Abstract
The eukaryotic group II chaperonin TRiC/CCT assists the folding of 10% of cytosolic proteins including many key structural and regulatory proteins. TRiC plays an essential role in maintaining protein homeostasis, and dysfunction of TRiC is closely related to human diseases including cancer and neurodegenerative diseases. TRiC consists of eight paralogous subunits, each of which plays a specific role in the assembly, allosteric cooperativity, and substrate recognition and folding of this complex macromolecular machine. TRiC-mediated substrate folding is regulated through its ATP-driven conformational changes. In recent years, progresses have been made on the structure, subunit arrangement, conformational cycle, and substrate folding of TRiC. Additionally, accumulating evidences also demonstrate the linkage between TRiC oligomer or monomer and diseases. In this review, we focus on the TRiC structure itself, TRiC assisted substrate folding, TRiC and disease, and the potential therapeutic application of TRiC in various diseases.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
Many proteins need assistance to fold into their native state and achieve functional activity after release from ribosome into the crowded environment of the cell (Fig. 19.1) (Kim et al. 2013). Correspondingly, cells have evolved a system of molecular chaperones and quality control machineries, often called the “protein homeostasis” network. Proteins from this network function to bind unfolded or misfolded polypeptides and induce their folding, sequester them, or facilitate their degradation (Kim et al. 2013; Willison 2018). The chaperonins, a critical group of molecular chaperones, are large double-ring complexes of 800–1000 kDa built of 7–9 subunits per ring, and each ring provides a central cavity for ATP-driven chaperonin-mediated protein folding cycle (Hartl and Hayer-Hartl 2002; Horwich et al. 2007; Willison 2018).
Proposed TRiC-mediated substrate folding pathway. The scheme shows the downstream protein folding pathway from the ribosome. Hsp70 serves as the hub of the chaperone network, which cooperates with Hsp40s and nucleotide exchange factors (NEFs) to fold ~20% of the proteome. TRiC binds and helps fold ~10% of cytosolic proteins directly or with the assistance of cofactors (prefoldin, Hsp70) for efficient substrate delivery. TRiC-mediated substrate folding is regulated through its ATP-driven conformational changes. Then substrate is released from the central cavity of TRiC to achieve its native state, which sometimes needs the help of cofactor, such as Phlp1. In addition, eukaryotes also engage the Hsp90 chaperone system to catalyze the activation of metastable proteins via the Hsp70 system and the co-chaperone Hop
Chaperonins have been classified into two groups (Horwich and Willison 1993), including group I chaperonins presented in prokaryotes (GroEL/GroES), mitochondria (Hsp60/Hsp10), and chloroplasts (Rubisco subunit binding protein), and group II chaperonins presented in archaea (thermosome, Mm-cpn) and eukaryotes (TCP-1 ring complex, TRiC, or chaperonin-containing TCP-1, CCT) (Fig. 19.2a–c). Group I chaperonin consists of a detachable co-chaperonin, while group II chaperonin has a build-in lid in the apical domain that can shut close the chamber (Fig. 19.2a–c). The archaea group II chaperonin has eight or nine subunits per ring, while the eukaryotic group II chaperonin has eight paralogous subunits. The eukaryotic group II chaperonin TRiC/CCT (~960 kDa) comprises of two hetero-oligomeric stacked rings with eight distinct subunits each in a certain arrangement. The paralogous subunits share about 23–35% and 27–39% sequence identity for yeast and bovine TRiC, respectively (Frydman et al. 1992; Ursic and Culbertson 1991). An individual TRiC subunit has three domains: an equatorial domain (E domain) that contains the ATP-binding site and forms the intra- and inter-ring contacts, an apical domain (A domain) that comprises the interaction sites with target proteins, and an intermediate domain (I domain) that connects the other two domains (Fig. 19.2d) (Ditzel et al. 1998; Waldmann et al. 1995; Klumpp et al. 1997).
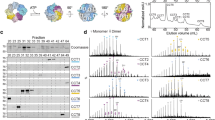
TRiC structural studies and subunit arrangement. a The crystal structures of GroEL (PDB ID: 1SS8, orange) and co-chaperone GroES (PDB ID: 1PCQ, cyan). b The crystal structure of archaea group II chaperonin Mm-cpn (PDB ID: 3RUQ). c The cryo-EM-structure-derived pseudo atomic model of yeast TRiC at open NPP state with different subunits in distinct colors (PDB ID: 5GW4). d The three domains of a TRiC subunit, with the key structural elements labeled. e A cartoon diagram illustrating TRiC ATPase cycle. f Available relative high-resolution structures of TRiC at different nucleotide states, including the cryo-EM structures of NPP-TRiC (EMDB: 9540), TRiC-AMP-PNP (EMDB: 9541), and TRiC-ATP-AlFx (EMDB: 5415), X-ray structures of TRiC-ATP-γS (PDB: 4B2T) and TRiC-ADP-BeF3 (PDB: 4V8R). g Illustration of using YISEL method to identify the location of CCT2 subunit in TRiC complex. The map of the closed yeast TRiC is in grey, with eGFP (dark green) tagged to CCT2 subunits (one in each ring). h Illustration of using YISPANL method to identify the location of CCT6 subunit in TRiC complex. The map of open yeast TRiC is in grey, with NZ-1 fab (sky blue) tagged to CCT6 subunits. i Determined TRiC subunit arrangement
Coordinated with its ATP-driven conformational cycle, TRiC can assist the folding of approximately 10% of cytosolic proteins, including the cytoskeleton proteins actin and tubulin, which are also redundant substrates of TRiC (Khabirova et al. 2014; Waldmann et al. 1995; Llorca et al. 1999, 2000), cell cycle regulator CDC20 (Camasses et al. 2003), and G-protein signaling related element Gβ (Plimpton et al. 2015). TRiC is also essential for the folding of many proteins involved in oncogenesis, such as p53, VHL tumor suppressor, and STAT3 (Trinidad et al. 2013; Kasembeli et al. 2014; McClellan et al. 2005), and for the folding of retinal developmental factors, including transducin α and PEX7 (Tables 19.1 and 19.2) (Hunziker et al. 2016; van den Brink et al. 2003). Hence, dysfunction of TRiC is closely related to cancer and neurodegenerative diseases such as Huntington’s, Alzheimer’s, and Parkinson’s diseases (Chen et al. 2014; Bassiouni et al. 2016).
Our knowledge of the TRiC complex has been summarized in a large number of superb reviews (Willison 2018a, b; Balchin et al. 2016; Lopez et al. 2015; Broadley and Hartl 2009; Dunn et al. 2001; Skjaerven et al. 2015; Fernandez-Fernandez et al. 2016). Our current review will mainly focus on the recent progresses on the structure and function of TRiC complex and TRiC related diseases.
TRiC Molecular Structure
Structural Studies of TRiC and Its ATP-Driven Conformational Cycle
Purification of such a large eukaryotic complex as TRiC is usually challenging. There are several established purification protocols for direct purification of TRiC from mammalian tissues or cells (Lewis et al. 1992; Frydman et al. 1992; Knee et al. 2013; Gao et al. 1992), as well as from yeast (Pappenberger et al. 2006; Zang et al. 2016). In addition, a method was also reported to reconstitute human TRiC by co-expressing its eight distinct subunits in BHK-21 cells (Machida et al. 2012).
Chaperonins are molecular machines that assist protein folding by undergoing ATP-dependent conformational changes that are coordinated in time and space owing to complex allosteric regulation (Fig. 19.2e). Since the early days, both negative staining-electron microscopy (NS-EM) and cryo-electron microscopy (cryo-EM) have been used to investigate the complex structure itself and the ATP-induced conformational changes of TRiC (Lewis et al. 1992; Frydman et al. 1992; Gao et al. 1992; Llorca et al. 1998). Moreover, small angle X-ray scattering (SAXS) analysis also suggested that ATP hydrolysis is needed for the lid closure of TRiC (Meyer et al. 2003).
Furthermore, reference-free 2D analysis of cryo-EM data showed the asymmetric nature among the eight distinct subunits of bovine TRiC (Rivenzon-Segal et al. 2005). A cryo-EM structural study revealed that the inter-domain motions lead to lid closure in bovine TRiC, which is radically different from those of group I chaperonins despite their overall structural similarity (Booth et al. 2008). Our previous asymmetric cryo-EM 3D reconstructions on TRiC in different nucleotide states demonstrated that TRiC changes its intra- and inter-ring subunit interaction pattern during the ATPase cycle, with all the subunits in each ring highly asymmetric in the apo state, whereas all the nucleotide-containing states tend to be more symmetrical. Moreover, the eight subunits in each ring form four subunits pairs, including CCT2-CCT4, CCT1-CCT3, CCT6-CCT8, and CCT7-CCT5; and there exists a two fold axis between its two rings, resulting in two homotypic subunit interactions across the rings (Cong et al. 2010, 2012).
We recently reconstructed two cryo-EM structures of yeast TRiC in a newly captured nucleotide partially preloaded (NPP) state (termed NPP-TRiC) and in the ATP-bound state at ~4.6 Å resolution (Zang et al. 2016). Our NPP-TRiC map showed that CCT2 subunit-pair forms an unexpected Z-shaped feature (red subunit pair, Fig. 19.2c), which conformation is different from that of bovine TRiC in the apo state, whose CCT2 subunit is intrinsically dynamic with its A domain mostly missing (Cong et al. 2010, 2012). ATP binding could induce a dramatic conformational change of CCT2 subunit, which could be either unbent or stabilized for yeast or bovine TRiC, respectively, constructing a typical inward tilting conformation as the other subunits. Strikingly, for yeast TRiC, CCT3, CCT6, and CCT8 subunits have preloaded nucleotide from the environment of the yeast cell (Zang et al. 2016). Overall, TRiC has evolved into a complex that is structurally divided into two sides and reveals a staggered ATP binding mechanism. Interestingly, there are extra densities blocking the two chambers of yeast TRiC in both NPP-TRiC and TRiC-AMP-PNP maps as well as in the bovine apo-TRiC and TRiC-AMP-PNP maps, which may be correlated to the N- and C- termini and might be involved in the complex assembly and allosteric regulation (Cong et al. 2012; Zang et al. 2016).
