers have documented their findings in order to better understand as to how the microbial populations cause the solubilization of insoluble P (Illmer and Schinner 1995; Khan et al. 2007, 2009; Buch et al. 2008). Of the various strategies adopted by microbes, the involvement of low molecular mass organic acids (OA) secreted by microorganisms has been a well-recognized and widely accepted theory as a principal means of P-solubilization, and various studies have identified and quantified organic acids and defined their role in the solubilization process (Maliha et al. 2004; Khan et al. 2010; Marra et al. 2012). The OA produced by many P-solubilizers, for example, bacterial cultures (Table 2.2) or fungi (Table 2.3), in the natural environment or under in vitro conditions chelate mineral ions or decrease the pH to bring P into solution (Pradhan and Shukla 2005). Consequently, the acidification of microbial cells and their surrounding leads to the release of P-ions from the P-mineral by H+ substitution for Ca2+ (Goldstein 1994; Mullen 2005; Trivedi and Sa 2008). The efficiency of solubilization, however, depends on the kind of organic acids released into the medium and their concentration. Furthermore, the quality of the acid is more important for P-solubilization than the total amount of acids produced by phosphate solubilizing (PS) organisms (Scervino et al. 2010a, b). Additionally, the simultaneous production of different organic acids by the PS strains may contribute to the greater potential for solubilization of insoluble inorganic phosphates (Marra et al. 2012).
There are also reports which suggest that insoluble P could be transformed into soluble forms of P without OA production by microbes (Asea et al. 1988; Illmer and Schinner 1992; Chen et al. 2006). For example, Altomare et al. (1999) while investigating the P-solubilizing ability of plant growth-promoting and biocontrol fungus Trichoderma harzianum T-22 did not produce OA under in vitro condition suggesting that the insoluble P could be solubilized by mechanisms other than acidification process also. The fungal-solubilizing activity was credited both to chelation and to reduction processes, which may be useful in the management of phytopathogens. Apart from the OA theory, some of the inorganic acids (Reyes et al. 2001; Richardson 2001) such as HCl (Kim et al. 1997), nitric acid and sulphuric acids (Dugan and Lundgren 1965) produced by chemoautotrophs and the H+ pump, for example, in Penicillium rugulosum, have also been reported to solubilize the insoluble P (Reyes et al. 1999). The inorganic acids so released convert TCP to di- and monobasic phosphates with the net result of an enhanced availability of the element to plants.
2.2.1 Mineralization: Enzymatic Degradation of Complex Organic P Compounds
Organic P compounds undergo mineralization, and the resulting P is taken up as nutrient by plants. In this regard, numerous soil microbes or rhizosphere microflora possess the ability to transform organic P into soluble forms of P (Tarafdar and Claassen 1988; Rodriguez et al. 2006). This mineralization process is mediated by the enzymes especially phosphatases (Tarafdar et al. 1988; Yadav and Tarafdar 2003; Aseri et al. 2009) and phytases (Maougal et al. 2014), released by the soil microbes. The enzyme phosphatases (e.g. acid and alkaline phosphatases) released exterior to the cell (exo-enzymes) are non-specific in nature and use organic P as a substrate to convert it into inorganic form (Beech et al. 2001). Of the two phosphatases, acid phosphatases (To-O et al. 2000), a widely distributed enzyme and commonly found in fungi (To-O et al. 1997; Abd-Alla and Omar 2001), for example, Colletotrichum graminicola (Schadeck et al. 1998a, b), are considered as the principal mechanism for mineralization of soil organic P (Hilda and Fraga 1999) where it catalyses the release of inorganic P from organic P compounds such as inositol hexaphosphate (Nozawa et al. 1998; Tarafdar and Gharu 2006; Yadav and Tarafdar 2007, 2011). However, the degradation of organic P mediated by phosphatases varies greatly among different fungi (Guimarães et al. 2006). Another attractive application of P-dissolving enzymes is the mineralization of soil organic P through phytate degradation mediated by the enzyme phytase, which specifically causes release of P from phytic acid. Phytate is a major component of organic P in soil. Though the ability of plants to obtain P directly from phytate is very limited, the growth and P nutrition of Arabidopsis plants supplied with phytate was improved significantly when they were genetically transformed with the phytase gene [phyA] derived from Aspergillus niger (Richardson 2001). This led to the increase in P nutrition to such an extent that the growth and P content of the plant was equivalent to control plants supplied with inorganic P. Similar increase in utilization of inositol P by plants in the presence of microbial communities including P-solubilizing fungus (A. niger) capable of producing phytase is reported (Richardson 2001; Vassilev et al. 2007). Phosphonatases (Kumar et al. 2013) and C–P lyases (Salimpour et al. 2010) are the other enzymes that cleave the C–P of organophosphonates. Once the inorganic or organic P compound is changed to soluble P, it can now easily be used up as P nutrient by plants, algae, cyanobacteria and autotrophic bacteria and thereafter could be immobilized into organic cellular macromolecules, for example, DNA, RNA and ATP. Considering the critical impact of such enzymes in dissolution of complex organic compounds into usable form of P, it is highly desirable to develop the bacterial/fungal inoculants with high phosphatase and phytase activity which in turn could possibly be of great practical value in sustainable crop production.
2.3 Physiological Functions of Phosphate-Solubilizing Microorganisms
Phosphate-solubilizing microorganisms increase the overall performance of plants by providing mainly soluble P to plants in different production systems. However, they also benefit plants by other mechanisms (Fig. 2.1). Indeed, PSM exhibit multifunctional properties (Vassileva et al. 2010; Yadav et al. 2011; Khan et al. 2013); for example, they are known to synthesize siderophores (Hamadali et al. 2008; Wani et al. 2008a; Tank and Saraf 2003; Viruel et al. 2011) and IAA and gibberellic acid (Sattar and Gaur 1987; Souchie et al. 2007; Viruel et al. 2011). Phosphate-solubilizing bacteria such as Gram-negative P. fluorescens, P. aeruginosa and Chromobacterium violaceum also secrete antibiotics (Lipping et al. 2008; Taurian et al. 2010) and provide protection to plants against soilborne pathogens (biocontrol) (Khan et al. 2002; Vassilev et al. 2006; Singh et al. 2010). Other physiological traits of PSM involve the release of cyanide, a secondary metabolite which is ecologically important (Wani et al. 2007b) and gives a selective advantage to the producing strains (Rudrappa et al. 2008; Badawi et al. 2011). Besides strict P-solubilizers, a few genera of rhizobia, for example, Bradyrhizobium and Rhizobium, have also been found to solubilize P and secrete IAA (Pandey and Maheshwari 2007; Badawi et al. 2011). There are numerous PS bacteria that possess the ability to synthesize a key enzyme, 1-aminocyclopropane-1-carboxylate (ACC) deaminase (Glick et al. 2007), which hydrolyses ACC [the immediate precursor of plant hormone ethylene (C2H4)] to NH3 and α-ketobutyrate and thus mitigate the inhibitory effects of C2H4. Some of the compounds synthesized by PS bacteria with possible effect on plant growth are listed in Tables 2.4 and 2.5.
An illustration depicting functional diversity among PS bacteria [Adapted from Khan et al. (2013)]
2.3.1 Some Examples of Positive Plant Growth Regulators Synthesized by PSM
Plant growth regulators (PGRs) are the substances that influence physiological processes of plants at very low concentrations and modify or control one or more specific metabolic events of a plant (Danova et al. 2012; Sane et al. 2012). According to the Environmental Protection Agency (EPA), the plant regulators have been defined as “any substance or mixture of substances intended, through physiological action, to accelerate or retard the rate of growth or maturation, or otherwise alter the behaviour of plants or their produce”. Such compounds produced by the plant or by PGPR are called plant hormones (Davies 1995; Karadeniz et al. 2006). Broadly, on the basis of chemical structures and their subsequent effects on plants, plant growth-regulating substances have been divided into five general groups: (1) auxins, (2) gibberellins, (3) cytokinins, (4) ethylene and (5) a group called inhibitors, which includes abscisic acid (ABA), phenolics and alkaloids (Frankenberger and Arshad 1995; Ferguson and Lessenger 2006). The production of auxins (Glick 1995; Wani et al. 2007b, 2008b; Ahemad and Khan 2012) and ethylene (Sasek et al. 2012), for example, by PSM, is considered a common microbiological trait, while the synthesis of cytokinins by bacteria, for example, Paenibacillus polymyxa (Raza et al. 2008), is less common (Timmusk et al. 1999). The gibberellin secretion at high concentrations is, however, very rare (Solano et al. 2010). Generally, majority (>80 %) of the soil bacteria are capable of secreting auxins especially IAA, indole butyric acid or similar compounds via tryptophan metabolism (Solano et al. 2010; Legault et al. 2011). A few examples of the phytohormones secreted by PGPR including PS bacteria (Table 2.4) and PS rhizobia (Table 2.5) and other compounds and their direct or indirect impact on plant growth and development are reviewed and discussed briefly in the following section.
