Abstract
The expression of a gene is characterised by its transcription factors and the function processing them. If the transcription factors are not affected by gene products, the regulating function is often represented as a combinational logic circuit, where the outputs (product) are determined by current input values (transcription factors) only, and are hence independent on their relative arrival times. However, the simultaneous arrival of transcription factors (TFs) in genetic circuits is a strong assumption, given that the processes of transcription and translation of a gene into a protein introduce intrinsic time delays and that there is no global synchronisation among the arrival times of different molecular species at molecular targets.
In this paper, we construct an experimentally implementable genetic circuit with two inputs and a single output, such that, in presence of small delays in input arrival, the circuit exhibits qualitatively distinct observable phenotypes. In particular, these phenotypes are long lived transients: they all converge to a single value, but so slowly, that they seem stable for an extended time period, longer than typical experiment duration. We used rule-based language to prototype our circuit, and we implemented a search for finding the parameter combinations raising the phenotypes of interest.
The behaviour of our prototype circuit has wide implications. First, it suggests that GRNs can exploit event timing to create phenotypes. Second, it opens the possibility that GRNs are using event timing to react to stimuli and memorise events, without explicit feedback in regulation. From the modelling perspective, our prototype circuit demonstrates the critical importance of analysing the transient dynamics at the promoter binding sites of the \({\mathsf {DNA}}\), before applying rapid equilibrium assumptions.
Tatjana Petrov’s research was supported by SNSF Advanced Postdoc. Mobility Fellowship grant number \(P300P2\_161067\), the Ministry of Science, Research and the Arts of the state of Baden-Württemberg, and the DFG Centre of Excellence 2117 ‘Centre for the Advanced Study of Collective Behaviour’ (ID: 422037984). Claudia Igler is the recipient of a DOC Fellowship of the Austrian Academy of Sciences. Thomas A. Henzinger’s research was supported in part by the Austrian Science Fund (FWF) under grant Z211-N23 (Wittgenstein Award).
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The fundamental conceptual breakthroughs related to how a gene is turned on and off, have inspired a large body of theoretical and experimental work on gene regulation, including the explanation of stochastic switching between lysis and lysogeny of phage [25], all the way to more complex logic gate formalisms that attempt to abstract more complex biological behaviour. Synthetic biology enthusiasts often use analogies with how electronic circuits are manipulated by computers [13, 24], and have demonstrated success in engineering simple genetic circuits that are encoded in DNA and perform their function in vivo. However, such digital (in the sense that the expression states are encoded through Boolean values) and combinational design (in the sense that the output is a pure function of present input only, different to the sequential design) quickly becomes infeasible in experiment, because the cellular environment is resource-limited and highly crosstalk-prone. The effective engineering of biological systems needs to take into account the intrinsic properties of the biological medium, so as not to fight against the principles of tinkering that characterise biology [16], but rather to make use of them. Significant conceptual challenges remain related to the still unsatisfactory quantitative but also qualitative understanding of the underlying processes [20]. Understanding time-dependent phenomena is fundamental in this complex picture of the cell that unravels itself at the molecular scale, especially since cells do not have computer-like clocking mechanisms, beyond circadian and cell cycle ones. A major question emerges as to what are the macroscopic effects of small delays in the arrival times of different molecules at molecular targets.
Gene expression in a single cell is modelled by a stochastic process which captures the stochastic switching among possible configurations at the \({\mathsf {DNA}}\) (the architectural configuration of which is often termed promoter logic, e.g. shown in Fig. 4), and their effect on the copy number of other species involved in regulation, such as mRNA, proteins and transcription factors (TFs). The switching mechanism depends on the binding affinities of the TFs and RNA polymerase to their respective binding sites as well as the concentrations of those proteins in the cell. Stochastic dynamics of such gene regulatory process typically has a single equilibrium, as a consequence of reversibility of reactions occurring at the \({\mathsf {DNA}}\) binding sites. Sometimes, the transient regime of the distribution among \({\mathsf {DNA}}\) configurations is rapid and robust to possible delays in arrival of TFs. In such cases, it is satisfactory to use the statistical thermodynamics model, which has shown unquestionable success (e.g. [1, 37]). It estimates the probability of being in any of the possible \({\mathsf {DNA}}\) binding configurations from their relative binding energies (Boltzmann weights) and the protein concentrations, both of which can often be experimentally accessed. While this model takes into account the stochasticity inherent to the \({\mathsf {DNA}}\) binding configurations (unlike the also widely used deterministic limit [18]), it neglects the transient probabilities in the promoter logic before the equilibrium is reached. The question arises: In which ways does the transient regime at the \({\mathsf {DNA}}\) (promoter) affect the shape and duration of observable protein dynamics? Can it happen that the observable transients move towards the unique equilibrium so slowly, that they are mistaken for steady state dynamics?
In this paper, as a proof-of-concept, we construct a prototype genetic circuit based on two different transcription factors that regulate the same gene, without feedback. Our circuit demonstrates that, for gene regulation, qualitatively distinct transients may take extraordinarily long times to disappear, and the observable phenotype in the transient can be highly sensitive to the order of arrival of TFs in the system. In particular, the transient phenotype may appear to be stable even though it is not, creating an effect of long lived transients[7]. Our prototype circuit is realistic, experimentally implementable in the sense that the mechanism can be implemented by the current technology and kinetic rate values are in realistic ranges. The behaviour of this circuit suggests that the genetic circuit can memorise the order of arrival of TFs, although there is no explicit feedback at the gene regulatory level.
2 Preliminaries and Background
A gene is expressed at a basal rate, whenever the RNA polymerase (\({\mathsf {RNAP}}\)) is bound to its promoter region at the \({\mathsf {DNA}}\). Activators are transcription factors (TFs) that bind to specific locations on the DNA, or to other TFs, and enhance the expression of gene g by promoting the binding of \({\mathsf {RNAP}}\). Repressors reduce the expression of gene g, by directly blocking the binding of \({\mathsf {RNAP}}\), or indirectly, by inhibiting the activators, or promoting direct repressors. The mechanism of how and at which rates the molecular species are interacting is transparently written in a list of reactions. Reactions are equipped with the stochastic semantics which is valid under mild assumptions [12].
Definition 1
A reaction system; is a pair \(({\mathsf S},\mathsf R)\), such that \({\mathsf S}=\{S_{1},\ldots ,S_{n}\}\) is a finite set of species, and \(\mathsf R=\{\mathsf {r}_1,\ldots ,\mathsf {r}_{r}\}\) is a finite set of reactions. The state of a system can be represented as a multi-set of species, denoted by \(\mathbf {x}=(x_1,...,x_{n})\in \mathbf N^{n}\). Each reaction is a triple \(\mathsf {r}_j\equiv ({\mathbf {a}}_{j},\varvec{\nu }_{j},c_j)\in \mathbf N^{n}\times \mathbf N^{n}\times {\mathbb R}_{\ge 0}\), written down in the following form:

The vectors \({\mathbf {a}}_{j}\) and \({\mathbf {a}}_{j}'\) are often called respectively the consumption and production vectors due to jth reaction, and \(c_j\) is the respective kinetic rate. If the jth reaction occurs, after being in state \(\mathbf {x}\), the next state will be \(\mathbf {x}' = \mathbf {x}+\varvec{\nu }_j\). This will be possible only if \(x_i\ge a_{ij}\) for \(i=1,\ldots ,n\).