There are biochemical investigations and computational analyses on the subunit specificity of TRiC in ATP usage. It has also been suggested the ATP binding affinity varies among the eight distinct subunits of TRiC. A recent report has shown that within one ring of bovine TRiC, only four subunits (CCT1, CCT2, CCT4, and CCT5) bind ATP at its physiological concentration (Reissmann et al. 2012). Additionally, it has also been reported that the most dramatic phenotypic effects are associated with mutations in the nucleotide-binding pockets of CCT4, CCT2, CCT5, and CCT7 of yeast TRiC; however, similar mutations do not markedly affect CCT3, CCT6, and CCT8 (Amit et al. 2010), which is consistent with our observation that there are preloaded nucleotides in these three subunits (CCT3/6/8) (Zang et al. 2016). Moreover, equivalent mutations in the eight subunits of TRiC at the positions involved in ATP hydrolysis led to dramatically different phenotypes, indicating each subunit may have different function and play distinct role in TRiC ATPase hydrolysis (Amit et al. 2010). By measuring emissions from chaperonin-bound fluorescent nucleotides, it was suggested that TRiC either hydrolyzes no ATP or hydrolyzes about four ATPs in each of the two rings at the same time (Jiang et al. 2011). By measuring the amount of phosphate generated by ATP hydrolysis as a function of time, it has been indicated that TRiC is in an equilibrium with a large number of conformational states, and that “conformational selection” by ATP takes place before hydrolysis (Korobko et al. 2016). Diffracted X-ray tracking experiment by characterizing Chaetomium thermophilum TRiC variants containing ATPase-deficient subunits showed that ATP-induced motion in a ring is in an asymmetric manner (Yamamoto et al. 2017). All the above studies show that TRiC subunits consume ATP in an asymmetric manner, coordinated in time and space owing to complex allosteric regulations, and each subunit has its own functional specificity. However, the underling structural basis of this specificity and the evolutional benefit need further investigation.
TRiC Subunit Arrangement
To better understand the subunit specificity in ATP usage and allosteric regulation, as well as in substrate recognition and folding, it is essential to address the long-standing question regarding the subunit arrangement of the TRiC complex. In the early studies, TRiC micro-complexes (comprised of subsets of the constitutively expressed TRiC subunits) were examined and implied the unique topology of TRiC subunits (Liou and Willison 1997). In the past decade, extensive efforts have been made to determine the subunit arrangement of this complex molecular machine (Fig. 19.2f) (Dekker et al. 2011; Munoz et al. 2011; Cong et al. 2010; Liou and Willison 1997; Kalisman et al. 2012). Recently, the arrangement has been better determined by chemical cross-linking and mass spectrometry (XL-MS) (Kalisman et al. 2012; Leitner et al. 2012). Moreover, computational approaches have been used to allocate the subunits in the X-ray structures of yeast TRiC in the presence of ATP-BeFx and bovine TRiC in the presence of ATP-γS, which confirmed the XL-MS result (Kalisman et al. 2013). However, due to the structural similarity among TRiC subunits and resolution limitation, the conclusions have been inconsistent across studies (Dekker et al. 2011; Munoz et al. 2011; Cong et al. 2010; Liou and Willison 1997; Kalisman et al. 2012). Recently, combining the cryo-EM reconstruction of yeast TRiC at the open NPP state with our recently developed yeast internal-subunit eGFP labeling (YISEL) strategy and yeast inner-subunit PA–NZ-1 labeling (YISPANL) strategy, we unambiguously identified the subunit locations in the open-state TRiC (Fig. 19.2g–i) (Zang et al. 2018; Wang et al. 2018), and the subunit ordering determined in this way is in agreement with the previous XL-MS result (Kalisman et al. 2012; Leitner et al. 2012).
Structural Study on TRiC with Substrate
TRiC plays an important role in the maintenance of protein homeostasis. Therefore, capturing high-resolution structural information of TRiC in complex with substrate is of great significance for our understanding of the mechanism of how TRiC recognizes and folds substrate proteins through its ATP-driven conformational cycle. However, due to the relative low binding efficiency between TRiC and the pretreated unfolded substrates (by applying urea or GuHCl in vitro), and the potential conformational heterogeneity of bound substrate, so far the available structural information of TRiC-substrate remains limited.
Along this line, a number of efforts have been made to investigate the TRiC-substrate complex mainly using cryo-EM. Early cryo-EM studies of TRiC in complex with denatured actin (Llorca et al. 1999) or denatured tubulin (Llorca et al. 2000) in the open state indicated that actin binds below the helical protrusions with multiple TRiC subunits, while tubulin interacts directly with the helical protrusion of two TRiC subunits. Subsequently, structural and functional analyses of Huntingtin-TRiC complex uncovered the A domains of CCT1 and CCT4 involve in the inhibition of the aggregation process of Huntingtin, the causative agent of Huntington’s disease (Tam et al. 2006; Darrow et al. 2015; Sontag et al. 2013; Shahmoradian et al. 2013). Recently, the cryo-EM structure of cancer-related protein AML1-ETO in complex with human TRiC revealed the subunit-specific interaction, primarily through its DNA binding domain with two TRiC subunits (Fig. 19.3a) (Roh et al. 2016a, b).
Cryo-EM structures of TRiC in complex with substrate. a Cryo-EM structure of TRiC-AML1-175 (EMDB: 6227, the map is rendered according to the recommended threshold). TRiC density is colored in grey and AML1-175 in purple. Subunits are also labeled. b Cryo-EM structure of TRiC-actin (EMDB: 0017, the map is rendered according to the recommended threshold). TRiC density is colored in grey and actin density in orange
There are few clear definitions from the common sequence or structural features of TRiC substrates except for WD40-domain containing proteins, which have been described to be an important family of TRiC substrates (Lopez et al. 2015; Willison 2018b). The WD40-domain containing proteins usually consist of a 7-bladed-β-propeller and could coordinate multi-protein complex assemblies, where the repeating units serve as a rigid scaffold for protein interactions. A number of TRiC substrates have been found to have the WD40 β-propeller features such as TCAB1, CDC20, and Gβ (Freund et al. 2014; Kaisari et al. 2017; Plimpton et al. 2015; Miyata et al. 2014). To overcome the low binding efficiency problem, researchers purified the TRiC substrate, human Gβ, from insect cells. In this way, the potentially partially folded human Gβ may retrieve the intact insect TRiC from the cells. Then the cryo-EM structures of Gβ-TRiC and PhLP1-Gβ-TRiC complexes were reconstructed, revealing a mechanism for Gβ folding and Gβγ dimer assembly assisted by TRiC (Plimpton et al. 2015). Recently, it has been reported that TRiC could capture TAF5, assist its WD40 domain folding and subsequent handover to TAF6-TAF9 and ultimately holo-TFIID formation (Antonova et al. 2018).
There are also X-ray structures of actin-TRiC in the presence of ADP-BeFx in the closed state at 3.8 Å resolution, revealing residual density in one of the TRiC cavities as unfolded actin interacting with multiple TRiC subunits (Dekker et al. 2011), and of tubulin-TRiC with ATP-γS in the open state at 5.5 Å resolution, which also allocated part of the substrate density contacting with three TRiC subunits (Munoz et al. 2011). A recent study combining cryo-EM, fluorescence correlation spectroscopy (FCS), and hydrogen/deuterium exchange coupled to mass spectrometry (H/DX-MS) determined the conformational progression of actin as it is folded by bovine TRiC. This analysis revealved that actin binds to five TRiC subunits (CCT2/4/8/7/5), and captured the dynamic interaction locations through the ATP-driven TRiC-mediated actin folding cycle (Fig. 19.3b) (Balchin et al. 2018).
Key Structural Elements of Group II Chaperonin
To-date, many key structural elements of group II chaperonins involved in nucleotide consumption, subunit coordination, and substrate recognition have been delineated (Fig. 19.2d). The GDGTT motif, called the p-loop and located in the E domain of individual subunits of group II chaperonins, involves ATP binding and is strictly conserved in all chaperonins (Amit et al. 2010). The nucleotide-sensing loop (NSL) located in the I domain monitors the presence of the γ-phosphate of ATP (Pereira et al. 2012; Zang et al. 2016). The β-hairpin in the E domain of almost every archaea and eukaryotic group II chaperonin subunit has been suggested to play a role covering the entrance of the nucleotide pocket (Pereira et al. 2012; Zang et al. 2016). Between two adjacent subunits of group II chaperonins within a ring, there are contacts linking their E domains via a mixed 4-stranded β-sheet involving the N- and C- termini of one subunit and the stem-loop of the neighboring subunit that communicates with its nucleotide pocket (Zhang et al. 2010; Pereira et al. 2010; Ditzel et al. 1998). In addition, the proximal loop (PL) in the A domain recognizes substrate together with its neighboring α-helix 9 (H9), while the release loop for substrate (RLS) is responsible for evicting bound substrate into the TRiC chamber during TRiC ring closure (Lopez et al. 2015; Joachimiak et al. 2014). Still, it needs further examination on how these structural elements coordinate together to perform allosteric conformational transitions in ring closure and substrate folding.
