Abstract
In breast cancer diagnosis, fine needle aspiration biopsy is an important diagnostic tool. It is used to estimate cancer malignancy grade that is further required for treatment determination. In this paper we describe a scheme based on pattern recognition and image processing techniques for automatic breast cancer malignancy grading from cytological slides of fine needle aspiration biopsies. To determine a malignancy classification of the slide we propose to extract textural features of nuclei with an application of local binary patterns. Based on texture determination, we present an improved classification system for cancer malignancy grading.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
In recent years computerized cancer diagnosis plays a crucial role in cancer examination procedure. According to the Polish National Cancer Registry [1], in Poland alone, the number of reported cancer cases doubled in the last three decades. According to statistics, around 300 000 middle-aged women in European Union will be diagnosed with breast cancer. This makes it the highest oncological problem affecting developed countries. About 89 000 of cases will be fatal. To reduce this high ratio, a number of computer aided techniques were developed to make the process more reliable and faster. The aim here is to diagnose a cancer is an early development stage. Cancers in their early stages are more vulnerable to treatment and we can assume that most of the diagnosed cases will lead to a successful recovery. Conversely, most advanced cancers stages are usually almost impossible to treat.
To overcome the problem of late diagnosis, screening mammographic tests were introduced and when a suspicious region in the image is noted, a fine needle aspiration biopsy (FNA) is taken. FNA is a minimally invasive method to extract a small tissue sample of the questionable breast tissue. This procedure allows for the description of the type and malignancy grade of the cancer. Depiction of malignancy plays a crucial role in the determination of patient’s treatment.
Cancer malignancy is graded according to the three point scale that was first proposed by Bloom and Richardson in 1957 [4]. The proposed scheme was originally derived for assessment of malignancy from histopathological slides and today is very popular among pathologists. They use it for grading not only histological but also cytological tissue.
In literature we can find a numerous approaches to computerized cancer diagnosis [11]. Most of these approaches deal with the classification between benign and malignant cases [12] of histological tissue. To the best of authors knowledge, the first description of the computerized breast cytology classification problem was provided by Wolberg et al. in 1990 [21]. In this work, authors presented an application of a multi-surface pattern separation method applied to cancer diagnosis. Their idea was wildly propagated among researchers what led to the description of other breast cancer computer aided systems. In 2009, Malek et al. [15] described an application of active contours to nuclei segmentation and a fuzzy c-means classifier for classification of 80 malignant and 120 benign cases and in 2010, Niwas et al. provided a description of texture features for cancer classification [16]. In 2011, Jeleń et al. [13] described a neural network approach to breast cancer malignancy classification and in 2016, Jeleń et al. [12] provided a wide description of morphological features used for breast cancer classification. In that same work they provided an extended research on feature classification power and selection providing information about the best performing features.
Most recent approaches include a deep learning and convolutional neural network approaches. In 2017, Araújo et al. [3] used convolutional neural networks for feature extraction. In 2018, Kowal et al. [14] presented a similar study to this described in [12] on feature selection problem. Authors used convolutional neural networks for segmentation and have performed feature selection test on 500 images of benign and malignant cases.
For complete cancer diagnosis it is necessary to also determine the malignancy stage of cancer which is called a malignancy grading, and this is the problem that we are focusing on in this paper. The pre-screening process before taking an FNA results in the situation in which the biopsy slide being classified is nearly always malignant. Henceforth, in this work, we deal with a malignancy grading problem instead of malignancy diagnosis. In 2017, Alsaedi et al. [2] described six computer-aided grading frameworks assigning malignancy grades to cytological images of FNA biopsies of breast cancer.
In this paper we address an important issue of texture features that can be used for computer-aided grading of cancer malignancy. Texture is a very valuable source of information when analyzing structures in an image [9, 19] and therefore, a textural description of the nucleus is an important feature of the computerized breast cancer malignancy grading system. According to Bloom and Richardson textural analysis allows for the determination of the chromatin distribution and detection of mitotic cells.
The aim of the described study is to present an improved classification scheme that takes into consideration a textural description of nuclei in the slide and will determine the malignancy of breast cancer tissue. Additionally we will describe a set of morphological features that are used for comparison purposes.