Stochastic Semantics. The species multiplicities follow a continuous-time Markov chain (CTMC) \(\{X(t)\}_{t\ge 0}\), defined over the state space \( S=\{\mathbf {x}\mid \mathbf {x}\) is reachable from \(\mathbf {x}_0\) by a finite sequence of reactions from \(\{\mathsf {r}_1,\ldots ,\mathsf {r}_{r}\}\}\). In other words, the probability of moving to the state \(\mathbf {x}+\varvec{\nu }_j\) from \(\mathbf {x}\) after time \(\varDelta \) is
with \(\lambda _{j}\) the propensity of jth reaction, assumed to follow the principle of mass-action: \(\lambda _{j}(\mathbf {x})= c_j\prod _{i=1}^{n}{x_i\atopwithdelims ()a_{ij}} \). The binomial coefficient \({x_i\atopwithdelims ()a_{ij}}\) reflects the probability of choosing \(a_{ij}\) molecules of species \(S_i\) out of \(x_i\) available ones.
Example 1
(basal gene expression). Basal gene expression with \({\mathsf {RNAP}}\) binding can be modelled with four reactions, where the first reversible reaction models binding between the promoter site at the \({\mathsf {DNA}}\) and the polymerase, and the second two reactions model the protein production and degradation, respectively:

The state space of the underlying CTMC \(S\cong \{\texttt {0},\texttt {1}\}\times \{0,1,2,\ldots \}\), such that \(s_{(\texttt {1},x)}\in S\) denotes an active configuration (where the \({\mathsf {RNAP}}\) is bound to the \({\mathsf {DNA}}\)) with \(x\in \mathbf N\) protein copy number, as depicted in Fig. 2.
Example 2
(adding repression). Repressor blocking the polymerase binding can be modelled by adding a reaction
In this case, there are three possible promoter configurations, that is, \(S\cong \{{\mathsf {DNA}},{\mathsf {DNA}}.{\mathsf {RNAP}},{\mathsf {DNA}}.R\}\times \{0,1,2,\ldots \}\), where \(D_{\texttt {0}}=\{{\mathsf {DNA}},{\mathsf {DNA}}.R\}\) are inactive promoter states.
Computing the Transient. Using the vector notation \(\mathbf {X}(t)\in \mathbf N^n\) for the marginal of process \(\{X(t)\}_{t\ge 0}\) at time t, we can compute this transient distribution by integrating the chemical master equation (CME). Denoting by \(p^{(t)}(\mathbf {x})=\mathsf {P}(\mathbf {X}(t)=\mathbf {x})\), the CME for state \(\mathbf {x}\in \mathbf N^{n}\) reads
The solution may be obtained by solving the system of differential equations, but, due to its high (possibly infinite) dimensionality, it is often statistically estimated by simulating the traces of \(\{X_t\}\), known as the stochastic simulation algorithm (SSA) in chemical literature [12]. As the statistical estimation often remains computationally expensive for desired accuracy, for the case when the deterministic model is unsatisfactory due to the low multiplicities of many molecular species [19], different further approximation methods have been proposed, major challenge to which remains the quantification of approximation accuracy (see [36] and references therein for a thorough review on the subject).
2.1 Transients in Gene Expression Without Feedback
We will further focus on regulation of single gene without feedback. This allows a circuit-view, where activators and repressors are inputs, and the average transient protein expression is the output. Since there is a single \({\mathsf {DNA}}\) molecule per cell, each state counts one copy of the current \({\mathsf {DNA}}\) configuration, and zero copies of all other \({\mathsf {DNA}}\) binding configurations. Hence, the expression state for a single gene of interest consists of two layers: the proteins that we see, and the regulatory configuration of the \({\mathsf {DNA}}\) (for example, two activators and polymerase are bound) (Fig. 3). Such two-layer model allows us to study the transient of coupled promoter state and protein count. In order to focus our analysis on the effect of input timing perturbations, yet to keep our model simple, we chose not to involve further mechanistic details, such as the steps involving \({\mathsf {mRNA}}\).
The following observation states that in general, when there is no feedback, computing the output does not require integrating the Master equation for the entire CTMC, but only for a CTMC controlling the switching among the \({\mathsf {DNA}}\) configurations (depicted left in Fig. 3).
Lemma 1
Let \(\{X(t)\}_{t\ge 0}\) be the CTMC for a model of single gene regulation without feedback, over the state space \(S=S_{\texttt {0}}\uplus S_{\texttt {1}}= (D_0\uplus D_1)\times \{0,1,\ldots \}\), where \(D_0=\{\mathsf {D}_{01},\mathsf {D}_{02},\ldots \}\) are inactive \({\mathsf {DNA}}\) configurations, and \(D_1=\{\mathsf {D}_{11},\mathsf {D}_{12},\ldots \}\) are active \({\mathsf {DNA}}\) configurations (\({\mathsf {RNAP}}\) bound). Let the reaction system;\(({\mathsf S},\mathsf R)\) be such that all reactions are of one of the following types (for some \(i\ge 0\) and \(j\ge 0\)):

Then, the average amount of protein in a population follows the differential equation
where \(\langle x_p(t)\rangle \) denotes the average amount of the protein molecules at time t, and process \(\mathbf {X}(t)|_{\mathsf {D}}\) is the projection of process \(\mathbf {X}(t)\) to states at the promoter, that is \(\mathbf {X}(t)|_{\mathsf {D}}=d\) if and only if (\(\mathbf {X}(t)\in \cup _{i\ge 0}(d,i)\)). In other words, \(\mathsf {P}(\mathbf {X}(t)|_{\mathsf {D}}\in \mathsf {D}_{\texttt {1}})\) denotes the marginal probability that the promoter is in active state (bound \({\mathsf {RNAP}}\)) at time t.
The proof is discussed in Appendix, Sect. 1.C.
Corollary 1
Let \(\pi _{\texttt {1}}=\lim _{t\rightarrow \infty }\mathsf {P}(\mathbf {X}(t)|_{\mathsf {D}}\in \mathsf {D}_{\texttt {1}})\) denote the probability of active promoter at stationarity. Then, whenever the initial probability equals that of the stationary, i.e. \(\mathsf {P}(\mathbf {X}(0)|_{\mathsf {D}}\in \mathsf {D}_{\texttt {1}}) = \pi _{\texttt {1}}\), the average protein dynamics follows the differential equation
When \({\mathsf {DNA}}\) is modelled with one binding site, the promoter can be in only two states, and the analytic solution to Eq. (2) is tractable. In general, as activators and repressors bind to different regions (operator sites) of the same \({\mathsf {DNA}}\) molecule, the respective number of regulatory configurations at the promoter grows combinatorially with the number of operator sites. For instance, one hypothesised mechanism in \(\lambda \)-phage, containing only three left and three right operators, leads to 1200 different \({\mathsf {DNA}}\) configurations [35]Footnote 1! In such cases, the simplification based on the argument of fast equilibrium is often employed, meaning that the transient protein dynamics is computed according to Eq. (3), thus neglecting the transient changes in probability distribution among the \({\mathsf {DNA}}\) regulatory configurations.