Structure of TRiC with Co-chaperone
Although TRiC does not need a co-chaperone to cover the central cavity forming a closed ring, it has co-chaperone or cofactors to deliver unfolded/partially folded substrates, or to accelerate the substrate folding process (Fig. 19.1). There are several cofactors for TRiC: (1) prefoldin, a hexameric chaperone, binds specifically to chaperonin and transfers target protein (mainly actin) to it (Vainberg et al. 1998); (2) phosducin-like protein, including three known families of phosducin-like proteins participating in Gβ-protein signaling, which have also been implicated in other process, such as the folding of actin and tubulin (Plimpton et al. 2015; Stirling et al. 2006; Humrich et al. 2005); (3) Hsp70, another heat-shock protein that was found to be involved in substrate delivery to TRiC (Cuellar et al. 2008; Melville et al. 2003). Therefore, understanding the interaction mechanism of TRiC with cofactors may provide a thorough picture of TRiC-assisted substrate folding. The cryo-EM map of the TRiC-PFD complex indicates that PFD (human) binds to both rings of TRiC (bovine) in a unique configuration, suggesting a substrate handoff mechanism from PFD to TRiC (Martin-Benito et al. 2002). The cryo-EM structure of bovine TRiC-Hsc70 also supports the notion that Hsc70 may deliver the unfolded proteins to the substrate receiving region of TRiC, and it has been postulated that in eukarya, TRiC and Hsc70 evolved as a concerted action that makes the folding task more efficient (Cuellar et al. 2008). Recently, a cryo-EM structure of insect TRiC in complex with the co-chaperone phosducin-like protein 1 (PhLP1, human) and substrate Gβ (human) was reported. This study indicated that PhLP1 binding could stabilize the Gβ fold, disrupting interactions with TRiC and releasing a PhLP1-Gβ dimer for assembly with Gγ (Plimpton et al. 2015). Still, there are remaining obscure questions regarding the role that co-chaperone plays in cooperating with TRiC that need to be addressed, such as in what situation co-chaperone binds substrate, and when and how co-chaperone delivers substrate to TRiC or helps release substrate from TRiC.
Cryo-ET of TRiC with Substrate
It is exciting to directly visualize locations of TRiC in the cell and to understand how TRiC assists substrate folding in situ. In recent years, cryo-electron tomography (cryo-ET) has emerged as a powerful tool for direct visualization of the molecular organization of cellular landscapes, with the potential to reach near-atomic resolution (Beck and Baumeister 2016; Mosalaganti et al. 2018; Guo et al. 2018). Cryo-ET has been used to visualize the interacting location of TRiC on the mutant huntingtin exon fibril, containing an expanded polyglutamine tract with 51 residues (mhttQ51) and resolved a 3D structure of TRiC interacting with mhttQ51, which provided a structural description for TRiC’s inhibition of mhttQ51 aggregation in vitro (Shahmoradian et al. 2013). Recently, using cryo-ET and sub-tomogram averaging, the structure and cellular interactions of poly-GA aggregates (poly-Gly-Ala aggregates resulting from aberrant translation of an expanded GGGGCC repeat in C9orf72) within intact neurons was visualized, in which TRiC molecules were also found to locate between poly-GA ribbons (Guo et al. 2018). These studies pave the way for further understanding of the function of TRiC in substrate folding or its role in hindering substrate protein aggregation.
Structure of Mutated TRiC and Homo-Oligomer of TRiC Single Subunit
TRiC consists of eight distinct subunits which bring extra complexity to its architecture and functional specificity. Each subunit of TRiC may play a distinct role in the assembly, allosteric coordination, and substrate processing of TRiC complex (Spiess et al. 2004). Some TRiC subunits are able to form a biologically active homo-oligomer when respectively expressed in Escherichia coli. It has been shown that human CCT4 and CCT5 could form biologically active homo-oligomer independently, and a cryo-EM reconstruction of CCT5 homo-oligomeric ring suggests an overall TRiC-like structure (Sergeeva et al. 2013). Intriguingly, similar to WT TRiC, the synthetic CCT5 homo-oligomer complex was found to be able to cap mHTT fibrils at their tips and inhibit mHTT aggregation, revealed by cryo-ET analysis (Darrow et al. 2015), and it is also able to promote the disassembly of the mitotic checkpoint complex (MCC) in the same fashion as TRiC (Kaisari et al. 2017). Moreover, two mutations, C450Y of CCT4 and H147R of CCT5, were identified in hereditary sensory neuropathies (HSNs) (Bouhouche et al. 2006; Lee et al. 2003). NS-EM study revealed that CCT4-C450Y forms few ring-shaped species, whereas WT CCT4, CCT5-H147R, and WT CCT5 can form similar ring structures (Sergeeva et al. 2014). Recently, the X-ray structures of human CCT5 and CCT5-H147R recombinant complexes were resolved, respectively, which demonstrate the mutation may impact the allosteric cooperativity of TRiC (Pereira et al. 2017).
Functions of TRiC
Substrate Folding Assisted by TRiC
Although the mechanism of TRiC-substrate recognition and binding need further investigation, it has been demonstrated that substrate motifs are recognized by a cleft formed between the H9 and PL in the A domains of TRiC individual subunits (Fig. 19.2d), and this cleft contains subunit-specific patterns of polar and hydrophobic residues (Joachimiak et al. 2014). Thus, the diversification of TRiC subunits, especially in the A domain regions, enables TRiC to recognize and bind diverse substrates (Yamamoto et al. 2017). Considering that there are usually multiple TRiC subunits involving in substrate binding (Lopez et al. 2015), this explains why TRiC can assist up to 10% of cytosolic proteins to fold in the cell. Moreover, TRiC plays an important role in the folding of newly synthesized proteins (Frydman et al. 1994; Yam et al. 2008), but it can also prevent the aggregation of proteins with polyglutamine regions (Kitamura et al. 2006; Tam et al. 2006), thus potentially contributing to the suppression of mis-folding in diseases such as Huntington’s, Parkinson’s, and Alzheimer’s diseases. Still, for the folding state changes of substrates along with the ATP-driven ring closure process, TRiC remains to be further explored.
TRiC and Diseases
TRiC and Neurodegenerative Diseases
Many neurodegenerative disorders, such as Alzheimer’s, Parkinson’s, and polyglutamine diseases, as well as amyotrophic lateral sclerosis (ALS), are characterized by conformational changes in proteins that result in misfolding, aggregation and intra- or extra-neuronal accumulation of amyloid fibrils, molecular chaperones including TRiC provide the first line of defense (Table 19.1) (Muchowski and Wacker 2005; Roh et al. 2016b). Recent studies have investigated the role of TRiC in neurodegenerative diseases (Voisine et al. 2010; Liebman and Meredith 2010). It has been biochemically shown that TRiC can inhibit several aggregation variants both in vitro and in vivo (Kitamura et al. 2006; Tam et al. 2006; Behrends et al. 2006) and in cooperation with the Hsp70 system (Behrends et al. 2006). Moreover, a cryo-ET analysis provided a structural description for TRiC’s inhibition of mhttQ51 aggregation in vitro (Shahmoradian et al. 2013).
Moreover, genetic defects in CSA, a 7-bladed WD40 protein, mostly give rise to Cockayne syndrome, which is characterized by premature aging, progressive mental and sensorial retardation, microcephaly, severe growth failure, and cutaneous photosensitivity. TRiC has been shown to interact with CSA through its WD40 domain, thereby regulating CSA stability (Pines et al. 2018). In addition, several individual subunits of TRiC have been reported to be related to hereditary sensory neuropathy, neuronal apoptosis or neurological defects (Lee et al. 2003; Bouhouche et al. 2006; Wu et al. 2015b; Matsuda and Mishina 2004) (described elsewhere in this chapter).
TRiC and Eye Related Diseases
It had been reported that several retinal developmental factors are the client of TRiC, such as myosin, the cause of Usher syndrome (Bonné-Tamir et al. 1994; Adato et al. 1997), transducin α, the cause of Achromatopsia (Kohl et al. 2002), and peroxisomal targeting signal 2 receptor protein, the cause of Refsum disease (Srikakulam and Winkelmann 1999; Farr et al. 1997).
Several studies suggested an essential role of TRiC in ciliogenesis. The most abundant clients of TRiC are represented by cytoskeletal proteins from the actin and tubulin families (Gao et al. 1992; Grantham et al. 2006; Hanafy et al. 2004; Vinh and Drubin 1994; Stemp et al. 2005; Chen et al. 1994). It has been proven that TRiC plays an essential role in the biogenesis of vertebrate photoreceptor sensory cilia (Sinha et al. 2014). Additionally, suppressing TRiC activity in mouse photoreceptors results in the malformation of the retinal outer segment, a cellular compartment responsible for light detection, and triggers rapid retinal degeneration (Posokhova et al. 2011). The activity of TRiC is also essential for the assembly of the BBSome complex (Zhang et al. 2012), a complex thought to control trafficking of molecules into the cilium (Nachury et al. 2007; Loktev et al. 2008). As a result, knockdown of TRiC in zebrafish results in a Bardet-Biedl-like syndrome, a human genetic disorder resulting in obesity, retinal degeneration, polydactyly, and nephropathy (Seo et al. 2010).