2 Database Description
The database used in this study includes images of fine needle aspiration biopsy that were collected by dr. Artur Lipiński during an FNA examination at the Department of Pathology and Oncological Cytology of the Medical University of Wrocław, Poland. Slide preparation technique includes staining with Haematoxylin and Eosin, known as HE technique. The choice of the staining agents allows for visualizing a nuclei with purple and black dyes, cytoplasm with shades of pink and red blood cells with orange/red dyes. The setup consisted of an Olympus BX 50 microscope with CCD–IRIS camera mounted on the head of the microscope and a MultiScan Base 08.98 software. Such a system allowed for recording of images with the resolution of 96 dots per inch (dpi) and a size of 764 \(\times \) 572 pixels.
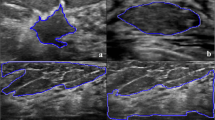
There are 218 images in the database that represent two classes of cancer malignancy, namely intermediate (G2) and high (G3) malignancy grades (see Fig. 1). The lack of low malignancy images is caused by the fact that these cases very rarely require FNA and in recent years there was only a few of them at the Medical University of Wrocław. There are 134 cases of intermediate and 84 of high malignancy. For all cases, breast tissue was surgically removed during follow-up biopsy and it was histopathologically graded using the Bloom–Richardson [4] grading scale which confirmed the FNA grading. Therefore all the cases in our database were histopathologically validated.
3 Methods
Recently a number of classification frameworks were presented [2]. All of them use shape descriptors to represent relevant Bloom-Richardson features. In [12] except for features definition, authors preformed a comparison of their discriminatory powers to choose a subset of the best performing features. According to the authors, there are two types of features required for proper malignancy classification. These features are described in Sect. 3.2. To correctly classify breast cancer malignancy we need to calculate features at two different magnification of the slide. Lower magnification will allow for the determination of cells behavior and their ability to form groups. This simply means if there are more loosely spread cells in the image, the more malignant the case should be. On higher magnification we will be able to determine cells morphology as well as textural differences between nuclei.
In this paper we propose to use only texture description to extract features from images of higher magnification images. In Sect. 3.1 we will describe Local Binary Patterns that were used as a texture measure. Last part of this section is devoted to the description of classification scheme that was used for the evaluation of the textural features performance.
3.1 Texture Description
Textural features are used to measure the texture information of the image [9, 19]. Here, the texture of the nucleus is taken into consideration. To extract textural features, a local binary patterns method was applied. This method was first proposed by He and Wang in 1990 [20]. Here, we use a variation of the method that was described by Ojala et al. in 2002 [17]. They described an efficient approach based on the local binary patterns and nonparametric discrimination of sample and prototype distributions to rotation invariant texture classification. Authors defined a texture Tx of a graylevel image in a local neighborhood as a joint distribution of gray levels as represented in Eq. 1.
where \(g_c\) is a gray level associated with the center of neighborhood, \(g_p\) for \(p\in \) {0, ..., P − 1} is a gray level associated with P pixels arranged in circular manner, equally spaced on a circle with a defined radius.
To obtain the Gray–Scale invariance authors subtracted the gray level value of the neighborhood center from gray level values of the neighborhood pixels and further scaling of the gray scale using only signs of the differences and not the exact values. This situation is represented by Eq. 2.
where
According Ojala et al. adding a \(2^p\) binomial factor to each scaling term, the characterization of the local spatial image texture structure can be rewritten by Eq. 4.
To further remove the effect of rotation, authors defined a following relation:
where ROR(x, i) is a circular bit-wise right shift on the P-bit number x i times, which means clockwise rotation of the neighborhood until maximal number of the most significant bits will be 0.
3.2 Morphological Features
As mentioned previously, there are two image magnifications that are used for breast cancer malignancy grading. Morphological feature extraction in this paper takes both of these types into consideration. The magnifications of images corresponds to the magnifications used during a cytological examination of a breast tissue. For the calculation of the low magnification features, images recorded with 100x magnification are used and for the calculation of high magnification features, images obtained with 400x magnification are used.