Fast equilibrium assumption is a prerequisite to applying a widely popular statistical thermodynamics model [38]. Assuming that the \({\mathsf {DNA}}\) regulatory configurations mix rapidly, this model allows to experimentally estimate the free energies of each promoter configuration, and then, subsequently, to derive the equilibrium constantsFootnote 2 for each of the reactions [28, 38]. As the absolute and precise values of kinetic rates are rarely available in practice, this method is powerful, because it allows to predict the dynamics of a genetic circuit from a scarcely available experimental data. However, the statistical thermodynamics model is applicable only when the assumption of rapid equilibrium at the promoter is valid.
In the following, we showcase a simple, experimentally realisable genetic circuit which demonstrates an interesting situation where the long transient at the promoter creates phenotypes that are qualitatively distinct from the phenotypes created when the promoter configurations start at the equilibrium. In particular, these phenotypes are long lived transients: they all converge to a single value, but so slowly, that they seem stable for an extended time period, longer than typical experiment duration.
3 Problem Statement
We focus on a single gene regulation without feedback, where activators and repressors are inputs, and the average protein expression is the output. Assuming that a fixed amount of activators and repressors are added to the system with a possible time lag, our reference scenarios are (Fig. 4 in Appendix):
-
\(\mathcal{X}_{A||R}\) in which activator and repressor are introduced together,
-
\(\mathcal{X}_{A\rightarrow R}(\varDelta )\) in which the activator is introduced \(\varDelta \) time units before the repressor,and
-
\(\mathcal{X}_{R\rightarrow A}(\varDelta )\) in which the repressor is introduced \(\varDelta \) time units before the activator.
Our goal is to construct a genetic circuit with the following requirements: (i) it is realistic, that is - experimentally implementable in the sense that the mechanism can be implemented by the current technology and kinetic rate values are in realistic ranges, (ii) the scenarios provide striking differences in the shape and duration of transient protein output. To quantify the latter, we introduce two quantitative measures:
-
amplitude, the maximum distance in phenotype of the scenario with delay from the scenario without delay, that is
$$ \alpha _{|s}:= \max _{t \ge t_0} |\langle x_p\rangle (t|s)-\langle x_p\rangle (t|\mathcal{X}_{A||R})|, $$where s refers to the scenario in question (\(\mathcal{X}_{A\rightarrow R}(\varDelta )\) or \(\mathcal{X}_{R\rightarrow A}(\varDelta )\)) and \(x_p(t|s)\) denotes the average protein number in a population at time t in scenario s, and
-
halflife, the time the system takes from the moment of reaching the amplitude, to the moment when the distance from the phenotype without delay disappears, that is
$$ t_{1/2\mid s}:=\arg \min _{t\ge t_{\alpha \mid s}}\{t\mid |\langle x_p\rangle (t|s)-x_p(t\mid \mathcal{X}_{A||R})|<\frac{1}{2}\alpha _{|s} \}, $$where \(t_{\alpha \mid s}\) denotes the moment when the amplitude is reached in scenario s.
In summary, the amplitude reflects how observable is the sensitivity to the delay among inputs, and the second measure, halflife, reflects how slow is the convergence to the real equilibrium after the amplitude has been observed. Long lived transients are characterised by a large amplitude relative to the basal expression and a half-life exceeding several cell division cycles.
4 Searching for Long Lived Transients
We develop and analyse models for two promoter architectures (drafted in Fig. 5):
-
Model without looping. A basic mechanism for activation and repression is assumed: repressor \(\mathsf {R}\) competes with \(\mathsf {RNAP}\), and the activator \(\mathsf {A}\) recruits the polymerase \({\mathsf {RNAP}}\) and binds independently of the repressor and the polymerase (configurations shown in Fig. 6).
-
Model with looping. In the model with looping (Fig. 5, right), two activators and two repressors can bind the \({\mathsf {DNA}}\); Binding of the second activator (resp. repressor) promotes looping of the \({\mathsf {DNA}}\) in the active (resp. repressed) state, thereby excluding binding of the other TF. This small mechanistic change leads to the blow-up of the state space of the CTMC to 23 states as a composition of two sub-models (Fig. 7).
Biological Context. The CTMC for the \({\mathsf {DNA}}\) switching of our prototype circuit with looping is inspired by the very well-characterized regulatory mechanisms of the lac operon and of the bacteriophage lambda genetic switch [17, 30].
Mathematical Context. In Markov chains, the time to be \(\epsilon \)-close to equilibrium, mixing time, varies depending on the initial distribution, the chain connectivity, and the rate parameters. In particular, long mixing times are prominent for chains with a large spectral gap of the underlying generator matrix, and can be guaranteed for chains with large connectivity diameter, suggesting that more states and sparse connectivities generally can prolong the mixing time [21]. Still, tightly estimating bounds on the mixing time for a given chain is an open problem, beyond the scope of this manuscript. Intuitively, \({\mathsf {DNA}}\) looping architecture is a good candidate for creating large mixing times, because the looped states are quick to reach when only activators or only repressors are present, but, once entered, they are then hard to exit (‘dynamically trapped states’).
4.1 Model Implementation
Implementation. The models are written and analysed within the rule-based modelling framework Kappa which allows us to represent the mechanistic model concisely and to run an efficient stochastic simulation algorithm [3, 11]. Source-code of the rule-based models is given in Appendix 1.B. Parameter exploration and additional output analysis were performed with Python.
Simulation. We simulated multiple samples of the stochastic model, and we statistically estimated the first two moments of protein expression. In the model with looping, we used 1000 individual cells for a time of 36000 s = 10 h, that equals around 20 average cell doubling times, where inputs are added from time point \(t_0\) = 5400 = 1.5 h (see Table 2 for all simulation parameters).
Kinetic Rates. All model parameters are in realistic ranges taken from the literature, given in Table 1 and further explained in the Appendix. The mechanism for the activator is inspired by the \(\lambda \)-phage. The mechanism for repressor is inspired by the \({\mathsf {lac}}\) operon. Further values that were tested to show the generality of our approach came from other well-characterized TFs such as \(\mathsf {CRP}\). The chosen parameter values were found in the literature, both for the scenario without looping [6, 15] and for the scenario with looping [32,33,34, 39, 40].
Parameter Search. We implemented a grid search of the viable parameter space (for different levels of eleven kinetic rate parameters, and the amounts of activator, repressor and \({\mathsf {RNAP}}\)), where we compute the average protein expression, amplitude and half-life for a subset of all parameter combinations. In our implementation, the user specifies a range for each parameter, and the models are executed, figures drawn for each possible parameter combination.
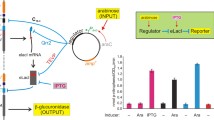
A small delay in arrival times of TFs can give rise to qualitatively opposite, stable transient phenotypes for a long period of time (a) Average protein level for a population of 1000 cells, in three input scenarios (full lines) and three reference scenarios (dotted lines). (b) 50 single cell traces (grey lines) and the respective average, for each of the six modes.