Gβ, as part of Gβγ (G protein βγ) and RGS9-Gβ5 heterodimers, plays an essential role in photo-transduction. Both are obligate dimers that rely on TRiC and its co-chaperone PhLP1 to form a complex from their nascent polypeptides (Tracy et al. 2015; Willardson and Howlett 2007). Moreover, two mutations T400P and R516H in CCT2 subunit has been reported to be related to Leber congenital amaurosis (LCA), a hereditary early-onset retinal dystrophy that is accompanied by severe macular degeneration (Minegishi et al. 2016).
TRiC and Cancer
TRiC has been found to be related to oncogenesis, probably through the interaction of TRiC with oncogenic clients which modulate the cancer cells (Roh et al. 2015). Studies have indicated that both TRiC and Hsp70 are required for correct folding of newly translated von Hippel-Lindau (VHL), which is coupled to assembly of a ternary complex with its partner proteins elongin BC (McClellan et al. 2005; Feldman et al. 2003; Melville et al. 2003; Feldman et al. 1999). Loss of VHL function is associated with a number of inherited and spontaneous tumors (Kaelin 2002). Moreover, TRiC also contributes to the folding and function of STAT3, a transcription factor for the transmission of peptide hormone signals from receptors on the extracellular membrane to nucleus, which is up-regulated in many pathological conditions, including cancer and inflammatory diseases (Kasembeli et al. 2014).
p53 is a transcription factor that mediates tumor suppressor responses. Correct folding of the p53 protein is essential for these activities, and point mutations that induce conformational instability of p53 are frequently found in cancers. It has been shown that folding of WT p53 is promoted by the interaction with TRiC. Depletion of TRiC in cells results in the accumulation of misfolded p53, leading to a reduction in p53-dependent gene expression. Although the most straightforward model for the role of TRiC is to assist in the correct folding of newly synthesized p53 (Malcikova et al. 2010), it is also possible that TRiC binding to p53 prevents its aggregation (Trinidad et al. 2013).
TRiC expression is closely correlated with cell growth and is markedly enhanced at early S phase of the cell cycle in mouse and human cultured cells (Yokota et al. 1999). Furthermore, specific TRiC subunits have been found to be up-regulated in the development of diverse cancers (Table 19.1). For instance, CCT1 and CCT2 have been reported to be essential for survival and proliferation of breast cancer (Guest et al. 2015). The expression levels of CCT3 mRNA and protein are up-regulated in hepatocellular carcinoma cells (HCC) cell lines, and over expression of CCT3 in the nuclei of cancerous cells is associated with HCC progression. CCT3 may be a target that affects the activation of STAT3 in HCC (Cui et al. 2015).
CCT8 has been reported to be dysregulated in several tumor tissues. In glioma, high expression of CCT8 is significantly associated with shorter overall survival. CCT8 can regulate the proliferation and invasion of glioblastomas (Qiu et al. 2015). Similarly, CCT8 expression is increased in hepatocellular carcinoma specimens, and knockdown of CCT8 can inhibit the proliferation of HCC (Huang et al. 2014). Also, CCT8 is highly expressed in proliferating germinal center cells and in progressive lymphomas. High expression of CCT8 is significantly associated with shorter overall survival in patients with diffuse large B-cell lymphoma (Yin et al. 2016). Further study is needed to address the following questions: whether the TRiC subunits in cancer act as a monomer, a homo-oligomer, or a TRiC complex; if it functions as a monomer, does it work as a chaperone with ATPase activity or just transiently interacts with a certain substrate? Hence, elucidating the roles of TRiC in oncogenesis may provide new strategies for related cancer therapy.
Potential Therapeutic Applications of TRiC
TRiC Subunit Can Serve as a Biomarker for Related Disease
Recently, accumulating evidences have suggested the relationship between specific TRiC subunits and disease, and several TRiC subunits have been found to be over-expressed in certain diseases (Table 19.1). Therefore, over-expressed specific TRiC subunit could be used as a clinical marker for related disease.
The expression of CCT5 along with RGS3 and YKT6 genes are up-regulated in p53-mutated breast tumors and are associated with a resistance to docetaxel, and this might be clinically useful in identifying the subset of breast cancer patients who may or may not benefit from docetaxel treatment. Treatment of human breast cancer cell line MCF-7 with siRNA specific for CCT5 resulted in a significant enhancement of docetaxel-induced apoptosis and this may provide new insights into the molecular mechanism of resistance to docetaxel (Ooe et al. 2007). It has been reported that CCT5 also shows higher expression in non-small cell lung cancer (NSCLC) tissues and could be used as a biomarker in the diagnosis of NSCLC in an early stage (Gao et al. 2017). Moreover, the expression of CCT2 has been shown to be elevated in urine of patients with type 2 diabetic mellitus in the hyperfiltration stage. Hyperfiltration in early diabetic nephropathy (DN) may be detectable by measuring urine CCT2, which can be a novel and valuable biomarker for clinical evaluation (Wu et al. 2015a). Increased CCT7 has been observed in fibrous diseases such as Dupuytren’s contracture (Satish et al. 2013) and skin contractive scars (Satish et al. 2010), and CCT7 has been indicated to be a unique potential marker for active fibroblasts (Bai et al. 2015).
Potential Application of TRiC in Preventing MHtt (Mutant Huntingtin) Aggregation
TRiC has been suggested to be able to prevent mHtt aggregation in cells (Behrends et al. 2006; Kitamura et al. 2006; Tam et al. 2006, 2009), and the underlying inhibition mechanism of TRiC against mhttQ51 aggregation in vitro has been investigated (Shahmoradian et al. 2013). Besides, the CCT5 homo-oligomer can cap mHtt fibrils in a similar fashion to the TRiC complex. These results suggest the option of exploring the use of the TRiC complex or CCT5 as a reagent to prevent mHtt aggregation, which may contribute to the development of HD therapy (Darrow et al. 2015). Moreover, it has been reported that the CCT1 subunit is sufficient to inhibit aggregation and reduce mHtt-mediated toxicity in mouse N2a neuronal cells (Tam et al. 2006), and the exogenous delivery of the 20 kDa recombinant apical domain of CCT1 (ApiCCT1) can inhibit aggregation of recombinant mHtt in vitro (Sontag et al. 2013). More remarkably, applying ApiCCT1 to the cortical compartment of Htt disease mouse model can rescue impaired anterograde transport of brain-derived neurotrophic factor (BDNF) (Zhao et al. 2016). Therefore, there is a potential application for ApiCCT1 to be explored in the treatment of HD.
TRiC Is a Potential Target for Therapeutic Intervention
Cryo-EM analysis revealed that the CCT2 subunit of TRiC interacts with β-tubulin (Llorca et al. 2001). Disrupting the constitutively associated β-tubulin-CCT2 complex can cause severe cell apoptosis in multidrug-resistant MES-SA/Dx5 cancer cells (Harker and Sikic 1985). This finding suggests that the β-tubulin-CCT2 complex may serve as an effective chemotherapeutic target for treating clinical tubulin-binding agent-resistant tumor (Lin et al. 2009).
Breast cancer is the leading cause of death in women between the ages of 35 and 54 (Fiorica 1992). The CCT2 subunit was identified as the intracellular target of CT20p, a peptide that displays cancer (include breast cancer) specific cytotoxicity. The susceptibility of breast cancer cell to CT20p would be increased when overexpressing CCT2. TRiC is thus a potential target for therapeutic intervention for breast cancer treatment (Bassiouni et al. 2016). SMADs are the main signal transducers for receptors of the TGF-β superfamily, which are critically important for regulating cell development and growth. CCT6 has been shown to be the SMAD2 blocker, and silencing CCT6 could result in efficient suppression of metastasis in vivo and significantly prolonged the survival of tumor-bearing mice (Ying et al. 2017).
It has been reported that the homozygous cct2-L394H-7del mutation in the zebrafish leads to a small eye phenotype, and injection of RNA encoding wild-type human CCT2 could rescue the small eye phenotype (Minegishi et al. 2018). Also, it has been suggested that two mutations, T400P and R516H, in the CCT2 subunit can evoke leber congenital amaurosis (LCA) (Minegishi et al. 2016). This indicates a potential novel direction for the treatment of the LCA eye disease.
Perspective
In recent years, accumulating evidence has suggested the linkage between TRiC and certain diseases; still, there is a long way to go to fully understand the underlying molecular mechanism. Future atomic-resolution structural details and biochemical analysis of the eukaryotic group II chaperonin TRiC/CCT, may offer the structural basis of its subunit specificity, and provide more mechanistic insights into the allosteric network of TRiC and how this network regulates TRiC conformational transitions. Additionally, how the coordinated conformational transitions of TRiC correlate with their productive substrate folding is yet to be further elucidated (Fig. 19.4). Moreover, with the development of computational method in cryo-EM, such as manifold embedding algorithm (Frank and Ourmazd 2016), the continuous conformational landscape of TRiC may be delineated, and its free-energy landscape could be mapped out. Furthermore, the in vivo study of TRiC in the native environment by cryo-ET and sub-tomogram averaging could enable us to directly visualize the process of TRiC-assisted substrate folding in the cell (Guo et al. 2018; Shahmoradian et al. 2013). Eventually, a combination of all the information on this complex macromolecular machine will facilitate therapeutic strategy development against TRiC-related diseases (Fig. 19.4).