2.3.1.1 Synthesis and Physiological Functions of Phytohormones
2.3.1.1.1 Synthesis of IAA
The synthesis of IAA by microbes (Fig. 2.2) involves one of the three pathways: (1) Indoleacetic acid formation via indole-3-pyruvic acid and indole-3-acetic aldehyde is found in the majority of bacteria like Erwinia herbicola; saprophytic species of the genera Agrobacterium and Pseudomonas; and certain representatives of Bradyrhizobium, Rhizobium, Azospirillum, Klebsiella and Enterobacter. (2) The conversion of tryptophan into indole-3-acetic aldehyde may involve an alternative pathway in which tryptamine is formed. This pathway is believed to operate in pseudomonads and azospirilla. (3) IAA biosynthesis via indole-3-acetamide formation is reported for phytopathogenic bacteria Agrobacterium tumefaciens, Pseudomonas syringae and E. herbicola and saprophytic pseudomonads like Pseudomonas putida and P. fluorescens. The genes controlling IAA synthesis via this pathway are also reported in symbiotic bacteria like Rhizobium spp., Bradyrhizobium spp. and Azospirillum spp., although the activity of the corresponding enzymes is either negligible or not detectable. Indoleacetic acid biosynthesis that involves tryptophan conversion into indole-3-acetonitrile is found in plants, Alcaligenes faecalis, and possibly the cyanobacterium Synechocystis sp., and the tryptophan-independent pathway, more common in plants, is also found in microorganisms (azospirilla and cyanobacteria). However, the synthesis of IAA using this pathway is reported to be insignificant, and the mechanisms are largely unknown. Many bacteria are known to synthesize auxins using such pathways and help the plants to grow better. Bacteria in general form maximum amount of IAA during the steady-state stage of their growth while ammonium ions and glutamine inhibit IAA biosynthesis (Tsavkelova et al. 2006). The genes involved in IAA synthesis in bacterial strains may be plasmid or chromosomal borne. For example, pathogenic bacteria contain Ti plasmids that control the formation of the phytohormone, whereas in saprophytic microorganisms, auxin biosynthesis is governed by chromosomal genes (Tsavkelova et al. 2006). It is reported that 80 % of microorganisms isolated from the rhizosphere of various crops possess the ability to synthesize and release auxins as secondary metabolites (Loper and Schroth 1986). Of the various PGPR strains, bacteria belonging to the genera Azospirillum, Pseudomonas, Xanthomonas and Rhizobium as well as Alcaligenes, Enterobacter, Acetobacter and Bradyrhizobium have been shown to produce auxins which help in stimulating plant growth (Egamberdieva et al. 2007; Wani et al. 2007c; Kumar et al. 2008; Poonguzhali et al. 2008). However, the extent of IAA production by bacterial strains could be different due in part to the involvement of biosynthetic pathways, location of the genes, regulatory sequences and the presence of enzymes to convert active free IAA into conjugated forms. Moreover, the synthesis of IAA is also influenced by environmental factors (Patten and Glick 1996). Synthesis of IAA by Rhizobium spp. in the presence and absence of tryptophan has also been demonstrated (Wani et al. 2007c). In a similar study, Bent et al. (2001) reported that the concentration of indole compounds by three different strains, Paenibacillus polymyxa (L6), P. polymyxa (Pw-2) and Pseudomonas fluorescens (M20), increased with increasing rate of tryptophan (0–200 mg/ml) at different incubation interval.
Biosynthetic pathways of IAA in bacteria [Adapted from Patten and Glick (1996)]
2.3.1.1.1.1 Physiological Functions of IAA
The production of phytohormones such as auxins by microbial communities has been reported by various workers over the last 20 years (Giordano et al. 1999a, b; Rajkumar and Freitas 2008; Singh 2008; Ahemad and Khan 2012). Among plant hormones, auxins are the major plant growth regulators produced by PSM (Oves et al. 2013) and exhibit many physiological functions as shown in Fig. 2.3. Apart from varying physiological functions of IAA, the role of IAA in legume–Rhizobium symbiosis is briefly discussed in the following section.
2.3.1.1.1.2 Role of Indoleacetic Acid in Legume–Rhizobium Symbiosis
Among nodule bacteria, rhizobial strains have been reported to produce auxins in variable amounts. For example, Vargas et al. (2009) in a study reported considerably lower frequency of auxin producers (23 %) among populations of clover nodulating R. leguminosarum bv. trifolii. The auxins so released by nodule bacteria are reported to affect nodulation, and accordingly, IAA synthesizing rhizobia have been found to produce more nodules than IAA-negative mutants (Boiero et al. 2007). The IAA produced by rhizobia may also induce root morphogenesis and consequently enhance its (1) size and weight, (2) branch numbers and patterns and (3) the surface area of roots as reported in non-legumes (Dazzo and Yanni 2006). Inoculation with auxin-producing bacteria may also result in the formation of adventitious roots (Solano et al. 2010). Furthermore, Noel et al. (1996) observed that the inoculation with IAA-producing strains of R. leguminosarum accelerated the germination of canola and lettuce. Similarly, Biswas et al. (2000) concluded that the inoculation of rice with R. leguminosarum bv. trifolii increased dry matter and grain production, besides an increment in N, P, K and Fe content in plant tissue. All these effects were ascribed due to the accumulation of IAA in the rhizosphere following rhizobial inoculation leading to some physiological changes in the root systems with consequent increase in nutrient uptake. In contrast, the overproduction of IAA in some cases by PGPR has been found to have deleterious impact on to plants (Schlindwein et al. 2008). For example, R. leguminosarum bv. trifolii strain TV-13 produced 171.1 mg/ml IAA in media enriched with tryptophan (Schlindwein et al. 2008), while strains of Bradyrhizobium sp. isolated from black wattle roots produced between 1.2 and 3.3 mg/ml IAA and increased the seedling vigour in relation to un-inoculated control plants. The variation in the amount of IAA produced by PGPR was, however, suggested due to differences in the composition of the growth medium and tryptophan concentration. In a follow-up study, Sridevi et al. (2008) observed that IAA production by rhizobia occurred only when tryptophan was added to YM and that the isolates produced the maximum amount of IAA in medium supplemented with 2.5 mg/ml tryptophan concentration.
2.3.1.1.2 Other Phytohormones
Like auxins, cytokinins influence both cell division and cell enlargement and also affect seed dormancy, flowering, fruiting and plant senescence (Ferguson and Lessenger 2006). Cytokinin production by PGPR (Boiero et al. 2007) is, however, less obvious compared to the production of auxins. This is probably due to the lack of methods used for cytokinin detection, and hence, reports on cytokinin synthesis by PGPR in general are scarce. Gibberellin is yet another growth regulator which (1) affects seed germination (Miransari and Smith 2009), (2) stimulates growth of plants (Guo et al. 2011) and (3) delays ageing (Ferguson and Lessenger 2006). The production of gibberellins at high concentrations is considered very rare and has been reported for two strains of Bacillus, isolated from the Alnus glutinosa rhizosphere (Solano et al. 2010). The concentration of gibberellins in nodules is, however, generally higher than in nearby root tissue as supported by the fact that rhizobia have the capacity to produce some amount of gibberellin-like substances. However, it is not known whether bacteria contribute significantly to the amount of gibberellins within the nodule or it is just imported from some remote host plant tissue (Dobert et al. 1992; Hedden and Thomas 2012). Despite all these contrasting facts, the role of gibberellin in Rhizobium–legume symbiosis that may have important implications in the endophytic colonization of non-legumes by rhizobia is adequately described. For example, A. caulinodans infects the semi-aquatic legume Sesbania rostrata via the intercellular crack entry, a process mediated by gibberellins. Considering that crack entry is the main process of endophytic colonization of non-legumes by rhizobia, the production of gibberellins by the bacterium is reported to facilitate this process (Lievens et al. 2005).
2.3.2 Negative Plant Growth Regulator
Abscisic acid is one of the strong inhibitor of growth and germination and promotes seed dormancy (Miransari and Smith 2009; Yang et al. 2009). Apart from these, ABA also helps plants to tolerate abiotic stresses. When plants are exposed to drought stress, the hormonal balance of plants changes and ABA content in the leaves increases, which reduce the level of cytokinin. This in turn elicits stomata closure (Yang et al. 2009). Cohen et al. (2009) in a similar study suggested that ABA produced along with gibberellins by PGPR strain significantly contributed to water stress alleviation of maize plants. Some rhizobial strains such as B. japonicum USDA110 also produce ABA (Boiero et al. 2007) and function in the same way as do the other PGPR (Zhang et al. 2012).
2.3.3 Growth Modulation Enzyme ACC Deaminase
Ethylene is a plant hormone which under normal conditions regulates many physiological processes, such as (1) seed germination, (2) root hair development and root elongation, (3) leaf and organ senescence, (4) leaf and petal abscission, (5) epinasty and (6) fruit ripening (Abeles et al. 1992; Frankenberger and Arshad 1995; Arshad and Frankenberger 2002; Siddikee et al. 2011). Also, ethylene regulates nod factor signalling and nodule formation and has primary functions in plant defence systems. Besides its physiological role in different developmental processes of plants, ethylene is also considered as a stress hormone, whose synthesis in plants is increased substantially by a number of biotic and abiotic stresses. At higher concentrations, ethylene, however, inhibits growth and development of plants (Grichko and Glick 2001). The enzyme 1-aminocyclopropane-1-carboxylate (ACC) deaminase (E.C. 4.1.99.4) which however mitigates the ethylene stress was first purified to homogeneity from Pseudomonas sp. strain ACP (Honma and Shimomura 1978), later on partially purified from P. chlororaphis 6G5 (Klee et al. 1991) and P. putida GR12-2 (Jacobson et al. 1994) and then purified to homogeneity from P. putida UW4 (Hontzeas et al. 2004). Enzyme ACC deaminase (a multimeric enzyme) has been found thereafter to be synthesized by a variety of PGPR (Belimov et al. 2005; Rajkumar et al. 2006; Madhaiyan et al. 2007; Mellado et al. 2007).