5 Results
In further text, by phenotype, we mean the average protein expression in a population of 1000 cells. All three scenarios \(\mathcal{X}_{A||R}\), \(\mathcal{X}_{A\rightarrow R}(\varDelta )\) and \(\mathcal{X}_{R\rightarrow A}(\varDelta )\) have the same phenotypes eventually. As a reference, we also analyse the scenarios where no TFs are input (\(\mathcal{X}_{basal}\)), only activators (\(\mathcal{X}_{A}\)) or only repressors are input (\(\mathcal{X}_{R}\)). We investigated the phenotype in the three scenarios for a large range of parameter combinations. (there are \(2^11>2000\) combinations when only two values for each parameter are set). We choose one parameter set as the reference parameter set (shown in Fig. 1a), where the phenotypes are symmetric with respect to the \(\mathcal{X}_{A||R}\) scenarios in the sense that the protein expression deviates in the same amount from the phenotype of \(\mathcal{X}_{A||R}\) and the rate of reaching the phenotype of \(\mathcal{X}_{R||A}\) is of the same scale.
A Small Delay in Arrival Times of TFs Can Give Rise to Qualitatively Opposite, Stable Transient Phenotypes for a Long Period of Time. In Fig. 1a, we plot the observable phenotype – the mean of protein expression for a given population of cells – in the three regimes of interest (full lines) and the three reference regimes (dotted lines). Three distinct transient phenotypes are observed:
-
for \(\mathcal{X}_{A||R}\), the expression is close to the level of basal expression,
-
the transient regime for the input \(\mathcal{X}_{A\rightarrow R}(1\min )\) shows high expression for multiple average cell doubling times, while
-
the transient regime for the input \(\mathcal{X}_{R\rightarrow A}(1\min )\) shows low expression for multiple average cell doubling times.
The transient for the input \(\mathcal{X}_{A||R}\) lasts roughly for one average cell doubling time (\(30\min \)), while both phenotypes for \(\mathcal{X}_{A\rightarrow R}(1\min )\) and \(\mathcal{X}_{R\rightarrow A}(1\min )\) last well over 10 average doubling times. Therefore, the delay in arrival times of TFs can result in long lived transient regimes with qualitatively opposite phenotypes (both differing significantly from the equilibrium phenotype), depending on which TF arrives first. Moreover, each of the phenotypes seems stable at the time-scale of multiple cell lifetimes. In other words, the small delays, hence two different histories of input, produce substantially different routes to the equilibrium, and the routes are so slow that they appear as steady state behavior at the timescale of most experiments.
In Fig. 1b, we see that individual cells exhibit ‘all-or-none’ behaviour: an individual cell either has high or low expression and the phenotype depends on whether the cell entered the active looped state or the repressed looped state. The expected time that a cell spends in one looped state is long. The protein expression for 50 randomly chosen single cells is displayed in Fig. 1b for each of the three regimes. In regime \(\mathcal{X}_{A||R}\), an individual cell either has high expression at around 400 proteins or low expression, being fully inhibited. The noise around the low expression value is not observable in the plot, because the low expression is fully inhibited most of the time. If the \({\mathsf {DNA}}\) unloops and subsequently loops towards a different regulatory state, eg from looped repressed to looped active state (or vice-versa), the protein expression will change from low to high expression (or vice-versa). In the taken time window (10 h), three (out of 50) displayed traces switching from the high to low expression level and one trace switching from low to high expression level. As expected, the average expression in a given population (thick line in respective color) follows a continuous line; It is saturated at around 270 protein molecules. In regime \(\mathcal{X}_{A\rightarrow R}\), all of the displayed 50 cells enter the active looped state before the repressors are input, but, due to the slow unlooping, the high expression profile is long-preserved, resulting in slow switching towards the low-expression state, and hence long transient time towards the average expression. In regime \(\mathcal{X}_{R\rightarrow A}\), even though repressors are input first, some cells are activated, but most of the cells are repressed. Similarly as in the profile \(\mathcal{X}_{A\rightarrow R}\), since DNA unlooping from the repressed state is slow, the transient of the average protein expression is also slow. The reference scenarios - \(\mathcal{X}_{basal}\), \(\mathcal{X}_{A}\) and \(\mathcal{X}_{R}\) show the expected behaviour.
Long Lived Transients are Robust to Changes in Kinetic Parameters. Are long lived transients a consequence of system regulatory architecture or a careful tuning of kinetic parameters? To tackle this question, we chose six different parameter combinations, listed in Table 1, and we reproduce the plot shown in Fig. 1a for each of the parameter combinations, each for three time delays - 1 min, 5 min and 15 min. \(\mathbf {p}_1\) is the reference parameter set (the plot shown in Fig. 1). Results, shown in Fig. 8, confirm that the long lived transients are preserved with the chosen parameter changes. Higher unlooping rate for either activators or repressors results in shortening the transient and moving the average expression level to lower and higher value respectively (\(\mathbf {p}_2\) and \(\mathbf {p}_3\)). While decreasing the number of activators does not change the phenotype much (\(\mathbf {p}_4\)), decreasing the number of repressors results in complete dominance of activation effect when both TFs are input simultaneously (\(\mathbf {p}_5\)). Still, the delay of activator input shows full repression profile for a long period of time. When \({\mathsf {RNAP}}\) rates are scaled so that the binding and unbinding rates are both ten times slower, the duration of transients shortens and the three input regimes show the same output after much shorter time (\(\approx \)10 h, \(\mathbf {p}_6\)).
To quantify the effect of long-lived transients, in Fig. 12 (up, model with looping), we see that the reference parameter set (\(\mathbf {p}_1\)) has a halflife longer than 20 hours no matter if the delay occurs in favour of the activator or repressor. The halflife decreases significantly in cases when the unlooping rate is decreased (one at a time - \(\mathbf {p}_2\) and \(\mathbf {p}_3\)), or when \({\mathsf {RNAP}}\) binding and unbinding rate is scaled down (\(\mathbf {p}_6\)), while the change in the number of activators/repressors reflect more on the amplitude than on the halflife (\(\mathbf {p}_4\), \(\mathbf {p}_5\)).
Long Lived Transients Are Not Observed in the Model Without Looping. We next inquire how changes in regulatory architecture affect the behaviour, i.e. is \({\mathsf {DNA}}\) looping essential for observing the long lived transients? We repeated the experiments on a model without looping. Phenotypes for six parameter combinations, listed in Table 1Footnote 3, are each plotted for three time delays (Fig. 10). For all parameter combinations, the amplitude and duration of transient regimes is clearly correlated with the duration of delay - the longer delays induce longer transient regimes. The transient phase is significantly shorter than in the model with looping (notice the different time-scale than in Figs. 1 and 8), but they still can last for several cell doubling times (for delays of 15 min up to 2.5 h or 5 average doubling times). However, they are not long lived transients, as the shape of transients clearly reveals that the steady-state regime is going to be reached later on, that is, the transients in this model would not be easily confused with the steady state. The observations above are indicating that looping is essential for creating the effect of long lived transients. \(\mathbf {p}_1\) is the reference parameter set, which we choose so that the level of expression when both activators and repressors are input is close to basal (the TFs neutralise each-other’s effect overall). As expected, decreasing the recruitment by the activator results in lower stationary expression (\(\mathbf {p}_2\)), increasing the number of repressors results in stronger repression (\(\mathbf {p}_3\)), weakening the repressor binding results in higher expression (\(\mathbf {p}_4\)), weak binding of repressor in combination is not affected by decreasing the recruitment by activator simultaneously (\(\mathbf {p}_5\)) and weak binding of repressor in combination with more repressor molecules results in low expression (\(\mathbf {p}_6\)).