Low Magnification Features (LM) – these features are defined based on the number of groups and their area (\(A_{100}\)). The first feature is calculated as a number of groups in the image that weren’t removed during the segmentation process and the area is calculated as the average number of nuclei pixels. The third feature describes nuclei dispersion within an image and is defined as a variation of cluster areas (\(A_c\)) which is determined by the following equation [12]:
High Magnification Features (HM) – the feature vector constructed by extraction of high magnification features includes 30 features calculated according to [12]. These features are divided into binary, momentum, histogram, textural and color based features.
Binary features that were calculated for set of nuclei \(N = \{ N_1,N_2,..., N_n \}\) from a binary image (\(\mathcal{I}\)), where N is defined as a collection of all connected components and the nucleus \(N_{i}\). Using this definition, the following features are extracted [12]:
-
Area (\(A_{400}\mathbf{)}\) – \(A_i\) is defined as the sum of all nuclei pixels of the nucleus \(N_i\).
-
Perimeter (\({Perim}\mathbf{)}\) – is a length of the nuclear boundary of a nucleus \(N_i\) that is approximated by a length of the polygonal approximation of the boundary.
-
Convexity (\({Conv}\mathbf{)}\) – is defined as a ratio of the nucleus area and the area of the minimal convex polygon that contains the nucleus, called a convex hull.
-
Eccentricity (\(Ecc\mathbf{)}\) – calculated as a ratio of the distance between focal points of an ellipse matched with a nucleus having the same second–moments as the segmented nuclei, and its major axis length.
-
Centroid (\((x-Ctr, y-Ctr)\mathbf{)}\) – For each nucleus, the centroid (\(Ctr_i\)) is a point \((\overline{x}_i, \overline{y}_i)\) called a center of mass of the extracted nucleus along each row (X) and column (Y).
-
Orientation (\(Or\mathbf{)}\) – For the coordinate system placed at the centroid \((\overline{x}_i, \overline{y}_i)\) of the nucleus the orientation is defined as
$$\begin{aligned} Or_i = tan(2\theta _i), \end{aligned}$$(6)where the angle \(\theta _i\) is measured counterclockwise from the x–axis.
-
Projection ((x-Prj, y-Prj)) – to calculate the feature \(Prj_i\) for each nucleus, projections along rows (x-Prj\(_i\)) and columns (x-Prj\(_i\)) are performed.
Momentum–based features use normalized central moments, \(\eta _{ij}\) to calculate rotation, scaling and translation invariant features as described in [19]. Using these \(\eta _{ij}\) values, seven momentum–based features, were calculated.
Another set of features is histogram based. The histogram is treated as a probability distribution function of grey level values from the red channel in the image and describes their frequencies in that image [12]. Based on this definition, five histogram-based features were calculated. Additionally statistical features of a texture were calculated based on the gray level co–occurrence matrix that describes the relationships between a pair of pixels and their grey levels [19]. Assuming that the distance between the pixels and the directions are given four textural features were extracted.
Last set of features is based on the spherical coordinate transform applied to the RGB image. Determining a histogram of the converted image 5 color–based features were computed.
3.3 Malignancy Classification
To classify the cytological FNA tissue we build four classifiers that take a feature vector (see Sect. 3.2) as an input and respond with a two element output vector \((1,0)^T\) for intermediate malignancy and \((0,1)^T\) for high malignancy. In the remainder of this section, classification methods are presented and, in the following section, their ability to classify malignancies is studied.
Decision Trees (Tree) – most of the traditional pattern recognition algorithms are based on the feature vectors that are real–valued and some kind of metric can be applied to them [6]. Tree classifiers on the other hand are able to solve classification problems that involve nominal data such as a list of attributes like fruit colors and sizes.
Decision trees are constructed in a way where the classes are held in the leaves of the tree and the decision rules are kept in the internal nodes including the root [18]. Classification with decision trees seeks a path from the root to the correct leaf creating a decision path. Here, we make use of the CART (Classification And Regression Trees) method described by Breiman et al. [5] that provides a general framework for decision tree construction. In general, the tree–growing process declares the node to be a leaf or finds another property that can be used to split the data represented at the node into subsets creating new nodes. This is process is run recursively until all the data is represented by the constructed tree.
Linear Discriminant Analysis (LDA) – is one the simplest classification algorithms that requires a construction of a, so called, decision boundary. This boundary is constructed as discriminant function of a form presented by [6]:
where w is a weight vector and \({\omega }_0\) is a threshold weight.
In a general way we can rewrite the the decision boundary as:
Here, \({\omega }_i\) represents the components of w. If we add to Eq. 8 terms related to products of pair of x, a definition of a quadratic discriminant function (Eq. 9) will be determined.
To train an LDA based classifier we need to calculate a decision boundary according to Eq. 9.
Support Vector Machines (SVM) – are used to separate two or more classes of patterns or data points by constructing a boundary between them [10]. An unknown point will be classified according to its orientation with respect to the boundary. To estimate the boundary between classes we support vectors from each class. This is performed as an iterative approach that minimizes an error function.
with the following restrictions:
where C and b are constants, w is the weight vector, \(\varepsilon _{i}\) is a bias value that deals with overlapping cases and \(\phi \) is a kernel function that transforms input data into the feature space. In this work we have used a linear kernel as described by Huang et al. in [10].
Classifiers Ensembles (Ensemble) – enhance a single–model classifiers offering hybrid approaches [8] for a better classification of two-class classification problems. Here, we use an AdaBoost. M2 approach, which is defined for a multi-class problems. The algorithm seeks an approximation of the Bayes classifier B(x), where x is a set of observations, based on a combination of other classifiers such as decision trees [7]. During training, weights of misclassified points are increased and a second classifier in the ensemble will use these new weights. This approach is called boosting. Such a procedure is iterated and can lead to a creation of numerous classifiers [22]. After training, each built classifier will be assigned with a score. Such a combination of classifiers will define an AdaBoost classifier as a classification model. All the classifiers in the final model are linearly connected.
4 Results and Discussion
In this paper we discuss a problem of malignancy classification of fine needle aspirates. For this purpose we have calculated textural features based on local binary patterns as described in Sect. 3.1. For our calculations we have used 8 pixels neighborhood. Additionally a set of morphological features proposed by Jeleń et al. [12] was extracted as described in Sect. 3.2.
Obtained feature vectors were presented as an input to four different classifiers to check their ability to distinguish between two malignancy classes: G2 and G3. Tests were performed using a 5–fold cross–validation method. Presented Table 1 classification results are averages over the 5 folds used for testing.
To perform a complete analysis of the texture description, we have calculated local binary patterns not only to a graylevel image (LBP features) but additionally for a combination of R, G and B channels. Table 1 summarizes obtained classification results for these features also. Further analysis shows that red and blue channel together provided the best classification of malignancy cases. Additionally we included morphological features of low magnification images to show the performance of a complete malignancy classification scheme.
From the results presented in this section we can see that classification based only on the morphological features from high magnification images provided the worst results. Textural features alone outperformed them but when low magnification features were taken into consideration, the proposed scheme was able to classify malignancy with accuracy around 90% or better. Inclusion of low magnification features to the local binary patterns calculated for a red and blue channel provided the best classification with an accuracy of 97.6% for an SVM classifier. From Table 1 we can also see that this classifier outperforms all other tested classifiers for textural features. Classification results obtained for morphological features clearly show that classifier ensembles perform much better.
5 Conclusions and Further Work
In this paper we conducted a research on malignancy grade classification based on textural features calculated with local binary patters. We have compared classification accuracies of texture features extracted for different color channels and graylevel image against accuracies obtained for classification of morphological features. As described in the previous section, the best performing scheme included local binary patterns with low magnification features yielding an accuracy of 97.6% when SVM classifier was used. The worst performing setup included only morphological features extracted form high magnification features. Considering the above, we can conclude that textural representation of fine needle aspirates would be very good choice for breast cancer malignancy classification.
In the further research the classification framework that includes images for all three malignancy classes, should be tested. Introducing the low malignancy class will make the classification more difficult and therefore even a more interesting problem. Also, it would be necessary to perform per nuclei analysis but the problem of occluding cells needs to be addressed and taken into consideration.
References
Polish National Cancer Registry, December 2019
Alsaedi, M., Fevens, T., Krzyżak, A., Jeleń, Ł.: Cytological malignancy grading systems for fine needle aspiration biopsies of breast cancer. In: 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp. 705–709 (2017)
Araújo, T., Aresta, G., Castro, E., Rouco, J., Aguiar, P., Eloy, C., Polónia, A., Campilho, A.: Classification of breast cancer histology images using convolutional neural networks. PLOS ONE 12(6), 1–14 (2017)
Bloom, H.J.G., Richardson, W.W.: Histological grading and prognosis in breast cancer. Br. J. Cancer 11, 359–377 (1957)
Breiman, L., Friedman, J., Stone, C.J., Olshen, R.A. (eds.): Classification and Regression Trees. Chapman & Hall, Boca Raton (1993)
Duda, R.O., Hart, P.E., Stork, D.G.: Pattern Classification, 2nd edn. Wiley Interscience Publishers, New York (2000)
Freund, Y., Schapire, R.E.: A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 55(1), 119–139 (1997)
Galar, M., Fernandez, A., Barrenechea, E., Bustince, H., Herrera, F.: A review on ensembles for the class imbalance problem: bagging-, boosting-, and hybrid-based approaches. IEEE Trans. Syst. Man Cybern. Part C Appl. Rev. 42(4), 463–484 (2012)
Haralick, R.M.: Statistical and structural approaches to texture. IEEE Trans. Syst. Man Cybern. 67(5), 786–804 (1979)
Huang, T.M., Kecman, V., Kopriva, I.: Kernel based algorithms for mining huge data sets: supervised, semi-supervised, and unsupervised learning. Springer (2006)
Irshad, H., Veillard, A., Roux, L., Racoceanu, D.: Methods for nuclei detection, segmentation, and classification in digital histopathology: a review-current status and future potential. IEEE Rev. Biomed. Eng. 7, 97–114 (2014)
Jeleń, Ł., Krzyżak, A., Fevens, T., Jeleń, M.: Influence of feature set reduction on breast cancer malignancy classification of fine needle aspiration biopsies. Comput. Biol. Med. 79, 80–91 (2016)
Jeleń, Ł., Lipiński, A., Detyna, J., Jeleń, M.: Grading breast cancer malignancy with neural networks. Bio-Algorithms Med. Syst. 2(14), 47–54 (2011)
Kowal, M., Skobel, M., Nowicki, N.: The feature selection problem in computer assisted cytology. Int. J. Appl. Math. Comput. Sci. 28(4), 759–770 (2018)
Malek, J., Sebri, A., Mabrouk, S., Torki, K., Tourki, R.: Automated breast cancer diagnosis based on GVF-snake segmentation, wavelet features extraction and fuzzy classification. J. Signal Process. Syst. 55(1–3), 49–66 (2009)
Niwas, S.I., Palanisamy, P., Sujathan, K.: Complex wavelet based texture features of cancer cytology images. In: 2010 International Conference on Industrial and Information Systems (ICIIS), pp. 348–353 (2010)
Ojala, T., Pietikainen, M., Maenpaa, T.: Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans. Pattern Anal. Mach. Intell. 24(7), 971–987 (2002)
Safavian, S.R., Landgrebe, D.: A survey of decision tree classifier methodology. IEEE Trans. Syst. Man Cybern. 21(3), 660–674 (1991)
Umbaugh, S.E.: Digital Image Processing and Analysis: Human and Computer Vision Applications with CVIPTools, 2nd edn. CRC Press, New York (2011)
Wang, L., He, D.-C.: Texture classification using texture spectrum. Pattern Recognit. 23(8), 905–910 (1990)
Wolberg, W.H., Mangasarian, O.L.: Multisurface method of pattern separation for medical diagnosis applied to breast cytology. Proc. Natl. Acad. Sci. U.S.A. 87, 9193–9196 (1990)
Zhu, J., Rosset, S., Zou, H., Hastie, T.: Multi-class adaboost. Stat. Interface 2, 349–360 (2006)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Jeleń, Ł. (2020). Texture Description for Classification of Fine Needle Aspirates. In: Korbicz, J., Maniewski, R., Patan, K., Kowal, M. (eds) Current Trends in Biomedical Engineering and Bioimages Analysis. PCBEE 2019. Advances in Intelligent Systems and Computing, vol 1033. Springer, Cham. https://doi.org/10.1007/978-3-030-29885-2_10
Download citation
DOI: https://doi.org/10.1007/978-3-030-29885-2_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-29884-5
Online ISBN: 978-3-030-29885-2
eBook Packages: EngineeringEngineering (R0)