2.3.3.1 How the Bacterial ACC Deaminase Works
Mechanistically, the ACC deaminase-producing plant growth-promoting bacteria first bind to the surface of a plant (usually seeds or roots), although these bacteria may also be found on leaves and flowers or within a plant’s internal tissues, i.e. as an endophyte (Glick et al. 1998). Along with other small molecular components of root exudates, some of the plant ACC (a non-ribosomal amino acid) is exuded from seeds, roots or leaves (Penrose et al. 2001) and may be taken up by the bacteria associated with these tissues and subsequently cleaved by ACC deaminase (Penrose and Glick 2003). The ACC, the immediate precursor of C2H4, when hydrolysed by ACC deaminase results in NH3 and α-ketobutyrate formation (Glick et al. 1998; Penrose and Glick 2003; Reed et al. 2005; Safronova et al. 2006), and hence, it strongly alleviates the stress induced by ethylene-mediated impact on plants by lowering the C2H4 levels in plants (Glick et al. 2007; Sessitsch et al. 2005; Sun et al. 2009). The bacteria utilize the NH3 so evolved from ACC as a source of N and thereby restrict the accumulation of C2H4 within the plant, which otherwise inhibits plant growth (Belimov et al. 2002). Thus, the decreased levels of C2H4 in turn allow the plants to grow better (Zahir et al. 2008). A model to explain how ACC deaminase promotes plant growth is depicted in Fig. 2.4. It has been observed that plants inoculated with PGPR containing ACC deaminase were dramatically more resistant to the deleterious effects of stress ethylene, synthesized under stressful conditions such as flooding (Grichko and Glick 2001), heavy metals (Burd et al. 1998; Grichko et al. 2000), presence of phytopathogens (Wang et al. 2000), drought and high saline conditions (Mayak et al. 2004). The net result of the cleavage of exuded ACC by bacterial ACC deaminase is that the bacterium is de facto acting as a sink for ACC. Additionally, plants growing in association with ACC deaminase-containing plant growth-promoting bacteria generally have longer roots and shoots and are more resistant to growth inhibition by a variety of ethylene-inducing stresses. Furthermore, the reduction of ethylene levels in plant tissues following ACC deaminase activity can cause significant morphological changes in root tissue, such as changes in root hair length and increases in root mass, accompanied by the consequent improvement in nutrient uptake. The morphological changes in plants are further increased when ACC deaminase action is coupled with the production of auxins by PGPR. The question arises, how bacterial ACC deaminase selectively reduces the deleterious ethylene levels (the second ethylene peak) without affecting the small first peak of ethylene that is thought to activate plant defence responses. In this regard, ACC deaminase is generally present in bacteria at a relatively low level until it is induced, and the induction of enzyme activity is a rather slow and complex process. Immediately following an abiotic or biotic stress, the pool of ACC in the plant is low as is the level of ACC deaminase in the associated bacterium. Stress induces the induction of ACC oxidase in the plant so that there is an increased flux through ACC oxidase resulting in the first (small) peak of ethylene that in turn induces the transcription of protective/defensive genes in the plant. At the same time, bacterial ACC deaminase is induced by the increasing amounts of ACC that ensue from the induction of ACC synthase in the plant so that the magnitude of the second, deleterious, ethylene peak is decreased significantly (typically by 50–90 %). Because ACC oxidase has a greater affinity for ACC than does ACC deaminase, when ACC deaminase-producing bacteria are present, plant ethylene levels are dependent upon the ratio of ACC oxidase to ACC deaminase. That is, to effectively reduce plant ethylene levels, ACC deaminase must function before any significant amount of ACC oxidase is induced. Thus, in the absence of some other mechanism, IAA-producing bacteria might all be expected to ultimately be inhibitory to plant growth. However, this is in fact not the case because as plant ethylene levels increase, the ethylene that is produced through feedback mechanism inhibits IAA signal transduction thereby limiting the extent that IAA can activate ACC synthase transcription (Pierik et al. 2006; Prayitno et al. 2006; Czarny et al. 2007; Stearns et al. 2012). In plants inoculated with PGPR that secrete both IAA and ACC deaminase, the level of ethylene does not increase compared to the plants inoculated only with IAA-secreting bacteria. In the presence of ACC deaminase, there is much less ethylene and subsequent ethylene feedback inhibition of IAA signals transduction so that the bacterial IAA can continue to promote both plant growth and increase ACC synthase transcription. However, in this case, a large portion of the additional ACC which is synthesized is hydrolysed by the bacterial ACC deaminase. Therefore, the use of such plant growth-promoting bacteria containing ACC deaminase may prove useful in developing strategies to facilitate plant growth in stressed soil environments.
A schematic model of how plant growth-promoting bacteria that both produce ACC deaminase and synthesize IAA may facilitate plant growth. The only enzyme shown in this scheme is ACC deaminase. SAM is converted to ACC by the enzyme ACC synthase; ACC is converted to ethylene by ACC oxidase. IAA biosynthesis, both in bacteria and in plants, is a complex multi-enzyme/protein process as is IAA signal transduction. ACC 1-aminocyclopropane-1-carboxylate, IAA indole-3-acetic acid, SAM S-adenosyl methionine [Adapted from Glick (2014)]
2.3.3.2 Role of ACC Deaminase in Nodulation
ACC deaminase-containing bacteria are relatively common in soil and have been found in a wide range of environments across the world. Indeed, the ability of bacteria to hydrolyse ACC has a competitive advantage over other soil inhabitants because it can use ACC as an N source (Jacobson et al. 1994). This hypothesis suggests that ACC may act as a unique/novel source of N for some soil bacteria. While searching for ACC deaminase positive rhizobial strains, it was found that amongst 13 different rhizobial strains, five strains displayed enzyme activity while seven strains had the acdS gene (Ma et al. 2003). Conclusively, it was reported that the Mesorhizobium strain only expressed this activity when the bacterium was present within a root nodule. In other investigation conducted in southern Saskatchewan, Canada, of the total 233 rhizobial strains isolated from soil samples collected from 30 different sites, nearly 12 % (27 strains) displayed the ACC deaminase activity (Duan et al. 2009). Similarly, ACC deaminase genes have been reported in chickpea Mesorhizobium isolates (Nascimento et al. 2012), B. japonicum E109, USDA110 and SEMIA5080 (Boiero et al. 2007). Rhizobial strains that express ACC deaminase are up to 40 % more efficient at forming nitrogen-fixing nodules than strains that lack this activity (Ma et al. 2003, 2004). However, strains of rhizobia that express ACC deaminase have only a low level of enzyme activity compared with free-living plant growth-promoting bacteria, i.e. typically around 2–10 %. Thus, free-living bacteria bind relatively non-specifically to plant tissues (mainly roots) and have a high level of ACC deaminase activity that can protect plants from different abiotic and biotic stresses by lowering ethylene levels throughout the plant. On the other hand, (symbiotic) rhizobia that generally bind tightly only to the roots of specific plants have a low level of enzyme activity which facilitates nodulation by locally lowering ethylene levels. It is not known whether the large differences in enzyme activity that are observed when comparing free-living bacteria with rhizobia are a consequence of differences in the amount of enzyme synthesized by one type of bacteria versus the other or of differences in the specific catalytic activity of the enzymes from the different types of bacteria. It has also been observed that some rhizobia reduces the plant ethylene levels mediated by ACC deaminase activity and enhances nodulation in host legumes (Zahir et al. 2008; Belimov et al. 2009) or modifies root system of non-legumes. For instance, strains of R. leguminosarum bv. viciae and Mesorhizobium loti increased the number of lateral roots in Arabidopsis thaliana because of this plant growth-promoting mechanism (Contesto et al. 2008). In addition to the more common mode of acdS transcriptional regulation, acdS genes from various strains of M. loti have been found to be under the transcriptional control of the nifA promoter that is normally responsible for activating the transcription of nif, nitrogen fixation genes (Kaneko et al. 2000; Sullivan et al. 2002; Uchiumi et al. 2004; Nukui et al. 2006; Nascimento et al. 2012). The consequence of this somewhat unusual mode of regulation is that, unlike ACC deaminases from other rhizobia, the M. loti ACC deaminase does not facilitate nodulation but, rather, is expressed within nodules. The result of this unusual regulation is, in M. loti, ACC deaminase may act to decrease the rate of nodule senescence. This is particularly important because of the fact that nitrogen fixation, a process that utilizes a very high level of energy in the form of ATP, could (perhaps inadvertently) activate stress ethylene synthesis resulting in premature nodule senescence.
2.3.4 Physiological Functions of Siderophores
Iron is essential for almost all life for processes such as respiration and DNA synthesis. Despite being one of the most abundant elements in the Earth’s crust, the bioavailability of iron in many environments such as the soil is limited by the very low solubility of the Fe3+ ion. In the aerobic environment, iron accumulates in common mineral phases such as iron oxides and hydroxides and hence becomes inaccessible to organisms. Microbes (e.g. bacteria and fungi) have, therefore, evolved a strategy to acquire iron by releasing siderophores (Greek: “iron carrier”), small (generally less than 1,000 molecular weight) high-affinity iron-chelating compounds, which scavenge iron from the mineral phases by forming soluble Fe3+ complexes that can be taken up by active transport mechanisms. Broadly, siderophores act as solubilizing agents for iron from minerals or organic compounds under conditions of iron starvation (Miethke and Marahiel 2007; Indiragandhi et al. 2008). There are more than 500 different siderophores which are produced mainly by Gram-positive and Gram-negative bacteria. Siderophores are highly electronegative and bind Fe (III), preferentially forming a hexacoordinated complex. The iron ligation groups have been tentatively classified into three main chemical types: (1) hydroxamate (e.g. aerobactin and ferrichrome), (2) catecholates/phenolates (e.g. enterobactin) and (3) hydroxyl acids/carboxylates (e.g. pyochelin). Some siderophores contain more than one of the three iron-chelating groups (Table 2.6). Siderophores are, however, usually classified by the ligands used to chelate the ferric iron. Citric acid can also act as a siderophore. The wide variety of siderophores may be due to evolutionary pressures placed on microbes to produce structurally different siderophores (Fig. 2.5). Siderophores are important for some pathogenic bacteria for their acquisition of iron. The strict homeostasis of iron leads to a free concentration of about 10−24 mol/l, and hence, there are great evolutionary pressures put on pathogenic bacteria to obtain this metal. For example, the anthrax pathogen Bacillus anthracis releases two siderophores, bacillibactin and petrobactin, to scavenge ferric iron from iron proteins.
2.3.4.1 Role of Siderophores in Biological Nitrogen Fixation
Siderophore produced by majority of PGPR (Rajkumar et al. 2010) including rhizobia (Ahemad and Khan 2012) has been suggested as one of the modes of growth promotion of nodulated legumes under field conditions wherein siderophores facilitate the uptake of iron (assimilation) from the environment (Kloepper and Schroth 1978; Katiyar and Goel 2004). The iron enzymes involved include nitrogenase, leghemoglobin, ferredoxin and hydrogenase with nitrogenase and leghemoglobin constituting up to 12 % and 30 % of total protein in the bacterial and infected plant cells, respectively (Verma and Long 1983). A nodulated legume has been found to have an increased demand for iron compared to that of a non-nodulated plant (Derylo and Skorupska 1993 ). For example, Pseudomonas sp. strain 267 enhanced symbiotic N2 fixation in clover under gnotobiotic conditions, produced fluorescent siderophores under low-iron conditions and secreted B group vitamins (Marek-Kozaczuk and Skorupska 2001). However, Tn5 insertion mutants of strain 267 defective in siderophore production did not differ from the wild type in promoting the growth of clover suggesting that the siderophore production had no effect on stimulating nodulation. In contrast, Gill et al. (1991) demonstrated that mutants of R. melioti that were unable to produce siderophores were able to nodulate the plants, but the efficiency of N2 fixation was less compared to the wild type, indicating the importance of iron in N2 fixation. In a similar study, Kluyvera ascorbata, a siderophore-producing PGPR, was able to protect plants from heavy metal toxicity (Burd et al. 1998).
2.3.5 Cyanogenic Compounds
Cyanide is yet another secondary metabolite produced during the early stationary growth phase (Knowles and Bunch 1986) by several PGPR, notably Pseudomonas spp. and Bacillus (Wani et al. 2007d), Chromobacterium (Faramarzi and Brand 2006) and Rhizobium spp. (Wani et al. 2008a, b) by oxidative decarboxylation pathway using glycine, glutamate or methionine as precursors (Curl and Truelove 1986). The cyanide so released by microbial communities in solution acts as a secondary metabolite and confers a selective advantage onto the producer strains (Vining 1990). Although cyanide is a phytotoxic agent capable of disrupting enzyme activity involved in major metabolic processes, its role as a biocontrol substance is overwhelming (Devi et al. 2007; Voisard et al. 1989). Hydrogen cyanide (HCN) among cyanogenic compounds effectively blocks the cytochrome oxidase pathway and is highly toxic to all aerobic microorganisms at picomolar concentrations. However, producer microbes, mainly pseudomonads, are reported to be resistant (Bashan and de-Bashan 2005).
2.3.6 Production of Lytic Enzymes
In high input modem agricultural practices, pesticides are frequently and inappropriately used to protect crop plants from damage by insects, disease and so on which today still destroy almost 33 % of all food crops. The use of pesticides in this respect is considered effective if they provide the desired biological results and are inexpensive. However, the indiscriminate use of pesticides has resulted in the adverse impact on soil fertility, human health and the environment. Therefore, one of the safest strategies involving microorganisms for controlling/managing plant pests often called as “biocontrol” holds great promise as an alternative to the use of synthetic agrichemicals. Biological control agents are generally considered more environmentally sound than the pesticides and other antimicrobial treatments. In this context, the antagonistic potential of microbes in particular has formed the base for effective applications of such organisms as an alternative to the chemical control measures against a range of fungal and bacterial plant pathogens. And hence, a variety of microbial compounds have been identified/extracted that have been found to inhibit/suppress the phytopathogenic growth leading thereby to the reduction in damage to plants (Helbig 2001; Yang et al. 2005; Raza et al. 2008). These microbially synthesized compounds include defence enzymes, such as chitinase, β-1,3-glucanase, peroxidase, protease and lipase (Bashan and de-Bashan 2005; Karthikeyan et al. 2006). Chitinase and β-1,3-glucanase degrade the fungal cell wall and cause lysis of fungal cell. Furthermore, chitin and glucan oligomers released during degradation of the fungal cell wall by the action of lytic enzymes act as elicitors that elicit various defence mechanisms in plants. Such enzymes produced by Pseudomonas stutzeri have demonstrated the lysis of the pathogen Fusarium sp. (Bashan and de-Bashan 2005). Peroxidase (PO) represents another component of an early response in plants to pathogen attack and plays a key role in the biosynthesis of lignin which limits the extent of pathogen spread (Bruce and West 1989). In bean, rhizosphere colonized by various bacteria induced PO activity (Zdor and Anderson 1992). In a study, a rapid increase in PO activity was recorded in coconut (Cocos nucifera L.) treated with a mixture of P. fluorescens, T. viride and chitin which contributed to induced resistance against invasion by Ganoderma lucidum, the causal agent of Ganoderma disease (Karthikeyan et al. 2006). These findings suggest that PGPR possessing the ability to synthesize hydrolytic enzymes can effectively be utilized for managing the plant diseases and can help to reduce the pesticide usage.
2.4 Conclusion
In intensive agricultural practices, P is supplied to plants through synthetic phosphatic fertilizers, which indeed is expensive and environment disruptive. Application of phosphate-solubilizing microorganisms as an alternative to chemically synthesized phosphatic fertilizer is therefore an urgent requirement in crop production systems. The success of this microbiological approach, however, depends on identification, preparation and delivery of multifunctional phosphate solubilizers to farm practitioners. Understanding the mechanistic basis of phosphate solubilization by soil dwellers is critical and requires multifaceted approach to uncover the hidden phosphate-solubilizing potentials of functionally diverse yet naturally abundant soil microflora. Moreover, the functional variations among phosphate solubilizers need to be identified. Once identified and physiologically characterized, phosphate-solubilizing microbes are likely to provide benefits to crops in sustainable agriculture.
References
Abd-Alla MH, Omar SA (2001) Survival of rhizobia/bradyrhizobia and a rock-phosphate-solubilizing fungus Aspergillus niger on various carriers from some agro-industrial wastes and their effects on nodulation and growth of faba bean and soybean. J Plant Nutr 24:261–272
Abeles FB, Morgan PW, Saltveit ME Jr (1992) Ethylene in plant biology. Academic, New York
Afzal A, Bano A, Fatima M (2010) Higher soybean yield by inoculation with N-fixing and P-solubilizing bacteria. Agron Sustain Dev 30:487–495
Ahemad M, Khan MS (2012) Effect of fungicides on plant growth promoting activities of phosphate solubilizing Pseudomonas putida isolated from mustard (Brassica campestris) rhizosphere. Chemosphere 86:945–950
Ahemad M, Zaidi A, Khan MS, Oves M (2009) Biological importance of phosphorus and phosphate solubilizing microorganisms—an overview. In: Khan MS, Zaidi A (eds) Phosphate solubilizing microbes for crop improvement. Nova, New York, pp 1–4
Akintokun AK, Akande GA, Akintokun PO, Popoola TOS, Babalola AO (2007) Solubilization of insoluble phosphate by organic acid producing fungi isolated from Nigerian soil. Int J Soil Sci 2:301–307
Altomare C, Norvell WA, Bjorkman T, Harman GE (1999) Solubilization of phosphates and micronutrients by the plant growth promoting and biocontrol fungus Trichoderma harzianum rifai 1295-22. Appl Environ Microbiol 65:2926–2933
Arshad M, Frankenberger WT Jr (2002) Ethylene: agricultural sources and applications. Kluwer, New York, p 342
Arwidsson Z, Johansson E, Kronhelm TV, Allard B, van Hees P (2010) Remediation of metal contaminated soil by organic metabolites from fungi I—production of organic acids. Water Air Soil Pollut 205:215–226
Asea PEA, Kucey RMN, Stewart JWB (1988) Inorganic phosphate solubilization by two Penicillium species in solution culture and soil. Soil Biol Biochem 20:459–464
Aseri GK, Jain N, Tarafdar JC (2009) Hydrolysis of organic phosphate forms by phosphatases and phytase producing fungi of arid and semi-arid soils of India. Am-Eurasian J Agric Environ Sci 5:564–570
Badawi FSF, Biomy AMM, Desoky AH (2011) Peanut plant growth and yield as influenced by co-inoculation with Bradyrhizobium and some rhizo-microorganisms under sandy loam soil conditions. Ann Agric Sci 56:17–25
Bashan Y, de-Bashan LE (2005) Bacteria. In: Hillel D, Hillel D (eds) Encyclopaedia of soils in the environment, vol 1. Elsevier, Oxford, pp 103–115
Beech IB, Paiva M, Caus M, Coutinho C (2001) Enzymatic activity and within biofilms of sulphate-reducing bacteria. In: Gilbert PG, Allison D, Brading M, Verran J, Walker J (eds) Biofilm community interactions: change or necessity? Boiline, Cardiff, pp 231–239
Belimov AA, Safranova VI, Mimura T (2002) Response of spring rape (Brassica napus) to inoculation with PGPR containing ACC-deaminase depends on nutrient status of plant. Can J Microbiol 48:189–199
Belimov AA, Hontzeas N, Safronova VI, Demchinskaya SV, Piluzza G, Bullitta S, Glick BR (2005) Cadmium-tolerant plant growth-promoting rhizobacteria associated with the roots of Indian mustard (Brassica juncea L. Czern.). Soil Biol Biochem 37:241–250
Belimov AA, Dodd IC, Hontzeas N, Theobald JC, Safronova VI, Davies WJ (2009) Rhizosphere bacteria containing 1-aminocyclopropane-1-carboxylate deaminase increase yield of plants grown in drying soil via both local and systemic hormone signalling. New Phytol 181:413–423
Bent E, Tuzun S, Chanway CP, Enebak S (2001) Alterations in plant growth and in root hormone levels of lodgepole pines inoculated with rhizobacteria. Can J Microbiol 47:793–800
Bianco C, Defez R (2010) Improvement of phosphate solubilization and Medicago plant yield by an indole-3-acetic acid-overproducing strain of Sinorhizobium meliloti. Appl Environ Microbiol 76:4626–4632
Bishop ML, Chang AC, Lee RWK (1994) Enzymatic mineralization of organic phosphorus in a volcanic soil in Chile. Soil Sci 157:238–243
Biswas JC, Ladha JK, Dazzo FB, Yanni YG, Rolfe BG (2000) Rhizobial inoculation influences seedling vigor and yield of rice. Agron J 92:880–886
Boiero L, Perrig D, Masciarelli O, Penna C, Cassán F, Luna V (2007) Phytohormone production by three strains of Bradyrhizobium japonicum and possible physiological and technological implications. Appl Microbiol Biotechnol 74:874–880
Brady NC, Weil RR (2002) The nature and properties of soils, 13th edn. Prentice Hall of India, New Delhi, 960
Bruce RJ, West CA (1989) Elicitation of lignin biosynthesis and isoperoxidase activity by pectic fragments in suspension cultures of cluster bean. Plant Physiol 91:889–897
Buch A, Archana G, Kumar GN (2008) Metabolic channelling of glucose towards gluconate in phosphate-solubilizing Pseudomonas aeruginosa P4 under phosphorus deficiency. Res Microbiol 159:635–642
Burd GI, Dixon DG, Glick BR (1998) A plant growth-promoting bacterium that decreases nickel toxicity in seedlings. Appl Environ Microbiol 64:3663–3668
Canbolat MY, Bilen S, Cakmakci R, Sahin F, Aydin A (2006) Effect of plant growth promoting bacteria and soil compaction on barley seedling growth, nutrient uptake, soil properties and rhizosphere microflora. Biol Fertil Soils 42:350–357
Chakraborty U, Chakraborty BN, Basnet M, Chakraborty AP (2009) Evaluation of Ochrobactrum anthropi TRS-2 and its talc based formulation for enhancement of growth of tea plants and management of brown root rot disease. J Appl Microbiol 107:625–634
Chandra S, Choure K, Dubey RC, Maheshwari DK (2007) Rhizosphere competent Mesorhizobium loti MP6 induces root hair curling, inhibits Sclerotinia sclerotiorum and enhances growth of Indian mustard (Brassica campestris). Braz J Microbiol 38:124–130
Chen YP, Rekha PD, Arun AB, Shen FT, Lai WA, Young CC (2006) Phosphate solubilizing bacteria from subtropical soil and their tricalcium phosphate solubilizing abilities. Appl Soil Ecol 34:33–41
Cohen AC, Travaglia CN, Bottini R, Piccoli PN (2009) Participation of abscisic acid and gibberellins produced by endophytic Azospirillum in the alleviation of drought effects in maize. Botany 87:455–462
Contesto C, Desbrosses G, Lefoulon C, Bena G, Borel F, Galland M, Gamet L, Varoquaux F, Touraine B (2008) Effects of rhizobacterial ACC deaminase activity on Arabidopsis indicate that ethylene mediates local root responses to plant growth-promoting rhizobacteria. Plant Sci 175:178–189
Curl EA, Truelove B (1986) The rhizosphere. Springer, Berlin
Czarny JC, Shah S, Glick BR (2007) Response of canola plants at the transcriptional level to expression of a bacterial ACC deaminase in the roots. In: Ramina A, Chang C, Giovannoni J, Klee H, Perata P, Woltering E (eds) Advances in plant ethylene research. Springer, Dordrecht, pp 377–382
Danova K, Todorova M, Trendafilova A, Evstatieva L (2012) Cytokinin and auxin effect on the terpenoid profile of the essential oil and morphological characteristics of shoot cultures of Artemisia alba. Nat Prod Commun 7:1075–1076
Davies PJ (1995) Plant hormones. Kluwer, Dorderecht
Dazzo FB, Yanni YG (2006) The natural rhizobium-cereal crop association as an example of plant-bacterial interaction. In: Uphoff N, Ball AS, Fernandes E, Herren H, Husson O, Laing M, Palm C, Pretty J, Sanchez P, Sanginga N, Thies J (eds) Biological approaches to sustainable soil systems. CRC, Boca Raton, FL, pp 109–127
Derylo M, Skorupska A (1993) Enhancement of symbiotic nitrogen fixation by vitamin-secreting fluorescent Pseudomonas. Plant Soil 54:211–217
Devi KK, Seth N, Kothamasi S, Kothamasi D (2007) Hydrogen cyanide-producing rhizobacteria kill subterranean termite Odontotermes obesus (rambur) by cyanide poisoning under in vitro conditions. Curr Microbiol 54:74–78
Dobert RC, Rood SB, Zanewich K, Blevins DG (1992) Gibberellins and the Legume-Rhizobium symbiosis: III. Quantification of gibberellins from stems and nodules of Lima Bean and Cowpea. Plant Physiol 100:1994–2001
Duan J, Müller KM, Charles TC, Vesely S, Glick BR (2009) 1-Aminocyclopropane-1-carboxylate (ACC) deaminase genes in Rhizobia from southern Saskatchewan. Microb Ecol 57:423–436
Dugan P, Lundgren DG (1965) Energy supply for the chemoautotroph Ferrobacillus ferrooxidans. J Bacteriol 89:825–834
Egamberdieva D, Kamilova F, Validov S, Gafurova L, Kucharova Z, Lugtenberg B (2007) High incidence of plant growth-stimulating bacteria associated with the rhizosphere of wheat grown on salinated soil in Uzbekistan. Environ Microbiol 10:1–9
Farajzadeh D, Yakhchali B, Aliasgharzad N, Bashir NS, Farajzadeh M (2012) Plant growth promoting characterization of indigenous Azotobacteria isolated from soils in Iran. Curr Microbiol 64:397–403
Faramarzi MA, Brand H (2006) Formation of water-soluble metal cyanide complexes from solid by Pseudomonas plecoglossicida. FEMS Microbiol Lett 259:47–52
Ferguson L, Lessenger JE (2006) Plant growth regulators. In: Lessenger JE (ed) Agricultural medicine. Springer, New York, pp 156–166
Frankenberger WT Jr, Arshad M (1995) Phytohormones in soil: microbial production and function. Dekker, New York, p 503
Gill PRJ, Barton LL, Scoble MD, Neilands JB (1991) A high-affinity iron transport system of Rhizobium meliloti may be required for efficient nitrogen fixation in planta. Plant Soil 130:211–271
Giordano W, Avalos J, Cerdá-Olmedo E, Domenech CE (1999a) Nitrogen availability and production of bikaverin and gibberellins in Gibberella fujikuroi. FEMS Microbiol Lett 173:389–393
Giordano W, Avalos J, Fernandez-Martín R, Cerdá-Olmedo E, Domenech C (1999b) Lovastatin inhibits the production of gibberellins but not sterol or carotenoid biosynthesis in Gibberella fujikuroi. Microbiology 145:2997–3002
Glick BR (1995) The enhancement of plant growth by free living bacteria. Can J Microbiol 41:109–117
Glick BR (2014) Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res 169:30–39
Glick BR, Penrose DM, Li J (1998) A model for the lowering of plant ethylene concentration by plant growth promoting bacteria. J Theor Biol 190:63–68
Glick BR, Todorovic B, Czarny J, Cheng Z, Duan J, McConkey B (2007) Promotion of plant growth by bacterial ACC deaminase. Crit Rev Plant Sci 26:227–242
Goldstein AH (1994) Involvement of the quinoprotein glucose dehydrogenase in the solubilization of exogenous phosphates by gram-negative bacteria. In: Torriani-Gorini A, Yagil E, Silver S (eds) Phosphate in microorganisms: cellular and molecular biology. ASM Press, Washington, DC, pp 197–203
Grichko VP, Glick BR (2001) Amelioration of flooding stress by ACC deaminase containing plant growth-promoting bacteria. Plant Physiol Biochem 39:11–17
Grichko VP, Filby B, Glick BR (2000) Increased ability of transgenic plants expressing the bacterial enzyme ACC deaminase to accumulate Cd, Co, Cu, Ni, Pb, and Zn. J Biotechnol 81:45–53
Guimarães LHS, Peixoto-Nogueira SC, Michelinl M, Rizzatti ACS, Sandrim VC, Zanoelo F, Aquino ACMM, Junior AB, de Lourdes M, Polizeli TM (2006) Screening of filamentous fungi for production of enzymes of biotechnological interest. Braz J Microbiol 37:474–480
Guo J, Ma C, Kadmiel M, Gai Y, Strauss S (2011) Tissue-specific expression of Populus C19 GA 2-oxidases differentially regulate above- and below- ground biomass growth through control of bioactive GA concentrations. New Phytol 192:626–639
Gupta A, Rai V, Bagdwal N, Goel R (2005) In situ characterization of mercury-resistant growth-promoting fluorescent pseudomonads. Microbiol Res 160:385–388
Gupta N, Sabat J, Parida R, Kerkatta D (2007) Solubilization of tricalcium phosphate and rock phosphate by microbes isolated from chromite, iron and manganese mines. Acta Bot Croat 66:197–204
Hamadali H, Hafidi M, Virolle MJ, Ouhdouch Y (2008) Rock phosphate solubilizing actinomycetes: screening for plant growth promoting activities. World J Microbiol Biotechnol 24:2565–2575
Hedden P, Thomas SG (2012) Gibberellin biosynthesis and its regulation. Biochem J 444:11–25
Helbig J (2001) Biological control of Botrytis cinerea Pers. ex Fr. in strawberry by Paenibacillus polymyxa (isolate 18191). J Phytopathol 149:265–273
Hilda R, Fraga R (1999) Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol Adv 17:319–339
Honma M, Shimomura T (1978) Metabolism of 1-aminocyclopropane-1-carboxylic acid. Agric Biol Chem 43:1825–1831
Hontzeas N, Zoidakis J, Glick BR, Abu-Omar MM (2004) Expression and characterization of 1 aminocyclopropane-1-carboxylate deaminase from the rhizobacterium Pseudomonas putida UW4: a key enzyme in bacterial plant growth promotion. Biochim Biophys Acta 703:11–19
Illmer P, Schinner F (1992) Solubilization of inorganic phosphates by microorganisms isolated from forest soil. Soil Biol Biochem 24:389–395
Illmer P, Schinner F (1995) Solubilization of inorganic calcium phosphates-solubilization mechanisms soil. Soil Biol Biochem 27:257–263
Indiragandhi P, Anandham R, Madhaiyan M, Sa TM (2008) Characterization of plant growth-promoting traits of bacteria isolated from larval guts of diamondback moth Plutella xylostella (Lepidoptera: Plutellidae). Curr Microbiol 56:327–333
Jacobson CB, Pasternak JJ, Glick BR (1994) Partial purification and characterization of ACC deaminase from the plant growth-promoting rhizobacterium Pseudomonas putida GR12-2. Can J Microbiol 40:1019–1025
Jain R, Saxena J, Sharma V (2012) Solubilization of inorganic phosphates by Aspergillus awamori S19 isolated from rhizosphere soil of a semi-arid region. Ann Microbiol 62:725–735
Kaneko T, Nakamura Y, Sato S, Asamizu E, Kato T, Sasamoto S et al (2000) Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res 7:331–338
Karadeniz A, Topeuoglu SF, Inan S (2006) Auxin, gibberellin, cytokinin and abscisic acid production in some bacteria. World J Microbiol Biotechnol 22:1061–1064
Karthikeyan M, Radhika K, Mathiyazhagan S, Bhaskaran R, Samiyappan R, Velazhahan R (2006) Induction of phenolics and defense-related enzymes in coconut (Cocos nucifera L.) roots treated with biocontrol agents. Braz J Plant Physiol 18:367–377
Katiyar V, Goel R (2004) Siderophore mediated plant growth promotion at low temperature by mutant of fluorescent pseudomonad. Plant Growth Regul 42:239–244
Khan MS, Zaidi A, Aamil M (2002) Biocontrol of fungal pathogens by the use of plant growth promoting rhizobacteria and nitrogen fixing microorganisms. Indian J Bot Soc 81:255–263
Khan MS, Zaidi A, Wani PA (2007) Role of phosphate solubilizing microorganisms in sustainable agriculture—a review. Agron Sustain Dev 27:29–43
Khan MS, Zaidi A, Wani PA, Ahemad M, Oves M (2009) Functional diversity among plant growth-promoting rhizobacteria. In: Khan MS, Zaidi A, Musarrat J (eds) Microbial strategies for crop improvement. Springer, Berlin, pp 105–132
Khan MS, Zaidi A, Ahemad M, Oves M, Wani PA (2010) Plant growth promotion by phosphate solubilizing fungi-current perspective. Arch Agron Soil Sci 56:73–98
Khan MS, Ahmad E, Zaidi A, Oves M (2013) Functional aspect of phosphate-solubilizing bacteria: importance in crop production. In: Maheshwari DK et al (eds) Bacteria in agrobiology: crop productivity. Springer, Berlin, pp 237–265
Kim KY, Jordan D, Krishnan HB (1997) Rahnella aquatilis, bacterium isolated from soybean rhizosphere, can solubilize hydroxyapatite. FEMS Microbiol Lett 153:273–277
Klee HJ, Hayford MB, Kretzmer KA, Barry GF, Kishmore GM (1991) Control of ethylene synthesis by expression of a bacterial enzyme in transgenic tomato plants. Plant Cell 3:1187–1193
Kloepper JW, Schroth MN (1978) Plant growth-promoting rhizobacteria on radishes. In: Fourth international conference on plant pathogen bacteria, Angers, France, vol 2. pp 879–882
Knowles CJ, Bunch AW (1986) Microbial cyanide metabolism. Adv Microb Physiol 27:73–111
Krishnan HB, Kang BR, Krishnan AH, Kil Kim KY, Kim YC (2007) Rhizobium etli USDA9032 engineered to produce a phenazine antibiotic inhibits the growth of fungal pathogens but is impaired in symbiotic performance. Appl Environ Microbiol 73:327–330
Kumar KV, Singh N, Behl HM, Srivastava S (2008) Influence of plant growth promoting bacteria and its mutant on heavy metal toxicity in Brassica juncea grown in fly ash amended soil. Chemosphere 72:678–683
Kumar P, Dubey RC, Maheshwari DK (2012) Bacillus strains isolated from rhizosphere showed plant growth promoting and antagonistic activity against phytopathogens. Microbiol Res 167:493–499
Kumar V, Singh P, Jorquera MA, Sangwan P, Kumar P, Verma AK, Sanjeev A (2013) Isolation of phytase-producing bacteria from Himalayan soils and their effect on growth and phosphorus uptake of Indian mustard (Brassica juncea). World J Microbiol Biotechnol 29:1361–1369
Legault GS, Lerat S, Nicolas P, Beaulieu C (2011) Tryptophan regulates thaxtomin A and indole-3-acetic acid production in Streptomyces scabiei and modifies its interactions with radish seedlings. Phytopathology 101:1045–1051
Lievens S, Goormachtig S, Den Herder J, Capoen W, Mathis R, Hedden P, Holsters M (2005) Gibberellins are involved in nodulation of Sesbania rostrata. Plant Physiol 139:1366–1379
Lipping Y, Jiatao X, Daohong J, Yanping F, Guoqing L, Fangcan L (2008) Antifungal substances produced by Penicillium oxalicum strain PY-1 potential antibiotics against plant pathogenic fungi. World J Microbiol Biotechnol 24:909–915
Loper JE, Schroth MN (1986) Influence of bacterial sources of indole-2-acetic acid on root elongation of sugar beet. Phytopathology 76:386–389
Ma W, Guinel FC, Glick BR (2003) The Rhizobium leguminosarum bv. viciae ACC deaminase protein promotes the nodulation of pea plants. Appl Environ Microbiol 69:4396–4402
Ma W, Charles TC, Glick BR (2004) Expression of an exogenous 1-aminocyclopropane-1-carboxylate deaminase gene in Sinorhizobium meliloti increases its ability to nodulate alfalfa. Appl Environ Microbiol 70:5891–5897
Madhaiyan M, Poonguzhali S, Sa T (2007) Metal tolerating methylotrophic bacteria reduces nickel and cadmium toxicity and promotes plant growth of tomato (Lycopersicon esculentum L.). Chemosphere 69:220–228
Maliha R, Samina K, Najma A, Sadia A, Farooq L (2004) Organic acid production and phosphate solubilization by phosphate solubilizing microorganisms under in vitro conditions. Pak J Biol Sci 7:187–196
Maougal RT, Brauman A, Plassard C, Abadie J, Djekoun A, Drevon JJ (2014) Bacterial capacities to mineralize phytate increase in the rhizosphere of nodulated common bean (Phaseolus vulgaris) under P deficiency. Eur J Soil Biol 62:8–14
Marek-Kozaczuk M, Skorupska A (2001) Production of B group vitamins by plant growth-promoting Pseudomonas fluorescens strain 267 and the importance of vitamins in the colonization and nodulation of red clover. Biol Fertil Soil 33:146–151
Marra LM, Soares CRFSS, de Oliveira SM, Ferreira PAAA, Soares BL, Carvalho RF, Lima JM, Moreira FM (2012) Biological nitrogen fixation and phosphate solubilization by bacteria isolated from tropical soils. Plant Soil 357:289–307
Mayak S, Tirosh S, Glick BR (2004) Plant growth promoting bacteria that confer resistance to water stress in tomatoes and peppers. Plant Physiol 166:525–530
Mehboob I, Zahir A, Arshad M, Tanveer A, Azam F (2010) Growth promoting activities of different rhizobium spp., in wheat. Pak J Bot 43:1643–1650
Mellado JC, Onofre-Lemus J, Santos PE, Martinez-Aguilar L (2007) The tomato rhizosphere, an environment rich in nitrogen fixing Burkholderia species with capabilities of interest for agriculture and bioremediation. Appl Environ Microbiol 73:5308–5319
Mendes GO, Dias CS, Silva IR, Júnior JIR, Pereira OL, Costa MD (2013) Fungal rock phosphate solubilization using sugarcane bagasse. World J Microbiol Biotechnol 29:43–50
Miethke M, Marahiel MA (2007) Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev 71:413–451
Miransari M, Smith D (2009) Rhizobial lipo-chitooligosaccharides and gibberellins enhance barley (Hordeum vulgare L.) seed germination. Biotechnology 8:270–275
Mullen MD (2005) Phosphorus in soils: biological interactions. In: Hillel D (ed) Encyclopedia of soils in the environment. Elsevier, Oxford, pp 210–215
Nascimento F, Brígido C, Alho L, Glick BR, Oliveira S (2012) Enhanced chickpea growth-promotion ability of a Mesorhizobium strain expressing an exogenous ACC deaminase gene. Plant Soil 353:221–230
Noel TC, Sheng C, Yost CK, Pharis RP, Hynes MF (1996) Rhizobium leguminosarum as a plant growth-promoting rhizobacterium: direct growth promotion of canola and lettuce. Can J Microbiol 42:279–283
Noordman WH, Reissbrodt R, Bongers RS, Rademaker ILW, Bockelmann W, Smit G (2006) Growth stimulation of Brevibacterium sp. by siderophores. J Appl Microbiol 101:637–646
Nozawa M, Hu HY, Fujie K, Tanaka H, Urano K (1998) Quantitative detection of Enterobacter cloacae strain HO-I in bioreactor for chromate wastewater treatment using polymerase chain reaction [PCR]. Water Res 32:3472–3476
Nukui N, Minamisawa K, Ayabe SI, Aoki T (2006) Expression of the 1-aminocyclopropane-1- carboxylate deaminase gene requires symbiotic nitrogen fixing regulator gene nifA2 in Mesorhizobium loti MAFF303099. Appl Environ Microbiol 72:4964–4969
Oves M, Khan MS, Zaidi A (2013) Chromium reducing and plant growth promoting novel strain Pseudomonas aeruginosa OSG41 enhance chickpea growth in chromium amended soils. Eur J Soil Biol 56:72–83
Pandey P, Maheshwari DK (2007) Two-species microbial consortium for growth promotion of Cajanus cajan. Curr Sci 92:1137–1142
Patten CL, Glick BR (1996) Bacterial biosynthesis of indole-3-acetic acid. Can J Microbiol 42:207–220
Penrose DM, Glick BR (2003) Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol Plant 118:10–15
Penrose DM, Moffatt BA, Glick BR (2001) Determination of 1-aminocyclopropane-1-carboxylic acid (ACC) to assess the effects of ACC deaminase-containing bacteria on roots of canola seedlings. Can J Microbiol 47:77–80
Pierik R, Tholen D, Poorter H, Visser EJW, Voesenek LACJ (2006) The Janus face of ethylene: growth inhibition and stimulation. Trends Plant Sci 1:176–183
Ponmurugan P, Gopi C (2006) In vitro production of growth regulators and phosphatase activity by phosphate solubilizing bacteria. Afr J Biotechnol 5:340–350
Poonguzhali S, Madhaiyan M, Sa T (2008) Isolation and identification of phosphate solubilizing bacteria from chinese cabbage and their effect on growth and phosphorus utilization of plants. J Microbiol Biotechnol 18:773–777
Pradhan N, Shukla LB (2005) Solubilization of inorganic phosphates by fungi isolated from agriculture soil. Afr J Biotechnol 5:850–854
Prayitno J, Rolfe BG, Mathesius U (2006) The ethylene-insensitive sickle mutant of Medicago truncatula shows altered auxin transport regulation during nodulation. Plant Physiol 142:168–180
Qing-xia S-j L, Jing-you X, Zhao-lin J, Xi-jun C, Yun-hui T (2011) Purification and characterization of chitinase produced by Sinorhizobium sp. strain L03. Chin J Biol Control 27:241–245
Rajkumar M, Freitas H (2008) Influence of metal resistant-plant growth-promoting bacteria on the growth of Ricinus communis in soil contaminated with heavy metals. Chemosphere 71:834–842
Rajkumar M, Nagendran R, Kui Jae L, Wang Hyu L, Sung Zoo K (2006) Influence of plant growth promoting bacteria and Cr (vi) on the growth of Indian mustard. Chemosphere 62:741–748
Rajkumar M, Ae N, Prasad MNV, Freitas H (2010) Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol 28:142–149
Rashid S, Charles TC, Glick BR (2012) Isolation and characterization of new plant growth-promoting bacterial endophytes. Appl Soil Ecol 61:217–224
Raza W, Yang W, Shen QR (2008) Paenibacillus polymyxa: antibiotics, hydrolytic enzymes and hazard assessment. J Plant Pathol 90:419–430
Reed MLE, Warner B, Glick BR (2005) Plant growth-promoting bacteria facilitate the growth of the common reed Phragmites australis in the presence of copper or polycyclic aromatic hydrocarbons. Curr Microbiol 51:425–429
Reyes I, Bernier L, Simard R, Antoun H (1999) Effect of nitrogen source on solubilization of different inorganic phosphates by bacterial strain of Penicillium rugulosum and two UV induced mutants. FEMS Microbiol Ecol 28:281–290
Reyes I, Baziramakenga R, Bernier L, Antoun H (2001) Solubilization of phosphate rocks and minerals by a wild-type strain and two UV induced mutants of Penicillium rugulosum. Soil Biol Biochem 33:1741–1747
Richardson AE (1994) Soil microorganisms and phosphorus availability. In: Pankhurst CE, Doube BM, Gupta VVSR, Grace PR (eds) Management of the soil biota in sustainable farming systems. CSIRO Publishing, Melbourne, pp 50–62
Richardson AE (2001) Prospects for using soil microorganisms to improve the acquisition of phosphorus by plants. Aust J Plant Physiol 28:897–906
Rodriguez H, Fraga R, Gonzalez T, Bashan Y (2006) Genetics of phosphate solubilization and its potential applications for improving plant growth-promoting bacteria. Plant Soil 287:15–21
Rudrappa T, Splaine RE, Biedrzycki ML, Bais HP (2008) Cyanogenic pseudomonads influence multitrophic interactions in the rhizosphere. PLoS One 3(4):e2073. doi:10.1371/journal.pone.0002073
Safronova VI, Stepanok VV, Engqvist GL, Alekseyev YV, Belimov AA (2006) Root associated bacteria containing1-aminocyclopropane-1-carboxylate deaminase improve growth and nutrient uptake by pea genotypes cultivated in cadmium supplemented soil. Biol Fertil Soils 42:267–272
Salimpour S, Khavazi K, Nadian H, Besharati H, Miransari M (2010) Enhancing phosphorous availability to canola (Brassica napus L.) using P solubilizing and sulfur oxidizing bacteria. Aust J Crop Sci 4:330–334
Sane D, Aberlenc-Bertossi F, Diatta LI, Gueye B, Daher A (2012) Influence of growth regulators on callogenesis and somatic embryo development in date palm (Phoenix dactylifera L) Sahelian cultivars. Sci World J 2012:837395
Sasek V, Novakova M, Jindrichova B, Boka K, Valentova O, Burketova L (2012) Recognition of avirulence gene AvrLm1 from hemibiotrophic ascomycete Leptosphaeria maculans triggers salicylic acid and ethylene signaling in Brassica napus. Mol Plant Microbe Interact 25:1238–1250
Sattar MA, Gaur AC (1987) Production of auxins and gibberellins by phosphate dissolving microorganisms. Zentralbl Microbiol 142:393–395
Scervino JM, Mesa MP, Mónica ID, Recchi M, Moreno NS, Godeas A (2010a) Soil fungal isolates produce different organic acid patterns involved in phosphate salts solubilization. Biol Fertil Soil 46:755–763
Scervino JM, Mesa MP, Mónica ID, Recchi M, Moreno NS, Godeas A (2010b) Soil fungal isolates produce different organic acid patterns involved in phosphate salts solubilization. Biol Fertil Soils 46:755763
Schadeck RJG, Buchi DF, Leite B (1998a) Ultrastructural aspects of Colletotrichum graminicola conidium germination, appressorium formation and penetration on cellophane membranes: focus on lipid reserves. J Submicro Cytol Pathol 30:555–561
Schadeck RJG, Leite B, Buchi DF (1998b) Lipid mobilization and acid phosphatase activity in lytic compartments during conidium dormancy and appressorium formation of Colletotrichum graminicola. Cell Struct Funct 23:333–340
Schlindwein G, Vargas LK, Lisboa BB, Azambuja AC, Granada CE, Gabiatti NC, Prates F, Stumpf R (2008) Influence of rhizobial inoculation on seedling vigor and germination of lettuce. Cienc Rural 38:658–664
Selvakumar G, Kundu S, Joshi P, Nazim S, Gupta AD, Mishra PK, Gupta HS (2008a) Characterization of a cold-tolerant plant growth-promoting bacterium Pantoea dispersa 1A isolated from a sub-alpine soil in the North Western Indian Himalayas. World J Microbiol Biotechnol 24:955–960
Selvakumar G, Mohan M, Kundu S, Gupta AD, Joshi P, Nazim S, Gupta HS (2008b) Cold tolerance and plant growth promotion potential of Serratia marcescens strain SRM (MTCC 8708) isolated from flowers of summer squash (Cucurbita pepo). Lett Appl Microbiol 46:171–175
Sessitsch A, Coenye T, Sturz AV, Vandamme P, Barka EA, Salles JF, Elsas JDV, Faure D, Reiter B, Glick BR, Wang-Pruski G, Nowak J (2005) Burkholderia phytofirmans sp. nov., a novel plant-associated bacterium with plant-beneficial properties. Int J Syst Evol Microbiol 55:1187–1192
Shahid M, Hameed S, Imran A, Ali S, Elsas JD (2012) Root colonization and growth promotion of sunflower (Helianthus annuus L.) by phosphate solubilizing Enterobacter sp. Fs-11. World J Microbiol Biotechnol 28:2749–2758
Sharma SB, Sayyed RZ, Trivedi MH, Gobi TA (2013) Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. Springerplus 2:587
Sheng XF, Xia JJ (2006) Improvement of rape (Brassica napus) plant growth and cadmium uptake by cadmium-resistant bacteria. Chemosphere 64:1036–1042
Shenoy VV, Kalagudi GM (2005) Enhancing plant phosphorus use efficiency for sustainable cropping. Biotechnol Adv 23:501–513
Shin W, Ryu J, Kim Y, Yang J, Madhaiyan M, Sa T (2006) Phosphate solubilization and growth promotion of maize [Zea mays L.] by the rhizosphere soil fungus Penicillium oxalicum. In: 18th World congress of soil science. 9–15 July, Philadelphia, PA
Siddikee MA, Glick BR, Chauhan PS, Yim WJ, Sa T (2011) Enhancement of growth and salt tolerance of red pepper seedlings (Capsicum annuum L.) by regulating stress ethylene synthesis with halotolerant bacteria containing ACC deaminase activity. Plant Physiol Biochem 49:427–434
Singh MV (2008) Micronutrient deficiencies in crops and soils in India. In: Alloway VJ (ed) Micronutrient deficiencies in global crop production. Springer Science Buisness Media, Berlin, pp 93–125
Singh N, Kumar S, Bajpai VK, Dubey RC, Maheshwari DK, Kang SC (2010) Biological control of Macrophomina phaseolina by chemotactic fluorescent Pseudomonas aeruginosa PN1 and its plant growth promotory activity in chir-pine. Crop Prot 29:1142–1147
Solano RB, García JAL, Garcia-Villaraco A, Algar E, Garcia-Cristobal J, Mañero FJG (2010) Siderophore and chitinase producing isolates from the rhizosphere of Nicotiana glauca Graham enhance growth and induce systemic resistance in Solanum ycopersicum L. Plant Soil 334:189–197
Song OR, Lee SJ, Lee YS, Lee SC, Kim KK, Choi YL (2008) Solubilization of insoluble inorganic phosphate by Burkholderia cepacia DA23 isolated from cultivated soil. Baraz J Microbiol 39:151–156
Souchie EL, Azcon R, Barea JM, Saggin-Júnior OJ, da Silva EMR (2007) Indoleacetic acid production by P-solubilizing microorganisms and interaction with arbuscular mycorrhizal fungi. Acta Sci Biol Sci 29:315–320
Sridevi M, Kumar KG, Mallaiah KV (2008) Production of catechol-type of siderophores by Rhizobium sp Isolate from stem nodules of Sesbania procumbens (Roxb) 3:282–287
Stajković O, Delić D, Jošić D, Kuzmanović D, Rasulić N, Knežević-Vukčević J (2011) Improvement of common bean growth by co-inoculation with Rhizobium and plant growth-promoting bacteria. Rom Biotechnol Lett 16:5919–5926
Stearns JC, Woody OZ, McConkey BJ, Glick BR (2012) Effects of bacterial ACC deaminase on Brassica napus gene expression measured with an Arabidopsis thaliana microarray. Mol Plant Microbe Interact 25:668–676
Sullivan JT, Trzebiatowski JR, Cruickshank RW, Guozy J, Brown SD et al (2002) Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J Bacteriol 184:3086–3095
Sun Y, Cheng Z, Glick BR (2009) The presence of a 1-aminocyclopropane-1-carboxylate (ACC) deaminase deletion mutation alters the physiology of the endophytic plant growth-promoting bacterium Burkholderia phytofirmans PsJN. FEMS Microbiol Lett 296:131–136
Tank N, Saraf M (2003) Phosphate solubilization, exopolysaccharide production and indole acetic acid secretion by rhizobacteria isolated from Trigonella foenum-graecum. Ind J Microbiol 43:37–40
Tarafdar JC, Claassen N (1988) Organic phosphorus compounds as a phosphorus source for higher plants through the activity of phosphatases produced by plant roots and microorganisms. Biol Fertil Soils 5:308–312
Tarafdar JC, Gharu A (2006) Mobilization of organic and poorly soluble phosphates by Chaetomium globosum. Appl Soil Ecol 32:273–283
Tarafdar JC, Rao AV, Bala K (1988) Production of phosphatases by fungi isolated from desert soils. Folia Microbiol 33:453–457
Tate KR (1984) The biological transformation of P in soil. Plant Soil 76:245–256
Taurian T, Anzuay MS, Angelini JG, Tonelli ML, Ludueña L, Pena D, Ibáñez F, Fabra A (2010) Phosphate-solubilizing peanut associated bacteria: screening for plant growth-promoting activities. Plant Soil 329:421–431
Timmusk S, Nicander B, Granhall U, Tillberg E (1999) Cytokinin production by Paenibacillus polymyxa. Soil Biol Biochem 31:1847–1852
Tittabutr P, Awaya JD, Li QX, Borthakur D (2008) The cloned 1-aminocyclopropane-1- carboxylate (ACC) deaminase gene from Sinorhizobium sp. strain BL3 in Rhizobium sp. strain TAL1145 promotes nodulation and growth of Leucaena leucocephala. Syst Appl Microbiol 31:141–150
To-O K, Kamasaka H, Kusaka K, Kuriki T, Kometani K, Okada S (1997) A novel acid phosphatase from Aspergillus niger KU-8 that specifically hydrolyzes C-6 phosphate groups of phosphoryl oligosaccharides. Biosci Biotechnol Biochem 61:1512–1517
To-O K, Kamasaka H, Kuriki T, Okada S (2000) Substrate selectivity in Aspergillus niger KU-8 acid phosphatase II using phosphoryl oligosaccharides. Biosci Biotechnol Biochem 64:1534–1537
Toro M (2007) Phosphate solubilizing microorganisms in the rhizosphere of native plants from tropical savannas: an adaptive strategy to acid soils? In: Velazquez C, Rodriguez-Barrueco E (eds) Developments in plant and soil sciences. Springer, The Netherlands, pp 249–252
Tripathi M, Munot HP, Shouche Y, Meyer JM, Goel R (2005) Isolation and functional characterization of siderophore producing lead and cadmium resistant Pseudomonas putida KNP9. Curr Microbiol 50:233–237
Trivedi P, Sa T (2008) Pseudomonas corrugata (NRRL B-30409) mutants increased phosphate solubilization, organic acid production, and plant growth at lower temperatures. Curr Microbiol 56:140–144
Tsavkelova EA, Klimova SY, Cherdyntseva TA, Netrusov AI (2006) Microbial producers of plant growth stimulators and their practical use: a review. Appl Biochem Microbiol 42:117–126
Uchiumi T, Oowada T, Itakura M, Mitsui H, Nukui N et al (2004) Expression islands clustered on symbiosis island of Mesorhizobium loti genome. J Bacteriol 186:2439–2448
Vargas WA, Mandawe JC, Kenerley CM (2009) Plant-derived sucrose is a key element in the symbiotic association between Trichoderma virens and maize plants. Plant Physiol 151:792–808
Vassilev N, Vassileva M, Nikolaeva I (2006) Simultaneous P-solubilizing and biocontrol activity of microorganisms: potentials and future trends. Appl Microbiol Biotechnol 71:137–144
Vassilev N, Vassileva M, Bravo V, Fernández-Serrano M, Nikolaeva I (2007) Simultaneous phytase production and rock phosphate solubilization by Aspergillus niger grown on dry olive wastes. Ind Crops Prod 26:332–336
Vassileva M, Serrano M, Bravo V, Jurado E, Nikolaeva I, Martos V, Vassilev N (2010) Multifunctional properties of phosphate-solubilizing microorganisms grown on agro-industrial wastes in fermentation and soil conditions. Appl Microbiol Biotechnol 85:1287–1299
Vazquez P, Holguin G, Puente ME, Lopez-Cortes A, Bashan Y (2000) Phosphate-solubilizing microorganisms associated with the rhizosphere of mangroves in a semiarid coastal lagoon. Biol Fertil Soils 30:460–468
Verma DPS, Long S (1983) The molecular biology of rhizobium–legume symbiosis. Int Rev Cytol Suppl 14:211–245
Vining LC (1990) Functions of secondary metabolites. Annu Rev Microbiol 44:395–427
Viruel E, Lucca ME, Siñeriz F (2011) Plant growth promotion traits of phosphobacteria isolated from Puna, Argentina. Arch Microbiol 193:489–496
Vivas A, Biro B, Ruiz-Lozano JM, Barea JM, Azcon R (2006) Two bacterial strains isolated from a Zn-polluted soil enhance plant growth and mycorrhizal efficiency under Zn toxicity. Chemosphere 52:1523–1533
Voisard C, Keel C, Haas D, Defago G (1989) Cyanide production by Pseudomonas fluorescens helps suppress black root of tobacco under gnotobiotic conditions. EMBO J 8:351–358
Vyas P, Gulati A (2009) Organic acid production in vitro and plant growth promotion in maize under controlled environment by phosphate-solubilizing fluorescent Pseudomonas. BMC Microbiol 9:174
Wang C, Knill E, Glick BR, Defago G (2000) Effect of transferring 1-aminocyclopropane-lcarboxylic acid (ACC) deaminase genes into Pseudomonas fluorescens strain CHAO and its gac A derivative CHA96 on their growth promoting and disease-suppressive capacities. Can J Microbiol 46:898–907
Wang X, Wang Y, Tian J, Lim BL, Yan X, Liao H (2009) Overexpressing AtPAP15 enhances phosphorus efficiency in soybean. Plant Physiol 151:233–240
Wani PA, Khan MS, Zaidi A (2007a) Synergistic effects of the inoculation with nitrogen fixing and phosphate-solubilizing rhizobacteria on the performance of field grown chickpea. J Plant Nutr Soil Sci 170:283–287
Wani PA, Khan MS, Zaidi A (2007b) Chromium reduction, plant growth-promoting potentials, and metal solubilizatrion by Bacillus sp. isolated from alluvial soil. Curr Microbiol 54:237–243
Wani PA, Khan MS, Zaidi A (2007c) Effect of metal tolerant plant growth promoting Bradyrhizobium sp. (vigna) on growth, symbiosis, seed yield and metal uptake by greengram plants. Chemosphere 70:36–45
Wani PA, Khan MS, Zaidi A (2007d) Co-inoculation of nitrogen-fixing and phosphate-solubilizing bacteria to promote growth, yield and nutrient uptake in chickpea. Acta Agron Hung 55:315–323
Wani PA, Khan MS, Zaidi A (2008a) Effect of heavy metal toxicity on growth, symbiosis, seed yield and metal uptake in pea grown in metal amended soil. Bull Environ Contam Toxicol 81:152–158
Wani PA, Khan MS, Zaidi A (2008b) Effect of metal-tolerant plant growth-promoting Rhizobium on the performance of pea grown in metal-amended soil. Arch Environ Contam Toxicol 55:33–42
Yadav RS, Tarafdar JC (2003) Phytase and phosphatase producing fungi in arid and semi-arid soils and their efficiency in hydrolyzing different organic P compounds. Soil Biol Biochem 35:1–7
Yadav BK, Tarafdar JC (2007) Availability of unavailable phosphate compounds as a phosphorus source for clusterbean (Cyamopsis tetragonoloba (L.) Taub.) through the activity of phosphatase and phytase produced by actinomycetes. J Arid Legum 4:110–116
Yadav BK, Tarafdar JC (2011) Penicillium Purpurogenum, unique P mobilizers in arid agro-ecosystems. Arid Land Res Manag 25:87–99
Yadav BK, Verma A (2012). Phosphate solubilization and mobilization in soil through soil microorganisms under arid ecosystems, the functioning of ecosystems. In: Ali M (ed) In Tech. ISBN: 978-953-51-0573-2, Available from http://www.intechopen.com/books/the-functioning-ofecosystems/phosphate-solubilization-and-mobilization-in-soil-through-microorganisms-under-arid-ecosystems
Yadav S, Kaushik R, Saxena AK, Arora DK (2011) Diversity and phylogeny of plant growth-promoting bacilli from moderately acidic soil. J Basic Microbiol 51:98–106
Yang J, Huang X, Tian B, Wang M, Niu Q, Zhang K (2005) Isolation and characterization of a serine protease from the nematophagous fungus, Lecanicillium psalliotae, displaying nematicidal activity. Biotechnol Lett 27:1123–1128
Yang J, Kloepper JW, Ryu CM (2009) Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci 14:1–4
Yi Y, Huang W, Ge Y (2008) Exo-polysaccharide: a novel important factor in the microbial dissolution of tricalcium phosphate. World J Microbiol Biotechnol 24:1059–1065
Zahir ZA, Munir A, Asghar HN, Shaharoona B, Arshad M (2008) Effectiveness of rhizobacteria containing ACC deaminase for growth promotion of peas (Pisum sativum) under drought conditions. J Microbiol Biotechnol 18:958–963
Zaidi A, Khan MS (2006) Co-inoculation effects of phosphate solubilizing microorganisms and Glomus fasciculatum on green gram-Bradyrhizobium symbiosis. Turk J Agric For 30:223–230
Zdor RE, Anderson J (1992) Influence of root colonizing bacteria on the defense responses of bean. Plant Soil 140:99–107
Zhang JJ, Liu TY, Chen WF, Wang ET, Sui XH (2012) Mesorhizobium muleiense sp. nov., nodulating with Cicer arietinum L. Int J Syst Evol Microbiol 62:2737–2742
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Khan, M.S., Zaidi, A., Ahmad, E. (2014). Mechanism of Phosphate Solubilization and Physiological Functions of Phosphate-Solubilizing Microorganisms. In: Khan, M., Zaidi, A., Musarrat, J. (eds) Phosphate Solubilizing Microorganisms. Springer, Cham. https://doi.org/10.1007/978-3-319-08216-5_2
Download citation
DOI: https://doi.org/10.1007/978-3-319-08216-5_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-08215-8
Online ISBN: 978-3-319-08216-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)