In the summary of characteristics of long lived transients for the model without looping (shown in Fig. 12 down), we see that, for a delay of \(5\min \), all parameter combinations achieve the amplitude at comparable scale as that in the case of model with looping. However, the maximal halflife in all tested parameter points is \(15\min \), a 100-fold difference with respect to the halflife of long lived transients in the model with looping, confirming that adding the looped configurations was essential for the effect of long-lived transients.
Phenotypes in Long Lived Transients can be Modulated by the Delay Between Inputs. We now comment on the dependency on the delay. In Fig. 11, the phenotypes in scenario \(\mathcal{X}_{A\rightarrow R}(\varDelta )\) are observably equivalent for all chosen values of delay. In particular, they transiently reach the same protein expression value as the scenario \(\mathcal{X}_A\) where only activator is present. Therefore, this scenario seems to be independent of delay timing between TFs as long as the delay occurs in favour of the activator.
On the other hand, the difference between phenotypes in scenario \(\mathcal{X}_{R\rightarrow A}(\varDelta )\) is different for delay \(\varDelta =1\min \) than for delays \(\varDelta \in \{5\min ,15\min \}\). While for all three delays, the effect of long lived transients can be observed (the slope of approaching the limit value is small), the phenotype (protein expression around which the transients seem to stabilise) is different. It appears that, unlike delays longer than \(5\min \), the delay of \(1\min \) is not long enough for the population to repress protein expression to a value as low as in the scenario \(\mathcal{X}_R\) (where only repressor is present). In other words, the lowest gene expression value for delay of \(1\min \) is never as small as in the scenario \(\mathcal{X}_R\). To investigate the dependency of the transient phenotype on the delay, we simulated the scenario for several delay values between 1 sec and 5 min, namely \(\varDelta \in \{1{\mathsf {s}},20{\mathsf {s}},40{\mathsf {s}},60{\mathsf {s}},120{\mathsf {s}},180{\mathsf {s}}\}\) and we computed the amplitude for the scenario \(\mathcal{X}_{R\rightarrow A}\). The plot in Fig. 11 demonstrates that the amplitude approaches the value of \(\mathcal{X}_R\) scenario exponentially fast with increasing delay time.
Plotting the phenotypes for scenario \(\mathcal{X}_{A\rightarrow R}(\varDelta )\) for delays between 1 s and 1 min shows that the same activated gene expression levels are observed even for delays as small as 1 s (plots not shown). The explanation for different sensitivity of transient phenotypes to the delay in scenarios \(\mathcal{X}_{A\rightarrow R}(\varDelta )\) and \(\mathcal{X}_{R\rightarrow A}(\varDelta )\) are the different mechanisms implementing the activation and repression. When activator is input first, it quickly binds both operator sites and the probability of being in the looped active state almost instantaneously increases to the maximum value (as fast as within 1 s), and then starts decreasing only very slowly towards the equilibrium as soon as the repressor is present as well. On the other hand, when the repressor is input first, it does not bind both operator sites as quickly, because it is competing with the abundant \({\mathsf {RNAP}}\), even while the activator is not in the system. Only if there is enough time for the repressor to reach the looped repressed state with a probability nearly as high as in case of repressors only, the maximally repressed expression level will be observable. Otherwise, as soon as the activator is in the system, the probability of being in the looped repressed state starts shifting slowly towards the equilibrium point, and, consequently, the protein expression in the population starts increasing.
Long Lived Transients in Protein Expression Follow the Long Lived Transients (Mixing Times) in Promoter Activity. In Fig. 9, we plot the probability of the active regulatory configuration of the promoter for six different parameter combinations (listed in Table 1). Plots show the expected agreement with those in Fig. 8.
6 Discussion
Given that the processes of transcription and translation of a gene into a protein introduce intrinsic time delays and that there is no global synchronization among the arrival times of different molecular species at molecular targets, the simultaneous arrival of TFs in genetic circuits is a strong assumption. We subjected this assumption to a perturbation analysis, where the perturbed parameters are the relative arrival times of the TFs (different to the usual choices of perturbation parameters being the kinetic rates). We simulated a simple and realistic genetic circuit with two inputs and we showed that, in presence of small perturbations in the arrival of inputs (shorter than \(1\min \)), the circuit can exhibit three qualitatively distinct phenotypes which are stable for as long as any typical experiment would last (longer than 20 cell doubling-times). This has wide implications.
First, while our showcase example was constructed with the goal of demonstrating that long lived transients can appear in gene regulation, there are reasons to believe that many other gene regulatory schemes also exhibit long lived transients and implement multiple phenotypes by modulating the timing of inputs. To see this, consider that the number of potential phenotypes grows factorially with the number of inputs per gene as it is determined by the number of possible input orderings, meaning that, for instance, only 5 inputs would require us to analyse 5! = 120 different input scenarios. Moreover, our analysis indicates that long lived transients are possible in promoters with many configurations and certain states that are easy to reach but hard to exit. For instance, genomic regulation of the development of sea urchin embryo shows potential for long lived transients. The relevance of transient TF production has already been determined in this system [2, 44]: multiple TFs regulate a single gene which in turn has multiple targets, and there is clear differentiation between upstream and downstream components in the network. Therefore, considering long lived transients might clear up some puzzling observations like the discrepancy of TF interactions between endogenous promoters and minimal promoters controlled by three Endo16 regulatory modules [43].
Secondly, our proof-of-concept case study suggests that any modelling approach which assumes perfectly synchronous arrival of TFs or assumes rapid equilibrium at the promoter, may fail to explain a variety of phenotypes and raise false conclusions. To illustrate this point, think of an experimentalist who observes the system which seems equilibrated, but is a long lived transient (e.g. in our case study, a delay in favour of the activator occurs). Assuming that what she sees is an equilibrium, following the approach of statistical thermodynamics, she would proceed by estimating the free energies of binding configurations, but these estimates would be wrong, as the real equilibrium is much further away. Moreover, the obtained model would explain a single phenotype, and not the variety of quasi-stable phenotypes such as the ones we see in our showcase example. In summary, one cannot ignore the order of stimulating a cell, even when the GRN under consideration is assumed to be feedback-free. Similarly, one cannot assume what is observed towards the end of the life cycle is close to equilibrium even when the system seems relatively stable, e.g. growth at steady state in bulk, even when the stimulation was completed very early on. This opens further important questions such as how can an experimentalist who observes a stable phenotype for the chosen experiment duration, distinguish between a long lived transient and a real equilibrium? One immediate insight is the critical importance of experimentally measuring the kinetic rates as accurately as possible, and taking the timing of inputs into account.
Finally, our case study opens the possibility that GRNs are exploiting event timing to perform desired behaviours - it suggests that the cell does not compute with equilibrium dynamics - as is widely assumed in the field (with the exception of ‘well behaved’ limit cycle behaviours or pulsatile behaviour [22]), but uses the transients to react to stimuli and to memorise events. The \({\mathsf {DNA}}\) may be encoding more behaviours and thus phenotypes than an understanding based on the conventional input to output mapping suggests. In particular, as our analysis of delay timing between TFs shows (Fig. 11), a whole range of different stable gene expression levels can be encoded in the event timing of inputs. More broadly, this aspect may provide an explanation to why an organism can display so many more phenotypes, though the number of genes is limited, as the complexity of the organism increases, e.g. number of genes in bacteria and human vary by a factor of only 4!
Our primary goal was to show that a simple gene regulation without feedback, with realistic parameters, can exhibit long lived transients. We hypothesised that the promoter architecture with looping will have the desired feature, and, in order to find the feature, we performed a search over the 11-dimensional parameter space, which allowed us to display and discuss a range of parameterisations showing interesting behaviour. One of the compelling questions for future work is formalisation and computation of robustness of a given promoter architecture wrt. property of long-lived transients, as well as its sensitivity to a specific (group of) parameters. To this end, we believe that the ideas of parameter synthesis for stochastic chemical reaction networks, extensively studied in [4, 9, 10] (where the properties of the CTMC assigned to general biochemical reaction networks are expressed in continuous signalling logic (CSL)), would be a useful starting point.
Related Work. The consequences of combined effects of time delay and intrinsic noise on gene regulation has been studied in [45]. In more recent works [23], the authors elucidate the importance of relative timing of TF activation in combinatorial gene regulation with pulsatile signals. Like Lin et al. [23], our work shows that relative timing between TFs may be used by the cell to implement responses to different environments and therefore has to be taken into consideration for modelling gene expression patterns. However, while the authors in [23] suggest that the phenotypes differ in pulsatile regulation patterns, our study reveals the existence of long lived transients. From a dynamical system point of view, the effect of long lived transients that we present here can be seen through the prism of general theoretical frameworks such as proposed in [31, 42], where the authors discuss how to detect and automatically compute the meta-stable states from only the topology and timescales of the network; It would be interesting to see how precisely these methods could be used to detect the long lived transients we showcase in this paper.
Of relevance for synthetic biology, our construction based on looping suggests a way to implement memory units, though they may be leaky, in the sense that the signal is slowly being lost. In a broader context, cellular memory refers to systems whose present phenotype is dependent on the history of input stimuli and therefore the trajectory by which it has been reached [8]. The molecular mechanisms associated with such memory effects are usually based on feedback loops (e.g. the E. coli lac operon), \({\mathsf {DNA}}\) methylation patterns (e.g. temperate phage, pilus synthesis, cell differentiation) or inversions catalysed by site-specific recombinases (e.g. the Salmonella Hin system or the E. coli Fim system) [8, 29]. The long lived transient behaviour observed in our simulations differs from the mentioned memory mechanisms as it is purely relying on dynamical trapping of the transcriptional state. Different to the usual references to cellular memory, the long lived transients require no stabilisation of the phenotype through strong (covalent) modification of the DNA or any kind of feedback of the output on the promoter state (which is generally considered necessary for cellular memory).
The nature of the observed long lived transient states confer an epigenetic nature to these states. Methylation of histones is widely used in eukaryotic gene regulation as a modulator of gene activity that confers memory and stability to gene expression states. However, unlike methylation that requires a sleuth of specialised proteins that expend energy in order to form covalent bonds of methyl groups to histones, the long lived transients arise simply as a dynamical property of the system.
Notes
- 1.
Models used in this paper will count 23 and 6 distinct \({\mathsf {DNA}}\) binding configurations.
- 2.
The ratio between the binding and unbinding rate.
- 3.
Notice that these six parameter combinations are different than those used for the model with looping.
References
Bintu, L., et al.: Transcriptional regulation by the numbers: applications. Curr. Opin. Genet. Devel. 15(2), 125–135 (2005)
Bolouri, H., Davidson, E.H.: Modeling transcriptional regulatory networks. Bioessays: News Rev. Mol. Cell. Devel. Biol. 24(12), 1118–1129 (2002)
Boutillier, P., et al.: The Kappa platform for rule-based modeling. Bioinformatics 34(13), i583–i592 (2018)
Brim, L., Češka, M., Dražan, S., Šafránek, D.: Exploring parameter space of stochastic biochemical systems using quantitative model checking. In: Sharygina, N., Veith, H. (eds.) CAV 2013. LNCS, vol. 8044, pp. 107–123. Springer, Heidelberg (2013). https://doi.org/10.1007/978-3-642-39799-8_7
Brunner, M., Bujard, H.: Promoter recognition and promoter strength in the Escherichia coli system. EMBO J. 6(10), 3139 (1987)
Buchler, N.E., Gerland, U., Hwa, T.: On schemes of combinatorial transcription logic. Proc. Nat. Acad. Sci. 100(9), 5136–5141 (2003)
Byers, R., Hansell, R., Madras, N.: Stability-like properties of population models. Theor. Popul. Biol. 42(1), 10–34 (1992)
Casadesús, J., D’Ari, R.: Memory in bacteria and phage. BioEssays 24(6), 512–518 (2002)
Češka, M., Dannenberg, F., Paoletti, N., Kwiatkowska, M., Brim, L.: Precise parameter synthesis for stochastic biochemical systems. Acta Informatica 54(6), 589–623 (2017)
Česka, M., Šafránek, D., Dražan, S., Brim, L.: Robustness analysis of stochastic biochemical systems. PLoS ONE 9(4), e94553 (2014)
Danos, V., Feret, J., Fontana, W., Harmer, R., Krivine, J.: Rule-based modelling of cellular signalling. In: Caires, L., Vasconcelos, V.T. (eds.) CONCUR 2007. LNCS, vol. 4703, pp. 17–41. Springer, Heidelberg (2007). https://doi.org/10.1007/978-3-540-74407-8_3
Gillespie, D.: Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81, 2340–2361 (1977)
Guet, C.C., Elowitz, M.B., Hsing, W., Leibler, S.: Combinatorial synthesis of genetic networks. Science 296(5572), 1466–1470 (2002)
Harada, Y., Funatsu, T., Murakami, K., Nonoyama, Y., Ishihama, A., Yanagida, T.: Single-molecule imaging of RNA polymerase-dna interactions in real time. Biophys. J. 76(2), 709–715 (1999)
Hermsen, R., Tans, S., Ten Wolde, P.R.: Transcriptional regulation by competing transcription factor modules. PLoS Comput. Biol. 2(12), e164 (2006)
Jacob, F.: Evolution and tinkering. Science 196(4295), 1161–1166 (1977)
Jacob, F., Monod, J.: Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3(3), 318–356 (1961)
Kurtz, T.G.: Solutions of ordinary differential equations as limits of pure jump Markov processes. J. Appl. Probab. 7(1), 49–58 (1970)
Kurtz, T.G.: Limit theorems for sequences of jump Markov processes approximating ordinary differential processes. J. Appl. Probab. 8(2), 344–356 (1971)
Kwok, R.: Five hard truths for synthetic biology. Nature 463(7279), 288–290 (2010)
Levin, D.A., Peres, Y., Wilmer, E.L.: Markov Chains and Mixing Times. American Mathematical Society (2009)
Levine, J.H., Lin, Y., Elowitz, M.B.: Functional roles of pulsing in genetic circuits. Science 342(6163), 1193–1200 (2013)
Lin, Y., Sohn, C.H., Dalal, C.K., Cai, L., Elowitz, M.B.: Combinatorial gene regulation by modulation of relative pulse timing. Nature 527(7576), 54–58 (2015)
Marchisio, M.A., Stelling, J.: Automatic design of digital synthetic gene circuits. PLoS Comput. Biol. 7(2), e1001083 (2011)
McAdams, H.H., Arkin, A.: It‘sa noisy business! genetic regulation at the nanomolar scale. Trends Genet. 15(2), 65–69 (1999)
Milo, R., Jorgensen, P., Moran, U., Weber, G., Springer, M.: Bionumbers–the database of key numbers in molecular and cell biology. Nucleic Acids Res. 38(suppl 1), D750–D753 (2010)
Müller-hill, B.: Lac Operon. Wiley Online Library (1996)
Myers, C.J.: Engineering Genetic Circuits. CRC Press (2009)
Nashun, B., Hill, P.W., Hajkova, P.: Reprogramming of cell fate: epigenetic memory and the erasure of memories past. EMBO J. 34(10), 1296–1308 (2015)
Ptashne, M.: A Genetic Switch: Phage Lambda Revisited, vol. 3. Cold Spring Harbor Laboratory Press Cold Spring Harbor, New York (2004)
Radulescu, O., Swarup Samal, S., Naldi, A., Grigoriev, D., Weber, A.: Symbolic dynamics of biochemical pathways as finite states machines. In: Roux, O., Bourdon, J. (eds.) CMSB 2015. LNCS, vol. 9308, pp. 104–120. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-23401-4_10
Saiz, L., Rubi, J.M., Vilar, J.M.G.: Inferring the in vivo looping properties of DNA. Proc. Nat. Acad. Sci. U.S.A. 102(49), 17642–17645 (2005)
Saiz, L., Vilar, J.M.: DNA looping: the consequences and its control. Curr. Opin. Struct. Biol. 16(3), 344–350 (2006). Nucleic acids/Sequences and topology Anna Marie Pyle and Jonathan Widom/Nick V Grishin and Sarah A Teichmann
Saiz, L., Vilar, J.M.: Stochastic dynamics of macromolecular-assembly networks. Mol. Syst. Biol. 2(1) (2006)
Santillán, M., Mackey, M.C.: Why the lysogenic state of phage \(\lambda \) is so stable: a mathematical modeling approach. Biophys. J. 86(1), 75–84 (2004)
Schnoerr, D., Sanguinetti, G., Grima, R.: Approximation and inference methods for stochastic biochemical kinetics–a tutorial review. J. Phys. A: Math. Theor. 50(9), 093001 (2017)
Segal, E., Widom, J.: From dna sequence to transcriptional behaviour: a quantitative approach. Nat. Rev. Genet. 10(7), 443–456 (2009)
Shea, M.A., Ackers, G.K.: The OR control system of bacteriophage lambda: a physical-chemical model for gene regulation. J. Mol. Biol. 181(2), 211–230 (1985)
Vilar, J.M., Leibler, S.: DNA looping and physical constraints on transcription regulation. J. Mol. Biol. 331(5), 981–989 (2003)
Vilar, J.M., Saiz, L.: Dna looping in gene regulation: from the assembly of macromolecular complexes to the control of transcriptional noise. Curr. Opin. Genet. Devel. 15(2), 136–144 (2005)
Vilar, J.M., Saiz, L.: Suppression and enhancement of transcriptional noise by DNA looping. Phys. Rev. E 89(6), 062703 (2014)
Vivek-Ananth, R., Samal, A.: Advances in the integration of transcriptional regulatory information into genome-scale metabolic models. Biosystems 147, 1–10 (2016)
Yuh, C.H., Bolouri, H., Davidson, E.H.: Cis-regulatory logic in the endo16 gene: switching from a specification to a differentiation mode of control. Devel. (Cambridge, England) 128(5), 617–629 (2001)
Zeller, R.W., Griffith, J.D., Moore, J.G., Kirchhamer, C.V., Britten, R.J., Davidson, E.H.: A multimerizing transcription factor of sea urchin embryos capable of looping DNA. Proc. Nat. Acad. Sci. 92(7), 2989–2993 (1995)
Zhu, R., Salahub, D.: Delay stochastic simulation of single-gene expression reveals a detailed relationship between protein noise and mean abundance. FEBS Lett. 582(19), 2905–2910 (2008)
Acknowledgements
We are very grateful to Moritz Lang, Tiago Paixao and Jakob Ruess, for their feedback during the manuscript preparation.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Appendices
Appendix 1.A Parameter Values
Table 1 lists the parameter ranges used for our case study example. We next explain the choice of each of the parameters with respect to their biological context.
1.A.1 Stochastic Scaling Constant
The stochastic scaling of rates and concentrations is done with a standard scaling rate for E. coli cell \(N= 10^9\) [26].
1.A.2 Protein Production and Degradation
The protein production is taken 0.5 molec.s\(^{-1}\) ([41], caption of Fig. 2) and the degradation rate is taken 0.001 s\(^{-1}\) (corresponding to the halflife of \(12\min \), consistent with [26]).
1.A.3 RNAP Rates
On rate, off rate and number of RNAP molecules are consistent with the orders of values reported in [5, 14, 35].
1.A.4 Activator
The activation mechanism is inspired by the activation of the \(\mathsf {PRM}\) promoter in the lysogenic state by protein \({\mathsf {CI}}\) in the regulation of \(\lambda \)-phage: \({\mathsf {CI}}\) competes with \({\mathsf {Cro}}\) to bind to the promoter sites, and, when bound, it recruits \({\mathsf {RNAP}}\) (increases \(\mathsf {PRM}\) activity). The mechanism with looping, explained at mechanistic detail level in [35], contains three left and three right operators, leading to 1200 different DNA binding states. We model a mechanism with two states for the activator without looping (‘bound’ or ‘not bound’) and with four binding states for the activator with looping (see Fig. 5). The on-rate, off-rate well as the number of activators is taken from [35] (page 82) When activator is bound, the recruitment of \({\mathsf {RNAP}}\) is increased by factor 10 or 50 ([34] and [27] respectivelly) (Table 3).
1.A.5 Repressor
The repression mechanism is inspired by the well-studied transcriptional regulation, there is a word missing after transcription of the \({\mathsf {lac}}\) operon, the repressor \({\mathsf {LacI}}\). We take the binding and unbinding rates for the repressor from ([39], Fig. 4).
1.A.6 Looping Rates
The stability of the looped state is incorporated in the model by scaling down the unlooping rate. We choose the scaling factors of 100 and 1000 based on the computation of the ratio of dissociation rates for the models with and without looping ([40], Table 1; parameter a in [39]). The mechnism proposed in, eg. [39] suggests that the looping increases the binding rate (due to increased local concentration of TFs), while leaving the unbinding rate unchanged. As the scaled on-rates may exceed theoretical limit for diffusion-limited reactions, in our model, we incorporate the same effect by leaving the binding rate identical, and scaling down the unlooping rate.
