Abstract
Advancements in musculoskeletal analysis have been achieved by adopting deep learning technology in image recognition and analysis. Unlike musculoskeletal modeling based on computational anatomy, deep learning-based methods can obtain muscle information automatically. Through analysis of image features, both approaches can obtain muscle characteristics such as shape, volume, and area, and derive additional information by analyzing other image textures. In this chapter, we first discuss the necessity of musculoskeletal analysis and the required image processing technology. Then, the limitations of skeletal muscle recognition based on conventional handcrafted features are discussed, and developments in skeletal muscle recognition using machine learning and deep learning technology are described. Next, a technique for analyzing musculoskeletal systems using whole-body computed tomography (CT) images is shown. This study aims to achieve automatic recognition of skeletal muscles throughout the body and automatic classification of atrophic muscular disease using only image features, to demonstrate an application of whole-body musculoskeletal analysis driven by deep learning. Finally, we discuss future development of musculoskeletal analysis that effectively combines deep learning with handcrafted feature-based modeling techniques.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Skeletal muscle
- Musculoskeletal analysis
- Musculoskeletal segmentation
- Surface muscle
- Deep muscle
- Random forest
- FCN-8s
- 2D U-Net
- 3D U-Net
Importance of Musculoskeletal Analysis and Skeletal Muscle Analysis
Musculoskeletal analysis is important in various situations. Muscles are divided into skeletal muscle, myocardium, and smooth muscle. In particular, the muscles that make up the heart are called the myocardium, while the muscles that make up the visceral organs are called the smooth muscles, and the smooth muscles have no muscle ganglion. Because skeletal muscle, which is the focus of this chapter, adheres to bone and is directly involved in exercise, it is a fundamental focal point in musculoskeletal analysis.
In the field of orthopedics, analysis of bone itself is also important, and there are various bone analysis methods ranging from model-based segmentation to segmentation by deep learning [1,2,3]. We previously described the importance of skeletal muscle segmentation for orthopedic intervention and proposed model-based skeletal muscle recognition [4]. However, compared to the number of approaches that use deep learning for analysis of bone regions, there are far fewer approaches that use deep learning for skeletal muscle segmentation. Therefore, this chapter proposes a method of muscle recognition and analysis using deep learning.
Diseases related to skeletal muscle include myopathy, a myogenic disease, which is distinguished from neuropathy, a neurogenic disease; both diseases affect muscle function. Although most symptoms of myopathy involve atrophy, differentiating them from atrophy that occurs normally with aging is an important and difficult problem. Additionally, amyotrophic lateral sclerosis (ALS) and other similar afflictions focused on in this chapter are atrophic muscular diseases in which differential diagnoses do not exist; moreover, progress inhibitors have recently been approved by the U.S. Food and Drug Administration (FDA) [5], and treatment plans must appropriately differentiate muscular diseases with treatable atrophy. The measurement of skeletal muscle itself has become an important problem, because such measurements facilitate treatment of muscular diseases with atrophy; in turn, these treatments help patients live longer, healthier lives. Therefore, the measurement of skeletal muscle was addressed in a field study on the health and lifestyle habits of the elderly in Japan, which aimed to identify effective measures for improving their overall health [6].
Although the measurement of skeletal muscle quantity is required in musculoskeletal analysis as described above, accurate measurement of skeletal muscle quantity and automatic measurement of the muscle at each position are achievable for only a limited number of muscles, representing an unsolved problem in whole-body muscle analysis. Because skeletal muscle exists throughout the body, it is depicted in medical images obtained through various modalities. In tomographic CT images and MRIs, it will be difficult to search cross sections in which skeletal muscle is not depicted. However, CT images and tomographic MRIs that depict skeletal muscle photographed for the purpose of observing lesions can be utilized in support of automatic muscle analysis. In addition, because skeletal muscle is depicted in various cross sections of tomographic images, it is useful in automatic recognition of adjacent skeletal muscle as preprocessing for organ and lesion detection systems in computer-aided diagnosis (CAD) systems.