Potential therapeutic development based on high-resolution cryo-EM structural studies of TRiC. Illustration of cryo-EM sample preparation, data collection, 3D reconstruction, model building, and structure-based drug development. The high-resolution reconstruction of an ensemble of TRiC structures in its substrate folding cycle regulated by its ATP-driven conformational cycle, will provide a complete picture of the mechanism of TRiC-assisted substrate folding. This structural information will enable us to design drugs to inhibit the allosteric transition at any desired step, and to block the reorganization of TRiC with specific substrate so as to prevent the development of related diseases such as cancer
Author Declaration
During the page proof process, a cryo-EM study on TRiC in complex with substrate mLST8 was published, revealing the function of TRiC in the folding of mLST8 and in the preparation for the assembly of mLST8 into mTOR complexes (Cuéllar et al. 2019). Moreover, another study resolved the cryo-EM structure of TRiC-PFD complex, established the structural and functional basis for TRiC and PFD cooperation in protein folding, essential for cellular proteostasis (Gestaut et al. 2019).
References
Adato A, Weil D, Kalinski H, Pel-Or Y, Ayadi H, Petit C, Korostishevsky M, Bonne-Tamir B (1997) Mutation profile of all 49 exons of the human myosin VIIA gene, and haplotype analysis, in Usher 1B families from diverse origins. Am J Hum Genet 61(4):813–821. https://doi.org/10.1086/514899
Amit M, Weisberg SJ, Nadler-Holly M, McCormack EA, Feldmesser E, Kaganovich D, Willison KR, Horovitz A (2010) Equivalent mutations in the eight subunits of the chaperonin CCT produce dramatically different cellular and gene expression phenotypes. J Mol Biol 401(3):532–543. https://doi.org/10.1016/j.jmb.2010.06.037
Antonova SV, Haffke M, Corradini E, Mikuciunas M, Low TY, Signor L, van Es RM, Gupta K, Scheer E, Vos HR, Tora L, Heck AJR, Timmers HTM, Berger I (2018) Chaperonin CCT checkpoint function in basal transcription factor TFIID assembly. Nat Struct Mol Biol. https://doi.org/10.1038/s41594-018-0156-z
Bai XC, Rajendra E, Yang G, Shi Y, Scheres SH (2015) Sampling the conformational space of the catalytic subunit of human gamma-secretase. Elife 4. https://doi.org/10.7554/elife.11182
Bakthavatsalam D, Soung RH, Tweardy DJ, Chiu W, Dixon RA, Woodside DG (2014) Chaperonin-containing TCP-1 complex directly binds to the cytoplasmic domain of the LOX-1 receptor. FEBS Lett 588(13):2133–2140. https://doi.org/10.1016/j.febslet.2014.04.049
Balchin D, Hayer-Hartl M, Hartl FU (2016) In vivo aspects of protein folding and quality control. Science 353(6294):aac4354. https://doi.org/10.1126/science.aac4354
Balchin D, Milicic G, Strauss M, Hayer-Hartl M, Hartl FU (2018) Pathway of actin folding directed by the eukaryotic chaperonin TRiC. Cell. https://doi.org/10.1016/j.cell.2018.07.006
Bassiouni R, Nemec KN, Iketani A, Flores O, Showalter A, Khaled AS, Vishnubhotla P, Sprung RW Jr, Kaittanis C, Perez JM, Khaled AR (2016) Chaperonin containing TCP-1 protein level in breast cancer cells predicts therapeutic application of a cytotoxic peptide. Clin Cancer Res 22(17):4366–4379. https://doi.org/10.1158/1078-0432.CCR-15-2502
Beck M, Baumeister W (2016) Cryo-Electron tomography: can it reveal the molecular sociology of cells in atomic detail? Trends Cell Biol 26(11):825–837. https://doi.org/10.1016/j.tcb.2016.08.006
Behrends C, Langer CA, Boteva R, Bottcher UM, Stemp MJ, Schaffar G, Rao BV, Giese A, Kretzschmar H, Siegers K, Hartl FU (2006) Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol Cell 23(6):887–897. https://doi.org/10.1016/j.molcel.2006.08.017
Berger J, Berger S, Li M, Jacoby AS, Arner A, Bavi N, Stewart AG, Currie PD (2018) In vivo function of the chaperonin TRiC in alpha-actin folding during sarcomere assembly. Cell Rep 22(2):313–322. https://doi.org/10.1016/j.celrep.2017.12.069
Bonné-Tamir B, Korostishevsky M, Kalinsky H, Seroussi E, Beker R, Weiss S, Godel V (1994) Genetic mapping of the gene for usher syndrome: linkage analysis in a large samaritan kindred. Genomics 20(1):36–42. https://doi.org/10.1006/geno.1994.1124
Booth CR, Meyer AS, Cong Y, Topf M, Sali A, Ludtke SJ, Chiu W, Frydman J (2008) Mechanism of lid closure in the eukaryotic chaperonin TRiC/CCT. Nat Struct Mol Biol 15(7):746–753. https://doi.org/10.1038/nsmb.1436
Bouhouche A, Benomar A, Bouslam N, Chkili T, Yahyaoui M (2006) Mutation in the epsilon subunit of the cytosolic chaperonin-containing t-complex peptide-1 (Cct5) gene causes autosomal recessive mutilating sensory neuropathy with spastic paraplegia. J Med Genet 43(5):441–443. https://doi.org/10.1136/jmg.2005.039230
Broadley SA, Hartl FU (2009) The role of molecular chaperones in human misfolding diseases. FEBS Lett 583(16):2647–2653. https://doi.org/10.1016/j.febslet.2009.04.029
Camasses A, Bogdanova A, Shevchenko A, Zachariae W (2003) The CCT Chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol Cell 12(1):87–100. https://doi.org/10.1016/s1097-2765(03)00244-2
Carr AC, Khaled AS, Bassiouni R, Flores O, Nierenberg D, Bhatti H, Vishnubhotla P, Perez JM, Santra S, Khaled AR (2017) Targeting chaperonin containing TCP1 (CCT) as a molecular therapeutic for small cell lung cancer. Oncotarget 8(66):110273–110288. https://doi.org/10.18632/oncotarget.22681
Chen X, Sullivan DS, Huffaker TC (1994) Two yeast genes with similarity to TCP-1 are required for microtubule and actin function in vivo. Proc Natl Acad Sci USA 91(19):9111–9115. https://doi.org/10.1073/pnas.91.19.9111
Chen L, Zhang Z, Qiu J, Zhang L, Luo X, Jang J (2014) Chaperonin CCT-mediated AIB1 folding promotes the growth of ERalpha-positive breast cancer cells on hard substrates. PLoS ONE 9(5):e96085. https://doi.org/10.1371/journal.pone.0096085
Coghlin C, Carpenter B, Dundas SR, Lawrie LC, Telfer C, Murray GI (2006) Characterization and over-expression of chaperonin t-complex proteins in colorectal cancer. J Pathol 210(3):351–357. https://doi.org/10.1002/path.2056
Cong Y, Baker ML, Jakana J, Woolford D, Miller EJ, Reissmann S, Kumar RN, Redding-Johanson AM, Batth TS, Mukhopadhyay A, Ludtke SJ, Frydman J, Chiu W (2010) 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement. Proc Nat Acad Sci USA 107(11):4967–4972. https://doi.org/10.1073/pnas.0913774107
Cong Y, Schroder GF, Meyer AS, Jakana J, Ma B, Dougherty MT, Schmid MF, Reissmann S, Levitt M, Ludtke SL, Frydman J, Chiu W (2012) Symmetry-free cryo-EM structures of the chaperonin TRiC along its ATPase-driven conformational cycle. EMBO J 31(3):720–730. https://doi.org/10.1038/emboj.2011.366
Counts JT, Hester TM, Rouhana L (2017) Genetic expansion of chaperonin-containing TCP-1 (CCT/TRiC) complex subunits yields testis-specific isoforms required for spermatogenesis in planarian flatworms. Mol Reprod Dev 84(12):1271–1284. https://doi.org/10.1002/mrd.22925
Cuellar J, Martin-Benito J, Scheres SH, Sousa R, Moro F, Lopez-Vinas E, Gomez-Puertas P, Muga A, Carrascosa JL, Valpuesta JM (2008) The structure of CCT-Hsc70 NBD suggests a mechanism for Hsp70 delivery of substrates to the chaperonin. Nat Struct Mol Biol 15(8):858–864. https://doi.org/10.1038/nsmb.1464
Cuéllar J, Ludlam WG, Tensmeyer NC, Aoba T, Dhavale M, Santiago C, Bueno-Carrasco MT, Mann MJ, Plimpton RL, Makaju A, Franklin S, Willardson BM, Valpuesta JM (2019) Structural and functional analysis of the role of the chaperonin CCT in mTOR complex assembly. Nat Commun 10(1)
Cui X, Hu ZP, Li Z, Gao PJ, Zhu JY (2015) Overexpression of chaperonin containing TCP1, subunit 3 predicts poor prognosis in hepatocellular carcinoma. World J Gastroenterol 21(28):8588–8604. https://doi.org/10.3748/wjg.v21.i28.8588
Cyrne L, Guerreiro P, Cardoso AC, RodriguesPousada C, Soares H (1996) The Tetrahymena chaperonin subunit CCT eta gene is coexpressed with CCT gamma gene during cilia biogenesis and cell sexual reproduction. FEBS Lett 383(3):277–283. https://doi.org/10.1016/0014-5793(96)00240-2