Appendix 1.B Kappa Models




Appendix 1.C Supplementary Theory and Proofs
1.C.1 Deterministic Limit
In the continuous, deterministic model of a chemical reaction network, the state \(\mathbf {z}(t) = (z_1,\ldots ,z_n)(t)\in {\mathbb R}^n\) is represented by listing the concentrations of each species. The dynamics is given by a set of differential equations in form
where \(k_j\) is a deterministic rate constant, computed from the stochastic one and the volume \(N\) according to \(k_j := c_jN^{|{\mathbf {a}}_{j}|-1}\) (\(|\mathbf {x}|\) denotes the 1-norm of the vector \(\mathbf {x}\)). The deterministic model is a limit of the stochastic model when all species in a reaction network are highly abundant [19]: by scaling the species multiplicities with the volume: \(Z_i(t) = X_i(t)/N\), adjusting the propensities accordingly, in the limit of infinite volume \(N\rightarrow \infty \), the scaled process \({\mathbf Z}(t)\) follows an ordinary differential Eq. (4).
1.C.2 Expected Output in the Transient
The CME implies that the expectation of the marginal distribution of \(\{X_t\}\) satisfies the equations
To check (5), observe a transition from \(\mathbf {x}\) to \(\mathbf {x}+\varvec{\nu }_j\). The term \(\lambda _{j}(\mathbf {x})\mathsf {P}(\mathbf {X}_t=\mathbf {x})\) appears exactly once when summing up for the state \(\hat{\mathbf {x}} = \mathbf {x}\) as the outflow probability, and exactly once when summing up for the state \(\hat{\mathbf {x}} = \mathbf {x}+\varvec{\nu }_j\), as the inflow probability. This gives the term \((\mathbf {x}+\varvec{\nu }_j)-\mathbf {x}=\varvec{\nu }_j\cdot \lambda _{j}(\mathbf {x})p^{(t)}(\mathbf {x})\). It is worth noting that, upon scaling the rate constants, the equations for \(\mathsf {E}(\mathbf {X}_t)\) are equivalent to (4) only if all rate functions are linear, that is, when all reactions are unimolecular.
1.C.3 Proof for Lemma 1
We first notice that the process \(\mathbf {X}(t)|_{\mathsf {D}}\) is indeed Markovian, because all states of \(\mathbf {X}(t)\) projected to the same state in \(\mathbf {X}(t)|_{\mathsf {D}}\) are behaviourally indistinguishable (bisimulation equivalent), due to rates between lumped states not depending on protein count. From (5), it follows that

where \(\langle x_{\mathsf {D}1j}(t)\rangle \) denotes the expected value of being in one of the active promoter configurations. The latter equals (2), since in every reachable state \(\mathbf {x}\in (D_{\texttt {0}}\uplus D_{\texttt {1}})\times \{0,1,\ldots \}\), exactly one \({\mathsf {DNA}}\) configuration takes value 1.
Appendix 1.D Supporting Figures
See Fig. 13.
Transitions of the CTMC underlying basal gene expression. The state space \(S\cong \{\texttt {0},\texttt {1}\}\times \{0,1,2,\ldots \}\), such that \(s_{\texttt {1},x}\) denotes an active configuration (where the \({\mathsf {RNAP}}\) is bound to the \({\mathsf {DNA}}\)) and \(x\in \mathbf N\) protein molecules.
Two prototype GRNs and their promoter logic: (a) Model without looping: regulatory architecture (promoter logic), (b) Model with looping: regulatory architecture (promoter logic). Mechanistic models are listed in (Appendix 1.B).
Model with looping: CTMC regulating the \({\mathsf {DNA}}\) configurations has 23 different states. It is naturally represented as a composition of two sub-models: (left) the switching among configurations with respect to activator binding to its main and auxiliary binding sites (\(\mathsf {O_{Am}}\) and \(\mathsf {O_{Ax}}\) respectively, and (right) the switching among configurations with respect to repressor binding to its main and auxiliary binding sites (\(\mathsf {O_{Am}}\) and \(\mathsf {O_{Ax}}\) respectively. The unlooping rates (thicker blue lines) are typically much weaker than the TF unbinding. Any combination of the states in the two sub-models can be observed (reachable), except the state where both repressor and activator are looped. (Color figure online)
The effect of kinetic parameters on the shape and duration of transients for a model with looping (for six parameter values listed in Table 1 and time delays of \(1\min \), \(5\min \) and \(15\min \) respectively).
Long lived transients in protein expression follow the long lived transients (mixing times) in promoter activity. We plot the statistically inferred probability of promoter logic being in the active state for six different parameter combinations, listed in Table 1 and time delays of \(1\min \), \(5\min \) and \(15\min \) respectively.
Long lived transients are not observed in the model without looping (for six parameter values listed in Table 1 and time delays of \(1\min \), \(5\min \) and \(15\min \) respectively).
In the reference parameter set, when there is no delay (\(\varDelta = 0\)), the phenotype in scenario \(\mathcal{X}_{R\rightarrow A}(\varDelta )\) is equal to the one in scenario \(\mathcal{X}_{A || R}\), visibly different than the phenotype \(\mathcal{X}_R\) (290 protein molecules). The difference of from the scenario \(\mathcal{X}_{A || R}\) (the characteristic we formally termed amplitude – see Sect. 5) exponentially grows as the delay increases, that is, it quickly approaches the phenotype of scenario \(\mathcal{X}_R\). The difference of \(\mathcal{X}_{R\rightarrow A}(\varDelta )\) from \(\mathcal{X}_R\) becomes observably negligible already for delays larger than \(\varDelta \) = 5 min = 300 s (difference of 10 molecules, \(0.035\%\) of the initial difference). We obtained the dependency by fitting the data obtained by simulating the system for \(\varDelta \in \{1,20,40,60,120,180,240,300,600,900\}\).
We define a more global measure of the effect of delay in arrival times of TFs: first, the range of values that can be observed with a delay \(\varDelta = 5\min \) by the measuring the maximum distance between phenotypes the scenarios \(\mathcal{X}_{A\rightarrow R}\) and \(\mathcal{X}_{R\rightarrow A}\) \(\beta : = \max _{t \ge t_0} |x_p(t|\mathcal{X}_{A\rightarrow R})-x_p(t|\mathcal{X}_{R\rightarrow A})|\), and secondly, the halflife of this range \(t_{1/2}: = \arg \min _{t\ge t_{\beta }}\{t\mid |x_p(t|\mathcal{X}_{A\rightarrow R})-x_p(t\mid \mathcal{X}_{R\rightarrow A})|<\frac{1}{2}\beta \}\).
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Guet, C., Henzinger, T.A., Igler, C., Petrov, T., Sezgin, A. (2019). Transient Memory in Gene Regulation. In: Bortolussi, L., Sanguinetti, G. (eds) Computational Methods in Systems Biology. CMSB 2019. Lecture Notes in Computer Science(), vol 11773. Springer, Cham. https://doi.org/10.1007/978-3-030-31304-3_9
Download citation
DOI: https://doi.org/10.1007/978-3-030-31304-3_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-31303-6
Online ISBN: 978-3-030-31304-3
eBook Packages: Computer ScienceComputer Science (R0)