Musculoskeletal Recognition by Handcrafted Features and Its Limitations
Most skeletal muscle recognition methods that have been developed to date rely on handcrafted features. In the Computational Anatomy Project [7], we are performing computer-aided organ and disease recognition using CT images, where we adopted a method based on automatic recognition of normal structure. That is, similar to an analysis conducted by a doctor, by detecting the normal structure of the human body as a model in a computer (computational anatomy model), it is possible to detect an abnormality in an unknown case. Because the main purpose of this project was to construct a CAD system targeting the organ area, skeletal muscle was treated as one of the normal structures. In particular, in CT images, the density value distribution of the skeletal muscle overlaps with the density value distribution of the organ region, and thus it is difficult to segment the skeletal muscle and the other organ region using only the density value. Therefore, a muscle modeling technique that employs a shape model and probability model is commonly used [8]. In the Computational Anatomy Project, we have also achieved some success using computational models and muscle recognition for analysis of superficial and deep muscles [9]. In this study, we focused on the anatomical attachment point of the muscle, i.e., the origin and the insertion, and realized automatic recognition of muscles according to their position using a technique that arranged the shape model on the basis of the origin and the insertion. Automatic recognition of skeletal muscle was realized in surface and deep muscles of the thoracicoabdominal region. However, as described above, variations in skeletal muscle result not only from individual differences, but also from inter-individual differences due to aging, daily activities, and the characteristics of organs. Therefore, only a limited number of regions in the case database can be used for constructing a computational anatomical model of skeletal muscle with high recognition accuracy. In particular, there were only a limited number of scenarios in which shapes were comparatively clear or the boundaries between adjacent organs and adjoining muscle were clear.
In recent years, the Computational Anatomy Project has been developed into the Multidisciplinary Computational Anatomy Project, with the purpose of achieving comprehensive understanding of the human body using medical image information over the four axes of space, time, function, and pathology [10]. Skeletal muscle recognition in the Multidisciplinary Computational Anatomy Project was designed with an emphasis on multi-axis awareness rather than computational anatomy. Muscle modeling ranges from micro to macro and considers the functional aspects of muscles [11]. In addition, in this multidimensional Computational Anatomy Project, we worked toward the automatic recognition and analysis of skeletal muscles with extended muscle regions and in contact with muscles and complex contour shapes [11]. In these computational anatomy modeling projects, nine regions (surface muscle: sternocleidomastoid muscle, trapezius muscle, supraspinous muscle, large pectoral muscle, intercostal muscle, oblique abdominal muscle, rectus abdominis muscle; deep muscle: psoas major muscle, iliac muscle) of skeletal muscle modeling-based segmentation were realized.
Figure 1 shows a conceptual diagram of skeletal muscle recognition by handcrafted feature modeling, which was realized during the Computational Anatomy Project and the Multidisciplinary Computational Anatomy Project. As Fig. 1 shows, we realized recognition and analysis of various muscles using torso CT images and whole-body CT images; however, the features of the muscles are one-dimensional points (landmarks; LM), two-dimensional running (muscle running), and three-dimensional shapes used to represent muscle features, i.e., the landmarks are acquired from the bone corresponding to the origin and insertion of the skeletal muscle, the running of the muscle is expressed by connecting LM on the bone, and the shape model based on gray values and probability distributions is arranged according to the running. In a technique based on handcrafted features, all the procedures from 1D to 3D worked sequentially until recognition according to the position of skeletal muscle was achieved. Therefore, it can be said that many dependent procedures must be performed to obtain initial information such as muscle quantity and intramuscular fat quantity, which are necessary for the site analysis of skeletal muscle. In general, the accuracy of each procedure greatly affects the accuracy of the final result.
As described above, when using handcrafted features in the recognition of skeletal muscle according to site, the 2D cross section or the shape of the muscle became a limited region with an easy-to-model shape. We then tackled recognition and analysis of the muscle using machine learning and deep learning techniques in the later stage of the multiple calculation anatomy project. The following sections describe recognition and analysis of muscles with more complicated shapes through deep learning technology.
Skeletal Muscle Segmentation Using Deep Learning
This section describes skeletal muscle segmentation using deep learning. As described previously, the problem of segmenting skeletal muscles in CT images is essentially the same as the problem of automatic recognition of organs. However, as described above, when the recognition of skeletal muscle by region is considered, the shape is complicated, and difficulties result from the juxtaposition of the organ region and skeletal muscle as well as the connection between skeletal muscles. In addition, very large individual differences in muscle mass cause difficulties in model-based skeletal muscle recognition. Prior to discussing skeletal muscle segmentation by deep learning, this section introduces the segmentation of erector spinae using random forest, the well-known machine learning algorithm.
The cross-sectional area of the erector spinae muscle is smaller in patients with COPD than in healthy individuals, and it has been found that this cross-sectional area indicates the prognosis of COPD patients [12]. However, the erector spinae is a very large group of muscles originating from the sacrum and located along the left and right vertebral columns. Therefore, manual measurement of the transverse area of the erector spinae is a time-consuming and error-prone task. We performed 3D recognition of the erector spinae muscle using the iterative random forest method and multiple sources of information [13]. Here, the original image and the probability map of the erector spinae, which is repeatedly refined, were used as sources, and improvements in recognition accuracy and high-speed segmentation were realized. During the learning process, three classifiers are trained. Classifier 1 is trained using low-resolution appearance features extracted from downsampled CT images. Then, the appearance features and probability map obtained by classifier 1 are combined to create trained classifier 2. Concurrently, the probability map obtained from classifier 1 is upsampled, and learned classifier 3 is obtained together with high-resolution appearance features. Thus, the final erector spinae segmentation results were obtained by combining classifiers 1 and 2, which are learned with low-resolution data, and classifier 3, which is trained with high-resolution data. Ten training cases were randomly selected from torso CT images from 20 cases. These were evaluated in 10 test cases, in which the average Dice coefficient (DC) was 93.0 ± 2.1% and the Jaccard similarity coefficient (JSC) was 87.0 ± 3.5%. Thus, highly accurate segmentation of the erector spinae muscle was achieved with 10 learning images and very limited data. Figure 2 shows the recognition result using proposed iterative random forest method. It was shown that machine learning-based segmentation according to the position of the skeletal muscle could be a robust technique for analyzing large and complex muscles for which shape models are difficult to generate.
The following describes deep learning-based automatic recognition of the erector spinae in the cross sections of the 12 thoracic vertebrae. As stated above, the cross-sectional areas of the erector spinae muscle in the 12 thoracic sections indicate the prognosis of COPD patients. Therefore, we segmented the erector spinae muscle in the cross sections of the 12 thoracic vertebrae, and performed the segmentation of the muscle using deep learning and two-dimensional images [14]. The FCN-8s fully convolutional network (FCN) was used in order to utilize the results of the middle layer, in which detailed shape features are expected to be found. One-thousand correct images from 40 cases were prepared for learning of the erector spinae muscle in the cross sections of the 12 thoracic vertebrae; because the erector spinae muscle exists in the back, the learning was concentrated in the back half of the original image. Segmentation accuracy was evaluated using test images from 29 cases, and the average concordance rate of JSC was 82.4%. Figure 3 shows the segmentation of the erector spinae muscle in the cross section of the 12th thoracic vertebra. From top to bottom, the figure shows the original image from the top, the recognition result obtained by the model-based method, and the recognition result obtained by FCN-8s. Blue indicates a correct result, yellow indicates over-extraction, and red indicates unextracted areas. Because the boundary with the latissimus dorsi muscle is unclear, the conventional model-based method causes over-extraction during region expansion. With deep learning, it is proven that the target region is recognized even in these boundary areas.
Through deep learning of 2D images as shown above, automatic segmentation of skeletal muscle in a two-dimensional section could distinguish the boundary with the latissimus dorsi muscle, which could not be distinguished in conventional muscle modeling. Three-dimensional recognition of erector spinae by deep learning was carried out in a subsequent experiment.