Darrow MC, Sergeeva OA, Isas JM, Galaz-Montoya JG, King JA, Langen R, Schmid MF, Chiu W (2015) Structural mechanisms of mutant huntingtin aggregation suppression by the synthetic chaperonin-like CCT5 complex explained by cryoelectron tomography. J Biol Chem 290(28):17451–17461. https://doi.org/10.1074/jbc.M115.655373
Dekker C, Roe SM, McCormack EA, Beuron F, Pearl LH, Willison KR (2011) The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins. EMBO J 30(15):3078–3090. https://doi.org/10.1038/emboj.2011.208
Ditzel L, Lowe J, Stock D, Stetter KO, Huber H, Huber R, Steinbacher S (1998) Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT. Cell 93(1):125–138. https://doi.org/10.1016/s0092-8674(00)81152-6
Dunn AY, Melville MW, Frydman J (2001) Review: cellular substrates of the eukaryotic chaperonin TRiC/CCT. J Struct Biol 135(2):176–184. https://doi.org/10.1006/jsbi.2001.4380
Erdo F, Trapp T, Mies G, Hossmann KA (2004) Immunohistochemical analysis of protein expression after middle cerebral artery occlusion in mice. Acta Neuropathol 107(2):127–136. https://doi.org/10.1007/s00401-003-0789-8
Farr GW, Scharl EC, Schumacher RJ, Sondek S, Horwich AL (1997) Chaperonin-mediated folding in the eukaryotic cytosol proceeds through rounds of release of native and nonnative forms. Cell 89(6):927–937. https://doi.org/10.1016/S0092-8674(00)80278-0
Feldman DE, Thulasiraman V, Ferreyra RG, Frydman J (1999) Formation of the VHL-elongin BC tumor suppressor complex is mediated by the chaperonin TRiC. Mol Cell 4(6):1051–1061
Feldman DE, Spiess C, Howard DE, Frydman J (2003) Tumorigenic mutations in VHL disrupt folding in vivo by interfering with chaperonin binding. Mol Cell 12(5):1213–1224
Fernandez-Fernandez MR, Sot B, Valpuesta JM (2016) Molecular chaperones: functional mechanisms and nanotechnological applications. Nanotechnology 27(32):324004. https://doi.org/10.1088/0957-4484/27/32/324004
Fiorica JV (1992) Breast disease. Curr Opin Obstet Gynecol 4(6):897–903
Fontanella B, Birolo L, Infusini G, Cirulli C, Marzullo L, Pucci P, Turco MC, Tosco A (2010) The co-chaperone BAG3 interacts with the cytosolic chaperonin CCT: new hints for actin folding. Int J Biochem Cell Biol 42(5):641–650. https://doi.org/10.1016/j.biocel.2009.12.008
Frank J, Ourmazd A (2016) Continuous changes in structure mapped by manifold embedding of single-particle data in cryo-EM. Methods 100:61–67. https://doi.org/10.1016/j.ymeth.2016.02.007
Freund A, Zhong FL, Venteicher AS, Meng Z, Veenstra TD, Frydman J, Artandi SE (2014) Proteostatic control of telomerase function through TRiC-mediated folding of TCAB1. Cell 159(6):1389–1403. https://doi.org/10.1016/j.cell.2014.10.059
Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall JS, Tempst P, Hartl FU (1992) Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO J 11(13):4767–4778. https://doi.org/10.1002/j.1460-2075.1992.tb05582.x
Frydman J, Nimmesgern E, Ohtsuka K, Hartl FU (1994) Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature 370(6485):111–117. https://doi.org/10.1038/370111a0
Gao YJ, Thomas JO, Chow RL, Lee GH, Cowan NJ (1992) A cytoplasmic chaperonin that catalyzes beta-actin folding. Cell 69(6):1043–1050. https://doi.org/10.1016/0092-8674(92)90622-J
Gao HJ, Zheng M, Sun SJ, Wang HW, Yue ZG, Zhu Y, Han XC, Yang JQ, Zhou YQ, Cai YR, Hu WN (2017) Chaperonin containing TCP1 subunit 5 is a tumor associated antigen of non-small cell lung cancer. Oncotarget 8(38):64170–64179. https://doi.org/10.18632/oncotarget.19369
Gestaut D, Roh SH, Ma B, Pintilie G, Joachimiak LA, Leitner A, Walzthoeni T, Aebersold R, Chiu W, Frydman J (2019) The chaperonin TRiC/CCT associates with prefoldin through a conserved electrostatic interface essential for cellular proteostasis. Cell 177(3):751–765.e15
Grantham J, Brackley KI, Willison KR (2006) Substantial CCT activity is required for cell cycle progression and cytoskeletal organization in mammalian cells. Exp Cell Res 312(12):2309–2324. https://doi.org/10.1016/j.yexcr.2006.03.028
Guenther MG, Yu JJ, Kao GD, Yen TJ, Lazar MA (2002) Assembly of the SMRT-histone deacetylase 3 repression complex requires the TCP-1 ring complex. Genes Dev 16(24):3130–3135. https://doi.org/10.1101/gad.1037502
Guest ST, Kratche ZR, Bollig-Fischer A, Haddad R, Ethier SP (2015) Two members of the TRiC chaperonin complex, CCT2 and TCP1 are essential for survival of breast cancer cells and are linked to driving oncogenes. Exp Cell Res 332(2):223–235. https://doi.org/10.1016/j.yexcr.2015.02.005
Guo Q, Lehmer C, Martinez-Sanchez A, Rudack T, Beck F, Hartmann H, Perez-Berlanga M, Frottin F, Hipp MS, Hartl FU, Edbauer D, Baumeister W, Fernandez-Busnadiego R (2018) In situ structure of neuronal C9orf72 Poly-GA aggregates reveals proteasome recruitment. Cell. https://doi.org/10.1016/j.cell.2017.12.030
Hanafy KA, Martin E, Murad F (2004) CCTeta, a novel soluble guanylyl cyclase-interacting protein. J Biol Chem 279(45):46946–46953. https://doi.org/10.1074/jbc.M404134200
Harker WG, Sikic BI (1985) Multidrug (pleiotropic) resistance in doxorubicin-selected variants of the human sarcoma cell line MES-SA. Cancer Res 45(9):4091–4096
Horwich AL, Willison KR (1993) Protein folding in the cell: functions of two families of molecular chaperone, hsp 60 and TF55-TCP1. Philos Trans R Soc Lond B Biol Sci 339(1289):313–325; discussion 325–316. https://doi.org/10.1098/rstb.1993.0030
Huang X, Wang X, Cheng C, Cai J, He S, Wang H, Liu F, Zhu C, Ding Z, Huang X, Zhang T, Zhang Y (2014) Chaperonin containing TCP1, subunit 8 (CCT8) is upregulated in hepatocellular carcinoma and promotes HCC proliferation. APMIS 122(11):1070–1079. https://doi.org/10.1111/apm.12258
Humrich J, Bermel C, Bunemann M, Harmark L, Frost R, Quitterer U, Lohse MJ (2005) Phosducin-like protein regulates G-protein betagamma folding by interaction with tailless complex polypeptide-1alpha: dephosphorylation or splicing of PhLP turns the switch toward regulation of Gbetagamma folding. J Biol Chem 280(20):20042–20050. https://doi.org/10.1074/jbc.M409233200
Hunziker M, Barandun J, Petfalski E, Tan D, Delan-Forino C, Molloy KR, Kim KH, Dunn-Davies H, Shi Y, Chaker-Margot M, Chait BT, Walz T, Tollervey D, Klinge S (2016) UtpA and UtpB chaperone nascent pre-ribosomal RNA and U3 snoRNA to initiate eukaryotic ribosome assembly. Nat Commun 7:12090. https://doi.org/10.1038/ncomms12090
Jiang Y, Douglas NR, Conley NR, Miller EJ, Frydman J, Moerner WE (2011) Sensing cooperativity in ATP hydrolysis for single multisubunit enzymes in solution. Proc Natl Acad Sci USA 108(41):16962–16967. https://doi.org/10.1073/pnas.1112244108
Jiang XD, Mao WJ, Yang ZY, Zeng J, Zhang Y, Song Y, Kong Y, Ren SY, Zuo YF (2015) Silencing P2X7 receptor downregulates the expression of TCP-1 involved in lymphoma lymphatic metastasis. Oncotarget 6(39):42105–42117. https://doi.org/10.18632/oncotarget.5870
Joachimiak LA, Walzthoeni T, Liu CW, Aebersold R, Frydman J (2014) The structural basis of substrate recognition by the eukaryotic chaperonin TRiC/CCT. Cell 159(5):1042–1055. https://doi.org/10.1016/j.cell.2014.10.042
Kaelin WG Jr (2002) Molecular basis of the VHL hereditary cancer syndrome. Nat Rev Cancer 2(9):673–682. https://doi.org/10.1038/nrc885
Kaisari S, Sitry-Shevah D, Miniowitz-Shemtov S, Teichner A, Hershko A (2017) Role of CCT chaperonin in the disassembly of mitotic checkpoint complexes. Proc Natl Acad Sci USA 114(5):956–961. https://doi.org/10.1073/pnas.1620451114
Kalisman N, Adams CM, Levitt M (2012) Subunit order of eukaryotic TRiC/CCT chaperonin by cross-linking, mass spectrometry, and combinatorial homology modeling. Proc Natl Acad Sci USA 109(8):2884–2889. https://doi.org/10.1073/pnas.1119472109
Kalisman N, Schroder GF, Levitt M (2013) The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning. Structure 21(4):540–549. https://doi.org/10.1016/j.str.2013.01.017
Kasembeli M, Lau WC, Roh SH, Eckols TK, Frydman J, Chiu W, Tweardy DJ (2014) Modulation of STAT3 folding and function by TRiC/CCT chaperonin. PLoS Biol 12(4):e1001844. https://doi.org/10.1371/journal.pbio.1001844
Khabirova E, Moloney A, Marciniak SJ, Williams J, Lomas DA, Oliver SG, Favrin G, Sattelle DB, Crowther DC (2014) The TRiC/CCT chaperone is implicated in Alzheimer’s disease based on patient GWAS and an RNAi screen in Abeta-expressing Caenorhabditis elegans. PLoS One 9(7):e102985. https://doi.org/10.1371/journal.pone.0102985
Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU (2013) Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem 82(82):323–355. https://doi.org/10.1146/annurev-biochem-060208-092442
Kitamura A, Kubota H, Pack CG, Matsumoto G, Hirayama S, Takahashi Y, Kimura H, Kinjo M, Morimoto RI, Nagata K (2006) Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat Cell Biol 8(10):1163–1170. https://doi.org/10.1038/ncb1478
Klumpp M, Baumeister W, Essen LO (1997) Structure of the substrate binding domain of the thermosome, an archaeal group II chaperonin. Cell 91(2):263–270. https://doi.org/10.1016/S0092-8674(00)80408-0
Knee KM, Sergeeva OA, King JA (2013) Human TRiC complex purified from HeLa cells contains all eight CCT subunits and is active in vitro. Cell Stress Chaperones 18(2):137–144. https://doi.org/10.1007/s12192-012-0357-z
Kohl S, Baumann B, Rosenberg T, Kellner U, Lorenz B, Vadalà M, Jacobson SG, Wissinger B (2002) Mutations in the cone photoreceptor G-protein alpha-subunit gene GNAT2 in patients with achromatopsia. Am J Hum Genet 71(2):422–425. https://doi.org/10.1086/341835
Korobko I, Nadler-Holly M, Horovitz A (2016) Transient kinetic analysis of ATP hydrolysis by the CCT/TRiC chaperonin. J Mol Biol 428(22):4520–4527. https://doi.org/10.1016/j.jmb.2016.09.017
Koulikovska M, Podskochy A, Fagerholm P (2005) The expression pattern of the subunit of chaperonin containing T-complex polypeptide 1 and its substrate, alpha-smooth muscle actin, during corneal wound healing. Acta Ophthalmol Scand 83(5):543–548. https://doi.org/10.1111/j.1600-0420.2005.00482.x
Lee MJ, Stephenson DA, Groves MJ, Sweeney MG, Davis MB, An SF, Houlden H, Salih MAM, Timmerman V, de Jonghe P, Auer-Grumbach M, Di Maria E, Scaravilli F, Wood NW, Reilly MM (2003) Hereditary sensory neuropathy is caused by a mutation in the delta subunit of the cytosolic chaperonin-containing t-complex peptide-1 (Cct4) gene. Hum Mol Genet 12(15):1917–1925. https://doi.org/10.1093/hmg/ddg198
Leitner A, Joachimiak LA, Bracher A, Monkemeyer L, Walzthoeni T, Chen B, Pechmann S, Holmes S, Cong Y, Ma B, Ludtke S, Chiu W, Hartl FU, Aebersold R, Frydman J (2012) The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure 20(5):814–825. https://doi.org/10.1016/j.str.2012.03.007
Lewis VA, Hynes GM, Zheng D, Saibil H, Willison K (1992) T-complex polypeptide-1 is a subunit of a heteromeric particle in the eukaryotic cytosol. Nature 358(6383):249–252. https://doi.org/10.1038/358249a0
Liebman SW, Meredith SC (2010) Protein folding: sticky N17 speeds huntingtin pile-up. Nat Chem Biol 6(1):7–8. https://doi.org/10.1038/nchembio.279
Lin YF, Tsai WP, Liu HG, Liang PH (2009) Intracellular beta-tubulin/chaperonin containing TCP1-beta complex serves as a novel chemotherapeutic target against drug-resistant tumors. Cancer Res 69(17):6879–6888. https://doi.org/10.1158/0008-5472.CAN-08-4700
Liou AK, Willison KR (1997) Elucidation of the subunit orientation in CCT (chaperonin containing TCP1) from the subunit composition of CCT micro-complexes. EMBO J 16(14):4311–4316. https://doi.org/10.1093/emboj/16.14.4311
Liu BD, Larsson L, Caballero A, Hao XX, Oling D, Grantham J, Nystrom T (2010) The polarisome is required for segregation and retrograde transport of protein aggregates. Cell 140(2):257–267. https://doi.org/10.1016/j.cell.2009.12.031
Llorca O, Smyth MG, Marco S, Carrascosa JL, Willison KR, Valpuesta JM (1998) ATP binding induces large conformational changes in the apical and equatorial domains of the eukaryotic chaperonin containing TCP-1 complex. J Biol Chem 273(17):10091–10094. https://doi.org/10.1074/jbc.273.17.10091
Llorca O, McCormack EA, Hynes G, Grantham J, Cordell J, Carrascosa JL, Willison KR, Fernandez JJ, Valpuesta JM (1999) Eukaryotic type II chaperonin CCT interacts with actin through specific subunits. Nature 402(6762):693–696. https://doi.org/10.1038/45294valpuesta1999.pdf
Llorca O, Martin-Benito J, Ritco-Vonsovici M, Grantham J, Hynes GM, Willison KR, Carrascosa JL, Valpuesta JM (2000) Eukaryotic chaperonin CCT stabilizes actin and tubulin folding intermediates in open quasi-native conformations. EMBO J 19(22):5971–5979. https://doi.org/10.1093/emboj/19.22.5971
Llorca O, Martin-Benito J, Gomez-Puertas P, Ritco-Vonsovici M, Willison KR, Carrascosa JL, Valpuesta JM (2001) Analysis of the interaction between the eukaryotic chaperonin CCT and its substrates actin and tubulin. J Struct Biol 135(2):205–218. https://doi.org/10.1106/jsbi.2001.4359
Loktev AV, Zhang Q, Beck JS, Searby CC, Scheetz TE, Bazan JF, Slusarski DC, Sheffield VC, Jackson PK, Nachury MV (2008) A BBSome subunit links ciliogenesis, microtubule stability, and acetylation. Dev Cell 15(6):854–865. https://doi.org/10.1016/j.devcel.2008.11.001
Lopez T, Dalton K, Frydman J (2015) The mechanism and function of group ii chaperonins. J Mol Biol 427(18):2919–2930. https://doi.org/10.1016/j.jmb.2015.04.013
Machida K, Masutani M, Kobayashi T, Mikami S, Nishino Y, Miyazawa A, Imataka H (2012) Reconstitution of the human chaperonin CCT by co-expression of the eight distinct subunits in mammalian cells. Protein Expr Purif 82(1):61–69. https://doi.org/10.1016/j.pep.2011.11.010
Malcikova J, Tichy B, Damborsky J, Kabathova J, Trbusek M, Mayer J, Pospisilova S (2010) Analysis of the DNA-binding activity of p53 mutants using functional protein microarrays and its relationship to transcriptional activation. Biol Chem 391(2–3):197–205. https://doi.org/10.1515/BC.2010.027
Martin-Benito J, Boskovic J, Gomez-Puertas P, Carrascosa JL, Simons CT, Lewis SA, Bartolini F, Cowan NJ, Valpuesta JM (2002) Structure of eukaryotic prefoldin and of its complexes with unfolded actin and the cytosolic chaperonin CCT. EMBO J 21(23):6377–6386. https://doi.org/10.1093/emboj/cdf640
Masson N, Appelhoff RJ, Tuckerman JR, Tian YM, Demol H, Puype M, Vandekerckhove J, Ratcliffe PJ, Pugh CW (2004) The HIF prolyl hydroxylase PHD3 is a potential substrate of the TRiC chaperonin. FEBS Lett 570(1–3):166–170. https://doi.org/10.1016/j.febslet.2004.06.040
Matsuda N, Mishina M (2004) Identification of chaperonin CCT gamma subunit as a determinant of retinotectal development by whole-genome subtraction cloning from zebrafish no tectal neuron mutant. Development 131(9):1913–1925. https://doi.org/10.1242/dev.01085
Matus DQ, Li XY, Durbin S, Agarwal D, Chi Q, Weiss SJ, Sherwood DR (2010) In vivo identification of regulators of cell invasion across basement membranes. Sci Signal 3(120):ra35. https://doi.org/10.1126/scisignal.2000654
McClellan AJ, Scott MD, Frydman J (2005) Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell 121(5):739–748. https://doi.org/10.1016/j.cell.2005.03.024
Melville MW, McClellan AJ, Meyer AS, Darveau A, Frydman J (2003) The Hsp70 and TRiC/CCT chaperone systems cooperate in vivo to assemble the Von Hippel-Lindau tumor suppressor complex. Mol Cell Biol 23(9):3141–3151. https://doi.org/10.1128/mcb.23.9.3141-3151.2003
Meyer AS, Gillespie JR, Walther D, Millet IS, Doniach S, Frydman J (2003) Closing the folding chamber of the eukaryotic chaperonin requires the transition state of ATP hydrolysis. Cell 113(3):369–381. https://doi.org/10.1016/S0092-8674(03)00307-6
Minegishi Y, Sheng X, Yoshitake K, Sergeev Y, Iejima D, Shibagaki Y, Monma N, Ikeo K, Furuno M, Zhuang W, Liu Y, Rong W, Hattori S, Iwata T (2016) CCT2 mutations evoke leber congenital amaurosis due to chaperone complex instability. Sci Rep 6:33742. https://doi.org/10.1038/srep33742
Minegishi Y, Nakaya N, Tomarev SI (2018) Mutation in the Zebrafish cct2 gene leads to abnormalities of cell cycle and cell death in the retina: a model of CCT2-related leber congenital amaurosis. Invest Ophthalmol Vis Sci 59(2):995–1004. https://doi.org/10.1167/iovs.17-22919
Miyata Y, Shibata T, Aoshima M, Tsubata T, Nishida E (2014) The molecular chaperone TRiC/CCT binds to the Trp-Asp 40 (WD40) repeat protein WDR68 and promotes its folding, protein kinase DYRK1A binding, and nuclear accumulation. J Biol Chem 289(48):33320–33332. https://doi.org/10.1074/jbc.M114.586115
Monzo K, Dowd SR, Minden JS, Sisson JC (2010) Proteomic analysis reveals CCT is a target of Fragile X mental retardation protein regulation in Drosophila. Dev Biol 340(2):408–418. https://doi.org/10.1016/j.ydbio.2010.01.028
Mosalaganti S, Kosinski J, Albert S, Schaffer M, Strenkert D, Salome PA, Merchant SS, Plitzko JM, Baumeister W, Engel BD, Beck M (2018) In situ architecture of the algal nuclear pore complex. Nat Commun 9(1):2361. https://doi.org/10.1038/s41467-018-04739-y
Muchowski PJ, Wacker JL (2005) Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci 6:11. https://doi.org/10.1038/nrn1587
Munoz IG, Yebenes H, Zhou M, Mesa P, Serna M, Park AY, Bragado-Nilsson E, Beloso A, de Carcer G, Malumbres M, Robinson CV, Valpuesta JM, Montoya G (2011) Crystal structure of the open conformation of the mammalian chaperonin CCT in complex with tubulin. Nat Struct Mol Biol 18(1):14–19. https://doi.org/10.1038/nsmb.1971
Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peranen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, Jackson PK (2007) A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129(6):1201–1213. https://doi.org/10.1016/j.cell.2007.03.053
Noormohammadi A, Khodakarami A, Gutierrez-Garcia R, Lee HJ, Koyuncu S, Konig T, Schindler C, Saez I, Fatima A, Dieterich C, Vilchez D (2016) Somatic increase of CCT8 mimics proteostasis of human pluripotent stem cells and extends C. elegans lifespan. Nat Commun 7:13649. https://doi.org/10.1038/ncomms13649
Ooe A, Kato K, Noguchi S (2007) Possible involvement of CCT5, RGS3, and YKT6 genes up-regulated in p53-mutated tumors in resistance to docetaxel in human breast cancers. Breast Cancer Res Treat 101(3):305–315. https://doi.org/10.1007/s10549-006-9293-x
Pappenberger G, McCormack EA, Willison KR (2006) Quantitative actin folding reactions using yeast CCT purified via an internal tag in the CCT3/gamma subunit. J Mol Biol 360(2):484–496. https://doi.org/10.1016/j.jmb.2006.05.003
Pavel M, Imarisio S, Menzies FM, Jimenez-Sanchez M, Siddiqi FH, Wu X, Renna M, O’Kane CJ, Crowther DC, Rubinsztein DC (2016) CCT complex restricts neuropathogenic protein aggregation via autophagy. Nat Commun 7:13821. https://doi.org/10.1038/ncomms13821
Pereira JH, Ralston CY, Douglas NR, Meyer D, Knee KM, Goulet DR, King JA, Frydman J, Adams PD (2010) Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle. J Biol Chem 285(36):27958–27966. https://doi.org/10.1074/jbc.M110.125344
Pereira JH, Ralston CY, Douglas NR, Kumar R, Lopez T, McAndrew RP, Knee KM, King JA, Frydman J, Adams PD (2012) Mechanism of nucleotide sensing in group II chaperonins. EMBO J 31(3):731–740. https://doi.org/10.1038/emboj.2011.468
Pereira JH, McAndrew RP, Sergeeva OA, Ralston CY, King JA, Adams PD (2017) Structure of the human TRiC/CCT Subunit 5 associated with hereditary sensory neuropathy. Sci Rep 7(1):3673. https://doi.org/10.1038/s41598-017-03825-3
Pines A, Dijk M, Makowski M, Meulenbroek EM, Vrouwe MG, van der Weegen Y, Baltissen M, French PJ, van Royen ME, Luijsterburg MS, Mullenders LH, Vermeulen M, Vermeulen W, Pannu NS, van Attikum H (2018) TRiC controls transcription resumption after UV damage by regulating Cockayne syndrome protein A. Nat Commun 9(1):1040. https://doi.org/10.1038/s41467-018-03484-6
Plimpton RL, Cuellar J, Lai CW, Aoba T, Makaju A, Franklin S, Mathis AD, Prince JT, Carrascosa JL, Valpuesta JM, Willardson BM (2015) Structures of the Gbeta-CCT and PhLP1-Gbeta-CCT complexes reveal a mechanism for G-protein beta-subunit folding and Gbetagamma dimer assembly. Proc Natl Acad Sci USA 112(8):2413–2418. https://doi.org/10.1073/pnas.1419595112
Posokhova E, Song H, Belcastro M, Higgins L, Bigley LR, Michaud NA, Martemyanov KA, Sokolov M (2011) Disruption of the chaperonin containing TCP-1 function affects protein networks essential for rod outer segment morphogenesis and survival. Mol Cell Proteomics 10(1):M110 000570. https://doi.org/10.1074/mcp.m110.000570
Qian-Lin Z, Ting-Feng W, Qi-Feng C, Min-Hua Z, Ai-Guo L (2010) Inhibition of cytosolic chaperonin CCTzeta-1 expression depletes proliferation of colorectal carcinoma in vitro. J Surg Oncol 102(5):419–423. https://doi.org/10.1002/jso.21625
Qiu X, He X, Huang Q, Liu X, Sun G, Guo J, Yuan D, Yang L, Ban N, Fan S, Tao T, Wang D (2015) Overexpression of CCT8 and its significance for tumor cell proliferation, migration and invasion in glioma. Pathol Res Pract 211(10):717–725. https://doi.org/10.1016/j.prp.2015.04.012
Rademacher F, Kehren V, Stoldt VR, Ernst JF (1998) A Candida albicans chaperonin subunit (CaCct8p) as a suppressor of morphogenesis and Ras phenotypes in C-albicans and Saccharomyces cerevisiae. Microbiol-Uk 144:2951–2960. https://doi.org/10.1099/00221287-144-11-2951
Reissmann S, Joachimiak LA, Chen B, Meyer AS, Nguyen A, Frydman J (2012) A gradient of ATP affinities generates an asymmetric power stroke driving the chaperonin TRIC/CCT folding cycle. Cell reports 2(4):866–877. https://doi.org/10.1016/j.celrep.2012.08.036
Rivenzon-Segal D, Wolf SG, Shimon L, Willison KR, Horovitz A (2005) Sequential ATP-induced allosteric transitions of the cytoplasmic chaperonin containing TCP-1 revealed by EM analysis. Nat Struct Mol Biol 12(3):233–237. https://doi.org/10.1038/nsmb901
Roh SH, Kasembeli M, Bakthavatsalam D, Chiu W, Tweardy DJ (2015) Contribution of the type II chaperonin, TRiC/CCT, to oncogenesis. Int J Mol Sci 16(11):26706–26720. https://doi.org/10.3390/ijms161125975
Roh SH, Kasembeli M, Galaz-Montoya JG, Trnka M, Lau WC, Burlingame A, Chiu W, Tweardy DJ (2016a) Chaperonin TRiC/CCT modulates the folding and activity of leukemogenic fusion oncoprotein AML1-ETO. J Biol Chem 291(9):4732–4741. https://doi.org/10.1074/jbc.M115.684878
Roh SH, Kasembeli MM, Galaz-Montoya JG, Chiu W, Tweardy DJ (2016b) Chaperonin TRiC/CCT recognizes fusion oncoprotein AML1-ETO through subunit-specific interactions. Biophys J 110(11):2377–2385. https://doi.org/10.1016/j.bpj.2016.04.045
Saegusa K, Sato M, Sato K, Nakajima-Shimada J, Harada A, Sato K (2014) Caenorhabditis elegans chaperonin CCT/TRiC is required for actin and tubulin biogenesis and microvillus formation in intestinal epithelial cells. Mol Biol Cell 25(20):3095–3104. https://doi.org/10.1091/mbc.E13-09-0530
Satish L, Johnson S, Wang JH, Post JC, Ehrlich GD, Kathju S (2010) Chaperonin containing T-complex polypeptide subunit eta (CCT-eta) is a specific regulator of fibroblast motility and contractility. PLoS ONE 5(4):e10063. https://doi.org/10.1371/journal.pone.0010063
Satish L, O’Gorman DB, Johnson S, Raykha C, Gan BS, Wang JH, Kathju S (2013) Increased CCT-eta expression is a marker of latent and active disease and a modulator of fibroblast contractility in Dupuytren’s contracture. Cell Stress Chaperones 18(4):397–404. https://doi.org/10.1007/s12192-012-0392-9
Seixas C, Cruto T, Tavares A, Gaertig J, Soares H (2010) CCTalpha and CCTdelta chaperonin subunits are essential and required for cilia assembly and maintenance in Tetrahymena. PLoS ONE 5(5):e10704. https://doi.org/10.1371/journal.pone.0010704
Seo S, Baye LM, Schulz NP, Beck JS, Zhang Q, Slusarski DC, Sheffield VC (2010) BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc Natl Acad Sci USA 107(4):1488–1493. https://doi.org/10.1073/pnas.0910268107
Sergeeva OA, Chen B, Haase-Pettingell C, Ludtke SJ, Chiu W, King JA (2013) Human CCT4 and CCT5 chaperonin subunits expressed in Escherichia coli form biologically active homo-oligomers. J Biol Chem 288(24):17734–17744. https://doi.org/10.1074/jbc.M112.443929
Sergeeva OA, Tran MT, Haase-Pettingell C, King JA (2014) