Abstract
We present a new deep learning method, FML, that automatically computes linear measurements in a fetal brain MRI volume. The method is based on landmark detection and estimates their location reliability. It consists of four steps: 1) fetal brain region of interest detection with a two-stage anisotropic U-Net; 2) reference slice selection with a convolutional neural network (CNN); 3) linear measurement computation based on landmarks detection using a novel CNN, FMLNet; 4) measurement reliability estimation using a Gaussian Mixture Model. The advantages of our method are that it does not rely on heuristics to identify the landmarks, that it does not require fetal brain structures segmentation, and that it is robust since it incorporates reliability estimation. We demonstrate our method on three key fetal biometric measurements from fetal brain MRI volumes: Cerebral Biparietal Diameter (CBD), Bone Biparietal Diameter (BBD), and Trans Cerebellum Diameter (TCD). Experimental results on training \((N=164)\) and test \((N=46)\) datasets of fetal MRI volumes yield a 95% confidence interval agreement of 3.70 mm, 2.20 mm and 2.40 mm for CBD, BBD and TCD, in comparison to measurements performed by an expert fetal radiologist. All results were below the interobserver variability, and surpass previously published results. Our method is generic, as it can be directly applied to other linear measurements in volumetric scans and can be used in a clinical setup.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Magnetic resonance imaging (MRI) is increasingly used to assess fetal brain development. Clinical assessment of fetal brain development based on MRI is mainly subjective and is complemented with a few biometric linear measurements [17]. Three key biometric linear measurements currently performed on fetal brain MRI are the Cerebral Biparietal Diameter (CBD), the Bone Biparietal Diameter (BBD), and the Trans Cerebellum Diameter (TCD) [18]. These measurements are used to assess fetal development according to the gestational age. They are manually acquired on individual MR reference slices by a fetal radiologist following guidelines that indicate how to establish the scanning imaging plane, how to select the reference slice in the MR volume for each measurement, and how to identify the two endpoint landmarks of the linear measurement [9].
Various methods have been developed for computing biometric linear measurements in 2D ultrasound images, e.g., the biparietal diameter [13], the fetal head circumference [12], and the fetus femur length [13]. Recently, Avisdris et al. [3] describe an automatic method for computing fetal brain linear measurements in MRI scans. The method mimics the radiologist manual annotation workflow, relies on a fetal brain segmentation and is based on measurement specific geometric heuristics for identifying the anatomical landmarks of each linear measurement. While it yields acceptable measurements, its reliance on accurate fetal brain segmentation and ad hoc heuristics may not always be robust.
Methods for the automatic computation of linear measurements of a structure in volumetric scans have been proposed in the past. For example, Yan et al. [21] describe a deep learning method for the computation of the length and width of a lesion following the RECIST guidelines. The method uses the Mask-RCNN network [10] to detect and segment each lesion from which the linear measurements are computed. The training segmentation masks are obtained from the ground truth measurements by fitting an ellipse bounded by the long and short axes measurement endpoints. This method is specific to lesions and RECIST measurements and is not applicable to fetal brain measurements.
Automatic landmark detection in images is a common task in a variety of computer vision applications, e.g., face alignment [20], pose estimation and in medical image analysis [16, 22]. Two popular CNN-based methods consist of computing the spatial coordinates of each landmark by direct regression [22] or by heat map regression [16, 20]. In the latter, the network computes a heat map defined by a Gaussian function centered at the landmark coordinates whose co-variance describes the landmark location uncertainty. HRNet [20], a heat map regression network, achieves state of the art results in face landmark detection, human pose estimation, object classification and semantic segmentation.
Uncertainty estimation is an essential aspect of a variety of related tasks, e.g., classification, regression and segmentation with deep neural networks in computer vision [8] and medical image analysis [4]. Wang et al. [19] describes a test time augmentation (TTA) based uncertainty estimation method. TTA consists of generating similar new cases, computing the voxel predictions for each, and then obtaining the final voxel prediction on the original image by taking the mean or median value or voxel uncertainty by computing the entropy. This approach is not directly applicable to landmark detection. Payer et al. [15] describes a Gaussian-based uncertainty estimation method for landmark localization in hand X-ray images and in lateral cephalograms datasets. The method fits a Gaussian for each landmark from its predicted heat map. Their results show that the predicted uncertainties correlate with the landmark spatial error and the interobserver variability.
2 Method
We present a new deep learning method, called FML (Fetal Measurement by Landmarks), to automatically compute landmark-based linear measurements in a fetal brain MRI volume and to estimate their reliability. We demonstrate FML on 3 key fetal biometric measurements: CBD, BBD, TCD.
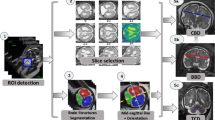
The method consists of four steps: 1) fetal brain region of interest (ROI) detection with a two-stage anisotropic U-Net; 2) reference slice selection with a 2D CNN; 3) linear measurement computation based on landmarks detection using a novel CNN, FMLNet; 4) measurement reliability estimation with a Bayesian score using a Gaussian Mixture Model (GMM).
2.1 Fetal Brain ROI Detection
The first step computes the fetal brain ROI in the fetal MRI volume with the method described in [6]. The ROI is a 3D axis-aligned bounding box that contains the fetal brain. The method uses a custom anisotropic 3D U-Net network trained with a Dice loss on a \(\times \)4 downsized fetal MRI volume. It outputs a coarse fetal brain segmentation from which a tight fitting ROI is computed.
2.2 Reference Slice Selection
The second step computes for each measurement, the reference slice of the fetal MRI volume on which the linear measurements will be performed from input fetal MRI volume with the method described in [3]. The method uses a slice-based 2D CNN that predicts for each slice in the ROI its probability to be a reference slice. It then selects the one with the highest probability. One CNN is trained for each measurement reference slice to detect the slice that was manually selected by the radiologist.
2.3 Linear Measurement Computation
The third step computes for each measurement the two anatomical landmarks of the measurement endpoints on the selected reference slice. The landmarks are computed using FMLNet, a variant of the HRNet network [20].
HRNet is a CNN type network whose key characteristic is that it maintains a high resolution representation of the input throughout network layers. This is achieved by connecting the high-to-low resolution convolution streams in parallel and by repeatedly spreading the intermediate results of each layer across the other layers at different resolutions.
We describe next the FMLNet architecture and its training and inference pipelines.
Diagram of the FMLNet architecture. FMLNet is CNN network that consists of four streams (rows) at subsequently lower resolutions: full, 1/2, 1/4, 1/8 (each in a different color). Each stream consists four convolutional blocks (dotted boxes); in each block, boxes represent feature maps and arrows correspond to layers. After each block, the feature maps are combined across streams (red and green arrows). At the end of the blocks (two upper right 4-box clusters), the features maps of all four resolutions are concatenated and combined (pink box). The outputs are the two landmark Gaussian heat maps, one for each measurement endpoint (top rightmost box with red ovals). (Color figure online)
FMLNet Architecture: Figure 1 shows the architecture of FMLNet. It is a CNN that combines the representations from four high-to-low resolution parallel streams into a single stream. The representations are then input to a two-layer convolution classifier. The first layer combines the feature maps of all four resolutions; the second layer computes a Gaussian heat map for each of the two landmark endpoint. One network is trained for each measurement with the Mean Squared Error (MSE) loss between the Gaussian maps created from the ground truth measurement landmarks and the predicted heat maps. At inference time, the two measurement landmark locations are defined by the coordinates of the pixel with the maximal value on each heat map.
FMLNet Training: Three FMLNet networks are trained, one for each of the linear measurements, CBD, BBD and TCD. The input is the reference slice image; the outputs are the two measurement endpoint locations on the image.
Two training time augmentations are used: 1) rotations around the image center at angles randomly sampled in the \([-180, 180]^o\) range; 2) image scaling at scales randomly sampled in the \([-5, +5]\%\) range. In addition, landmark class (left/right) reassignment (LCR) is performed on the resulting landmarks.
Landmark class reassignment is necessary because rotations may cause the left/right labeling of the two measurement landmarks to be inconsistent with the image coordinates, i.e. the left and right points may be switched, which will hamper the network training. This inconsistency is corrected by performing landmark class reassignment (Fig. 2). During each training epoch, the left/right assignment for each rotated image is verified with respect to the image coordinate system and if needed, is corrected by switching the left/right labels.
Note that these augmentations are different than the ones used in the original HRNet. Unlike faces, which are almost always vertical, the fetal brain can be in any orientation, so the full range of rotations (beyond flipping) should be accounted for. The network is trained for 200 epochs on a batch size of 16 images with the ADAM optimizer [14] with an initial learning rate of \(10^{-4}\) and a dropping factor of 0.2 in epochs 10, 40, 90, and 150.
FMLNet inference pipeline. The input is the reference slice of the measurement; the outputs are the measurement endpoints and the measurement value (blue line). The pipeline consists of: 1) test time augmentation of the reference slice image (TTA, illustrated with three augmentations); 2) landmarks prediction with FMLNet, and; 3) robust landmarks fusion. The dots on the images corresponds to the left (blue), right (green), unassigned (yellow) and outlier (red cross) landmark predictions. (Color figure online)
FMLNet Inference: the inference pipeline consists of three steps (Fig. 3): 1) test time augmentation (TTA) of the reference slice image; 2) landmarks location prediction with FMLNet; and 3) robust landmarks fusion (RLF).
-
1. Test time augmentation: new reference slice images are generated with a set of reversible spatial transformations \(T = \{t_i\}\) applied to the original image I. Rotation transformations are applied at a equally spaced angles in the \([0,360]^o\) range (in our case, \(a = 12\)). The result is a set of transformed images \(I' = \{I_i' = {t_i(I)}\}\).
-
2. Landmarks prediction with FMLNet: two measurement landmarks, \(L'_i = \{l'^{(k)}_i | k \in K \}\), \(K=\{left, right \}\) are computed for each image \(I_i'\) with the trained FMLNet. The resulting landmark predictions, \(L'_i\), are then mapped back to their original location on the reference slice image by applying the reverse spatial transformation \(t_i^{-1}\) to the corresponding image \(I'_i\). The result is a set of landmarks \(L = \{l^{(k)}_i | i \in [1..a], k \in K\}\) in the original image coordinates.
-
3. Robust landmarks fusion: the final landmark predictions, \(l^{(right)}, l^{(left)}\), are computed from the landmarks prediction set L with the Density Based Spatial Clustering of Applications with Noise (DBSCAN) [7] algorithm. First, DBSCAN clusters together a minimum of q points that are within a pre-defined distance of d between them (in our case, \(q=4\) and \(d=2\) pixels). Next, outlier points outside clusters are discarded, and two point clusters corresponding to the left and right measurement landmark endpoints are computed with the K-means algorithm [2]. Finally, the left and right landmark coordinates are obtained by computing the centroid of the points in each cluster.
2.4 Measurement Reliability Estimation
The fourth step estimates the reliability of the landmarks predictions using the set of landmark predictions L generated in the landmark prediction step of the FMLNet inference. It models the landmarks location distribution, irrespective of their landmark class (left/right), with a bi-modal Gaussian Mixture Model (GMM). The GMM Bayesian likelihood is computed to obtain an estimate of the prediction reliability. When likelihood value is low, the landmark locations are spatially dispersed, so their distribution is not bi-modal (two clusters). In this case, the measurement is labeled as unreliable and should be performed manually by an expert radiologist.
Formally, the GMM is defined as \( X \sim \sum _{k \in K}{\pi _k N(\mu _k , \varSigma _k)}\), where for each of the two landmark clusters k (left/right), \(\pi _{k}\) is the cluster probability, \(N(\mu _{k},\varSigma _{k})\) is a multivariate Gaussian distribution, \(\mu _{k} \in \mathbb {R}^{2}\) is the cluster mean location and \(\varSigma _{k} \in \mathbb {R}^{2 \times 2}\) is the cluster location covariance.
We estimate the GMM parameters, \(\varPhi = (\pi _{k}, \mu _{k}, \varSigma _{k})\), from the set of landmark predictions L by Expectation-Maximization (EM) [5]. The mean values of each cluster location are initialized with the final landmark locations, e.g., \(\mu _k = l^{(k)}\), and are as described in the robust landmark fusion inference step. To estimate the reliability of the predicted landmark points in L, we compute the GMM log-likelihood score:
where l is a point in L (the labels left/right are ignored), and \(N(l |\mu _{k},\varSigma _{k})\) is the Gaussian probability of cluster k for l.
3 Experimental Results
To evaluate our method, we conducted three studies on fetal MRI dataset.
Dataset and Annotations: The dataset consists of fetal brain MRI volumes acquired with the FRFSE protocol at the Sourasky Medical Center (Tel Aviv, Israel) as part of routine fetal assessment. The dataset includes 210 MRI volumes of 154 singleton pregnancies (cases) with mean gestational age of 32 weeks (std = 2.8, range 22–38). Of these, 113 volumes (87 cases) were diagnosed as normal, and 107 volumes (67 cases) as abnormal. To allow direct comparison, we use the same train/test splits of 164/46 volumes (121/33 disjoint cases) as in [3]. CBD, BBD and TCD measurements for all volumes were manually performed by a senior pediatric neuro-radiologist.
Studies: We conducted three studies. Study 1 evaluates the accuracy of the FMLNet method and the contribution of its various components. Study 2 analyzes the impact of selected reference slices. Study 3 evaluates the measurement reliability estimation.
In all studies, we use the following metrics: \(L_1\) difference, bias and agreement. For two sets of n measurements, \(M_1=\{m_i^1\}\), \(M_2=\{m_i^2\}\), \(m_i^1\) and \(m_i^2\) (\( 1 \le i \le n\)) are two values of the measurement, e.g., ground-truth and computed. The difference between two linear measurement sets \(M_1, M_2\) is defined as \(L_{1}(M_1, M_2) = 1/n\sum _{i=1}^{n}{|d_i|}\), where the difference between each measurement is \(d_i = m_i^1 - m_i^2\). For repeatability estimation, we use the Bland-Altman method [1] to estimate the bias and agreement between two observers. Agreement is defined by the 95\(\%\) confidence interval \(CI_{95}(M_1, M_2) = 1.96 \times \sqrt{1/n\sum _{i=1}^{n}{(L_{1}(M_1, M_2) - d_i)^2}}\). The measurements bias is defined as \(Bias(M_1,M_2) = 1/n\sum _{i=1}^{n}{d_i}\). These three metrics represent different aspects of algorithm performance.
Study 1: Accuracy Analysis and Ablation Study. We evaluate the accuracy of the FMLNet method and the contribution of its three main components: test time augmentation (TTA), robust landmark fusion (RLF) and landmark class reassignment (LCR). We compare its performance to that of HRNet, to the geometric method [3] and to the interobserver variability on the test dataset and its ground-truth values.
Table 1 shows the results. The original HRNet (row 2) performs poorly, as it yields high measurement agreement \(CI_{95}\) and difference \(L_1\) values. Removing each one of algorithmic components from FMLNet - TTA (row 3), RLF (row 4) or LCR (row 5), yields better results than standalone HRNet, but still not acceptable. FMLNet with all its components (row 6) yields the best results, which are always reliable (46 out of 46 for CBD, BBD, TCD). Using a two-sided t-test, no statistically significant difference was found in the performance between Normal (row 7) and Abnormal cases (row 8) in all measurements (\(p > 0.05\)).
Comparison of our method results with those of the previously published geometric method [3] with reliability estimation (row 10) shows that for all three measurements, FMLNet (row 6) performs better in terms of reliable measurements on the number of MR volumes N, agreement \(CI_{95}\) and difference \(L_1\). Specifically, for TCD, our method yields reliable measurements in all cases (\(N=46\)) with comparable agreement \(CI_{95}\), while the geometric method fails in 20% of the cases (9 out of 46). Using a two-sided t-test, in BBD and TCD our method performs significantly better (\(p < 0.01\)). These results shows superiority of our new method.
Study 2: Impact of Selected Reference Slices. Differences were detected between the reference slices selected by the radiologist and the slice selected by the algorithm on the test set of: 20/46 cases in CBD/BBD slices and 12/46 cases in TCD slices. Table 1 shows the measurements accuracy results. When analyzing the impact of selected reference slices on the accuracy of measurements (FMLNet - uses the slices selected by the algorithm vs FMLNet+ uses the slices selected by radiologist) - the main impact was on CBD measurements (almost doubles the variability), while the TCD and BBD accuracy remains similar. A possible explanation is that the TCD and BBD are measured on smooth contours (cerebellum and skull, respectively) while CBD is measured on a more complex contour, the brain sulcation and gyri. Furthermore, when normalizing the interobserver variability in the absolute value of measurements, the relative error variability results in 6\(\%\) for CBD and 4\(\%\) for BBD and TCD. For the geometric algorithm (Geometric vs Geometric+), no significant differences were observed when working on slices selected by a radiologist. These results show that improving the reference slice selection algorithm may improve the accuracy of the automatic measurements.
Study 3: Measurement Reliability Estimation. Table 1 shows the number of cases (column N) for each measurement for which the computed values are reliable.
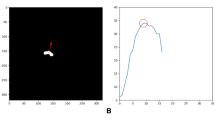
We evaluate the reliability estimation by computing the difference between the measurements computed and ground truth values, \(L_1\), and by establishing their correlation with the LLscore, on the test set cases. Figure 4 illustrates this correlation for the CBD measurement. For all three measurements, we observe that LLscore correlates well with \(L_1\). We establish from the plot graphs the threshold value of LLscore that adequately discriminates between reliable and unreliable computed measurements – set to \(-5.75\) for all three measurements.
CBD Measurement reliability estimation results. The plot graph (center) shows the log-likelihood score LLscore (horizontal axis) with respect to the measurement difference \(L_1\)(vertical axis) for the test set cases (blue dots). Representative images of unreliable (left) and reliable (right) measurements: (1) failure of brain ROI detection; (2) blurry image; (3) reliable predictions with outliers; and (4) reliable predictions. (Color figure online)
4 Conclusion
We have presented a new fully automatic method to compute landmark-based linear measurements in a fetal brain MRI volume, to estimate their reliability, and to identify the unreliable measurements that should be obtained manually for the case at hand. The computed reliability estimation value can be used to rank the measurement predictions for inspection and validation by a radiologist.
We demonstrate our method on three key fetal biometric measurements from fetal brain MRI scans, CBD, BBD, TCD, and show that it yields state-of-the-art results for all three measurements, within the interobserver variability. These results also comparable to the best reported results for fetal head circumference (HC) measurements in ultrasound images of the HC18 challenge [11] (mean \(L_1\) of 1.72 mm, \(CI_{95}\) of 3.16 mm).
The main novelties of our method are three-fold. First, it robustly handles the wide span of fetal brain orientations and correctly identifies left and right measurement landmark endpoints by test time augmentation, robust landmark fusion, and landmark class reassignment. Second, it directly computes landmarks by heat map regression, obviating the need for structure segmentation and its data annotation effort [3, 21]. Third, it computes landmarks location uncertainty estimation with a new method that combines test time augmentation [19] and landmark Gaussian-based uncertainty estimation [15] and that simultaneously computes the estimates on multiple landmarks with a GMM instead of using a single Gaussian for each landmark, thereby yielding a single reliability score.
The advantages of our method are that it only requires a small number (\(\sim \)150) of manual linear measurements for the training dataset, that it does not rely on heuristics to identify the landmarks, that it does not require fetal brain structures segmentation, and that it is robust since it incorporates reliability estimation. Note that our method is generic, i.e., it is not tailored to specific linear measurements, so it can be applied directly to other measurements.
References
Altman, D.G., Bland, J.M.: Measurement in medicine: the analysis of method comparison studies. J. R. Stat. Soc. Ser. D (Stat.) 32(3), 307–317 (1983)
Arthur, D., Vassilvitskii, S.: K-means++: the advantages of careful seeding. In: Proceedings of ACM-SIAM Symposium on Discrete Algorithms (2007)
Avisdris, N., et al.: Automatic linear measurements of the fetal brain with deep neural networks. Int. J. Comput. Assist. Radiol. Surg. (2021). https://doi.org/10.1007/s11548-021-02436-8
Ayhan, M.S., Berens, P.: Test-time data augmentation for estimation of heteroscedastic aleatoric uncertainty in deep neural networks. In: International Conference on Medical Imaging with Deep Learning (2018)
Dempster, A.P., Laird, N.M., Rubin, D.B.: Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. Ser. B 39(1), 1–22 (1977)
Dudovitch, G., Link-Sourani, D., Sira, L.B., Miller, E., Bashat, D.B., Joskowicz, L.: Deep learning automatic fetal structures segmentation in MRI scans with few annotated datasets. In: Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (2020)
Ester, M., Kriegel, H.P., Sander, J., Xu, X.: A density-based algorithm for discovering clusters in large spatial databases with noise. In: Proceedings of the International Conference on Knowledge Discovery and Data Mining (1996)
Gal, Y., Ghahramani, Z.: Dropout as a Bayesian approximation: representing model uncertainty in deep learning. In: Proceedings of International Conference on Machine Learning (2016)
Garel, C.: MRI of the Fetal Brain. Springer, Heidelberg (2004). https://doi.org/10.1007/978-3-642-18747-6
He, K., Gkioxari, G., Dollár, P., Girshick, R.: Mask R-CNN. In: Proceedings of International Conference on Computer Vision (2017)
van den Heuvel, T.L.: HC18 challange leaderboard (2021). https://hc18.grand-challenge.org/evaluation/challenge/leaderboard/. Accessed 29 June 2021
van den Heuvel, T.L., de Bruijn, D., de Korte, C.L., van Ginneken, B.: Automated measurement of fetal head circumference using 2d ultrasound images. PloS ONE 13(8), e0200412 (2018)
Khan, N.H., Tegnander, E., Dreier, J.M., Eik-Nes, S., Torp, H., Kiss, G.: Automatic detection and measurement of fetal biparietal diameter and femur length-feasibility on a portable ultrasound device. Open J. Obstetr. Gynecol. 7(3), 334–350 (2017)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. In: Proceedings of International Conference Learning Representations (2015)
Payer, C., Urschler, M., Bischof, H., Štern, D.: Uncertainty estimation in landmark localization based on Gaussian heatmaps. In: Sudre, C.H., et al. (eds.) UNSURE/GRAIL -2020. LNCS, vol. 12443, pp. 42–51. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-60365-6_5
Payer, C., štern, D., Bischof, H., Urschler, M.: Integrating spatial configuration into heatmap regression based CNNs for landmark localization. Med. Image Anal. 54, 207–219 (2019)
Prayer, D., et al.: ISUOG practice guidelines: performance of fetal magnetic resonance imaging. Ultrasound Obstetr. Gynecol. 49(5), 671–680 (2017)
Salomon, L., et al.: ISUOG practice guidelines: ultrasound assessment of fetal biometry and growth. Ultrasound Obstetr. Gynecol. 53(6), 715–723 (2019)
Wang, G., Li, W., Aertsen, M., Deprest, J., Ourselin, S., Vercauteren, T.: Aleatoric uncertainty estimation with test-time augmentation for medical image segmentation with convolutional neural networks. Neurocomputing 338, 34–45 (2019)
Wang, J., et al.: Deep high-resolution representation learning for visual recognition. IEEE Trans. Pattern Anal. Mach. Intell. 43, 3349–3364 (2019)
Yan, K., et al.: MULAN: multitask universal lesion analysis network for joint lesion detection, tagging, and segmentation. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11769, pp. 194–202. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32226-7_22
Zhang, J., Liu, M., Shen, D.: Detecting anatomical landmarks from limited medical imaging data using two-stage task-oriented deep neural networks. IEEE Trans. Pattern Anal. Mach. Intell. 26(10), 4753–4764 (2017)
Acknowledgments
This research was supported in part by Kamin Grants 72061 and 72126 from the Israel Innovation Authority.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Avisdris, N., Ben Bashat, D., Ben-Sira, L., Joskowicz, L. (2021). Fetal Brain MRI Measurements Using a Deep Learning Landmark Network with Reliability Estimation. In: Sudre, C.H., et al. Uncertainty for Safe Utilization of Machine Learning in Medical Imaging, and Perinatal Imaging, Placental and Preterm Image Analysis. UNSURE PIPPI 2021 2021. Lecture Notes in Computer Science(), vol 12959. Springer, Cham. https://doi.org/10.1007/978-3-030-87735-4_20
Download citation
DOI: https://doi.org/10.1007/978-3-030-87735-4_20
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-87734-7
Online ISBN: 978-3-030-87735-4
eBook Packages: Computer ScienceComputer Science (R0)