In order to recognize the erector spinae as a volume, three-dimensional deep learning was used. This method can obtain three-dimensional recognition results using plural two-dimensional cross sections; thus, it is referred to as a 2.5-dimensional method. Here, FCN-8s, which obtained good results in the simultaneous segmentation of multiple organs in torso CT images [15], was used [16]. Various architectures have recently been proposed for medical image segmentation based on machine learning. In particular, U-net is well known for its efficacy in the field of medical image segmentation, and features a decoder employing an architecture similar to that of its encoder. Our group achieved automatic segmentation of multiple organs from torso CT images using segmentation based on FCN and the voting principle [17]. In a comparison of segmentation methods based on FCN and U-net, we obtained results showing that FCN and voting-based methods are realistic methods for segmenting CT images [17]. In the 2.5-dimensional FCN, three anatomical sections are input, and the simultaneous probability is calculated from the recognition result in each section; the label value with the highest simultaneous probability is selected as the result. By using the simultaneous probability, the recognition result of each cross section is judged comprehensively. The average DC and average JC of the erector spinae recognition results were 89.9 ± 2.0% and 81.7 ± 3.2%, respectively, when evaluated by the leave-one-out method using the trunk CT images from 11 cases. Figure 4 shows the results of recognition of the erector spinae muscle using deep learning. In 10 cases similar to the three-dimensional recognition of the erector spinae muscle achieved by the random forest method described above, less segmentation was observed compared with the original random forest method; however, segmentation was successful, and boundaries with other skeletal muscles (such as the latissimus dorsi muscle) were recognized. Such boundaries were not recognized by the model-based method.
The results show that robust segmentation can be achieved with only 10 learning images. Skeletal muscle segmentation using deep learning is an effective technique that improves upon the manual 2D cross-sectional area measurements intended as an alternative measuring method to ease workloads incurred during automated 3D analysis. In particular, because the technique does not depend on the additional processing steps mentioned in the previous section, it can be said that high-quality segmentation results can be expected if a good training image is prepared. Moreover, high-quality segmentation results can be obtained in such a small number of cases, it is considered useful for efficient preparation of learning images needed for creating correct images used in developing deep learning-based segmentation algorithms.
Whole-Body Muscle Analysis Using Deep Learning
This section describes whole-body analysis of skeletal muscle using deep learning. As described above, the basic requirement for the analysis of skeletal muscle is the quantitative and automatic measurement of muscle mass, which is measured manually in current clinical situations. However, as previously mentioned, the technology for fully automated segmental analysis of skeletal muscle is still under development. Additionally, differentiating between diseases of the muscle itself and the quality of the muscle appears to be dependent on image analyses produced by the computer.
We have been working on whole-body muscle analysis using whole-body CT images. In particular, for whole-body CT images taken for diagnosis of amyotrophic lateral sclerosis (ALS), whole-body skeletal muscle recognition can be achieved using the active balloon model and skeletal muscle model [18]. We conducted muscle analysis [18] using texture analysis employing the Haralick’s features of the region. However, the recognition of skeletal muscle over the whole body differs significantly for individual body types, and highly accurate surface muscle recognition has not been realized yet. Therefore, the area used for research on discrimination of myopathy was limited to limbs in which whole-body skeletal muscle [18] could be recognized. In this section, we first introduce a method to automatically classify atrophic myopathy through deep learning using the upper extremities, and then introduce whole-body musculoskeletal segmentation by deep learning.
First, the automatic classification of atrophic muscular disease in the upper arm and lower arm using deep learning is shown. Here, as an initial challenge, the automatic classification of ALS, which is a neurogenic disease, and myopathy, which is myogenic atrophy, was carried out [19]. It is an important problem to separate ALS, which is an intractable disease for which a therapy has not been established, from other atrophic diseases for which treatment is possible. Moreover, it is important to test the deep learning approach for diseases such as ALS, which can only receive exclusion diagnoses. Here, the architecture of ResNet-50 was used. Drawing from five ALS cases and five myopathy cases, training images of 1678 upper arms and 1150 lower arms were prepared, and 171 upper and 130 lower arms were tested. The images were classified into ALS and myopathy. As a result, we obtained an average classification accuracy of 90.3% on the right forearm. This demonstrated the possibility of classifying diseases through deep learning with images of atrophic diseases, even when the differences were not recognized through visual observation. However, in order to fully diagnose ALS, it is necessary to evaluate more cases while considering the stage and type of the disease. In addition, methods for distinguishing the type of muscular atrophy and for analyzing the muscle region in more detail are necessary. This technique uses the results, which are roughly divided into 22 surface layer muscle regions [18] obtained by recognizing the body cavity through the active balloon model for whole-body skeletal muscle using conventional handcrafted characteristics, and differentiating the body cavity region from the whole body [18].
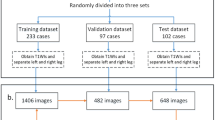
Using the above problems as a framework, we have been working toward fully automatic recognition of surface muscles in whole-body skeletal muscles [20]. As a preliminary experiment, the axial cross section was learned and recognized using 2D U-Net. When 50 cases were divided into training, test, and validation cases at a ratio of 8:1:1 according to the hold-out method, the segmentation results showed that DC was 81.7 ± 0.9% on average in three experiments. Figure 5 shows the results of whole-body skeletal muscle segmentation using 2D U-Net. Although only the axial CT slice was input, continuous recognition of surface muscle in the sagittal and coronal sections could be realized.
Similarly, bone segmentation was performed using 2D U-Net [21]. A related study has segmented bone from low-dose whole-body CT images using 2.5D U-Net [22]. Therefore, it is worth studying bone segmentation on whole-body plain CTs. The experiments were carried out using 17 whole-body CT images without contrast. A dataset consists of 12 training cases, two validation cases, and three test cases. We used whole-body CT images without contrast and trained 2-D U-Net with axial slices. The average dice coefficient of bone segmentation was 0.899, and this method was robust with regard to the position of hands. Figure 6 shows the segmentation result that bone segmentation by U-Net using only axial CT slices provides high performance with plain CT images. In the future, we will use this method for muscle segmentation to conduct bone classification using segmented bone images.
The whole-body musculoskeletal analysis presented in this section is still preliminary. However, deep learning has shown sufficient potential for classification and segmentation tasks, which are two major aspects of musculoskeletal muscle analysis. In the following section, we discuss the techniques needed to further advance whole-body musculoskeletal analysis.
Fusion of Deep Learning and Handcrafted Features in Skeletal Muscle Modeling
In the previous section, musculoskeletal segmentation and analysis using deep learning were described, and the possibility of musculoskeletal analysis by deep learning was shown.
In order to realize further advancements in musculoskeletal analysis in the future, deep learning must be effectively combined with the handcrafted features-based skeletal muscle modeling described at the beginning of this chapter. This is because correct images are required for deep learning. However, significant amounts of time are needed to paint all skeletal muscles and obtain a sufficient number of learning images, and the degree of difficulty in producing correct images is higher than for other organ regions. This section describes two approaches that aim to address these problems.
The first approach involves the simultaneous and automatic recognition of skeletal muscle, as well as the origin and stop position of skeletal muscle. Information about muscle and its adhesion position becomes important when conducting muscle analysis and muscle recognition while considering positions specified by handcrafted features. In particular, model-based methods provide information on the placement of muscle models. We carried out the simultaneous recognition of muscle and bone attachment positions on the muscle through deep learning. The 2.5-dimensional FCN employed in the automatic recognition of erector spinae was used. The mean DC of the erector spinae was 89.9 ± 2.0% and the mean JC was 81.7 ± 3.2% in 11 cases examined by the leave-one-out method. The average DC of the recognition results of the attachment area on the skeleton was 65.5 ± 3.3% and the average JC was 48.8 ± 3.7% [16]. Figure 7 shows the results of recognition of the attachment site between the erector spinae and the bone on the erector spinae. Because the bone attachment site area is small compared with the skeletal muscle area, the recognition accuracy is low; nevertheless, the attachment site of the muscle and bone is captured. Therefore, it is expected that the model can be applied to the analysis of muscle travel and the relation between muscle and disease.
Next, the challenge of classifying muscle groups obtained by deep learning through modeling the shape and running of muscle bundles [23] is shown. As described above, it is very difficult to create correct images from a sufficient number of cases for deep learning by separately painting muscle groups composed of multiple muscles. The results for the erector spinae muscle are shown here. Because the erector spinae muscle is composed of multiple muscles, it is labor intensive to prepare a learning image of the whole erector spinae muscle. We proposed a method to bundle three of the erector spinae muscles automatically recognized by deep learning (iliocostalis lumborum, iliocostalis thoracis, and longissimus thoracis); these particular muscles were selected because they are relatively distant from each other. The muscle bundle model is an ellipsoid connecting the beginning and end of each muscle, and the thickness of the ellipsoid is determined from the learning case. The erector spinae muscle can be segmented into three muscles by constructing a muscle bundle model containing the three muscles of the erector spinae muscle and arranging them into the recognized result of the erector spinae muscle using deep learning. Figure 8 shows the result of dividing the erector spinae muscle recognized by deep learning into three regions using the muscle bundle model. Green represents the erector spinae, yellow the iliocostalis lumborum, blue the longissimus thoracis, and orange the iliocostalis thoracis. Each modeled muscle bundle is convexly encapsulated and subdivided to the lower right. Because the erector spinae is composed of nine muscles, modeling of other muscles is also necessary. However, when we evaluated the ratio of the volume of each muscle bundle model to that of the erector spinae region in the convex hull region, the average Jaccard coefficient was 48.7 ± 6.8% in the right-hand region and 53.2 ± 4.2% in the left-hand region. However, as shown in the two-dimensional cross sections in Fig. 8, the positions of the muscles constituting the muscle group are shown, and for skeletal muscles for which preparing a correct individual muscle image is difficult, the recognition of the muscle through deep learning can be said to show the possibility of dividing muscles into regions according to position.
This section described the development of a new muscle analysis method that combines deep learning and muscle models. In the future, finer whole-body muscle analysis can be achieved by effectively using deep learning and modeling in conjunction with conventional handcrafted feature.
Conclusion
In this chapter, we described segmentation, recognition, and classification techniques we have developed to conduct musculoskeletal analysis using deep learning. Because musculoskeletal areas are large compared to areas occupied by organs, individual and intra-individual differences are also large, and it cannot be said that musculoskeletal analysis will be overwhelmingly simplified by the introduction of deep learning; however, handcrafted features also help improve the developed techniques. There is no doubt that deep learning techniques complement musculoskeletal analysis. As described in the latter half of the chapter, it is considered that musculoskeletal analysis can be markedly improved by effectively combining deep learning-based methods and handcrafted feature-based methods.
References
Yu W, Liu W, Tan L et al (2018) Multi-object model-based multi-atlas segmentation constrained grid cut for automatic segmentation of lumbar vertebrae from CT images, intelligent orthopaedics. Adv Exp Med Biol 1093:65–71. https://doi.org/10.1007/978-981-13-1396-7_5
Zeng G, Zheng G (2018) Deep learning-based automatic segmentation of the proximal femur from MR images, intelligent orthopaedics. Adv Exp Med Biol 1093:73–79. https://doi.org/10.1007/978-981-13-1396-7_6
Yu W, Zheng G (2018) Atlas-based 3D intensity volume reconstruction from 2D long leg standing X-rays: application to hard and soft tissues in lower extremity, intelligent orthopaedics. Adv Exp Med Biol 1093:105–112. https://doi.org/10.1007/978-981-13-1396-7_9
Kamiya N (2018) Muscle segmentation for orthopedic interventions, intelligent orthopaedics. Adv Exp Med Biol 1093:81–91. https://doi.org/10.1007/978-981-13-1396-7_7
Rothstein JD (2017) Edaravone: a new drug approved for ALS. Cell 171(4):725
Ministry of Health, Labour and Welfare, JAPAN, National Health Promotion Movement in the 21st Century (Healthy Japan 21)
Kobatake H, Masutani Y et al (2017) Computational anatomy based on whole body imaging: basic principles of computer-assisted diagnosis and therapy. Springer
Hanaoka S, Kamiya N, Sato Y et al (2017) Skeletal muscle, understanding medical images based on computational anatomy models. Springer, pp 165–171
Fujita H, Hara T, Zhou X et al (2014) Model construction for computational anatomy: progress overview FY2009-FY2013. In: Proceedings of the Fifth International Symposium on the Project “Computational Anatomy”, pp 25–35
Multidisciplinary Computational Anatomy and Its Application to Highly Intelligent Diagnosis and Therapy. http://wiki.tagen-compana.org
Fujita H, Hara T, Zhou X et al (2019) Function integrated diagnostic assistance based on multidisciplinary computational anatomy models -Progress Overview FY2014-FY2018-. In: Proceedings of the Fifth International Symposium on the Project “Multidisciplinary Computational Anatomy”, pp 115–128
Tanimura K, Sato S, Fuseya Y et al (2016) Quantitative assessment of erector spinae muscles in patients with chronic obstructive pulmonary disease. Novel chest computed tomography-derived index for prognosis. Ann Am Thorac Soc 13(3):334–341
Kamiya N, Li J, Kume M et al (2018) Fully automatic segmentation of paraspinal muscles from 3D torso CT images via multi-scale iterative random forest classifications. Int J Comput Assist Radiol Surg 13(11):1697–1706. https://doi.org/10.1007/s11548-018-1852-1
Kume M, Kamiya N, Zhou X et al (2017) Automated recognition of the erector spinae muscle based on deep CNN at the level of the twelfth thoracic vertebrae in torso CT images. In: Proceedings of the 36th JAMIT annual meeting
Zhou X, Takayama R, Wang S et al (2017) Deep learning of the sectional appearances of 3D CT images for anatomical structure segmentation based on an FCN voting method. Med Phys 44(10):5221–5233. https://doi.org/10.1002/mp.12480
Kamiya N, Kume M, Zheng G et al (2019) Automated recognition of erector spinae muscles and their skeletal attachment region via deep learning in torso CT images. Comput Methods Clin Appl Musculoskelet Imaging:1–10. https://doi.org/10.1007/978-3-030-11166-3_1
Zhou X, Ito T, Takayama R et al (2016) Three-dimensional CT image segmentation by combining 2D fully convolutional network with 3D majority voting. In: Proceedings of the Workshop on the 2nd Deep Learning in Medical Image Analysis (DLMIA) in MICCAI 2016, LNCS 10008, pp 111–120
Kamiya N, Ieda K, Zhou X et al (2017) Automated analysis of whole skeletal muscle for muscular atrophy detection of ALS in whole-body CT images: preliminary study. In: Proceedings of the SPIE Medical Imaging 2017, Computer-Aided Diagnosis, 10134, 1013442-1-1013442-6. https://doi.org/10.1117/12.2251584
Kamiya N, Oshima A, Asano E et al (2019) Initial study on the classification of amyotrophic diseases using texture analysis and deep learning in whole-body CT images. In: Proceedings of the SPIE 11050, International Forum on Medical Imaging in Asia 2019, 110500X. https://doi.org/10.1117/12.2518199
Oshima A, Kamiya N, Zhou X et al (2019) Automated segmentation of surface muscle in whole-body CT images using 2D U-Net: preliminary study. In: Proceedings of the IEEE EMBC2019, ThPOS-32.34, p 71
Wakamatsu Y, Kamiya N, Zhou X et al (2019) Bone segmentation in whole-body CT images using 2D U-Net. In: Proceedings of the IEEE EMBC2019, ThPOS-32.35, p 72
Klein A, Warszawski J, Hillengaß J et al (2019) Automatic bone segmentation in whole-body CT images. Int J Comput Assist Radiol Surg 14(1):21–29
Kume M, Kamiya N, Zhou X et al (2019) Development of representation method of muscle running using attachment region of the spinal column erector muscle in the torso CT images. IEICE Tech Rep 118(412):39–40
Acknowledgments
The authors would like to thank all the members of the Fujita Laboratory in the Graduate School of Medicine, Gifu University, for their collaboration. We especially thank Ms. Oshima and Mr. Wakamatsu in the School of Information Science and Technology, Aichi Prefectural University for providing binary code and testing systems. This work was supported in part by a JSPS Grant-in-Aid for Scientific Research on Innovative Areas (Multidisciplinary Computational Anatomy, #26108005 and #17H05301) and for Challenging Exploratory Research (#16K15346), Japan.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Kamiya, N. (2020). Deep Learning Technique for Musculoskeletal Analysis. In: Lee, G., Fujita, H. (eds) Deep Learning in Medical Image Analysis . Advances in Experimental Medicine and Biology, vol 1213. Springer, Cham. https://doi.org/10.1007/978-3-030-33128-3_11
Download citation
DOI: https://doi.org/10.1007/978-3-030-33128-3_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-33127-6
Online ISBN: 978-3-030-33128-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)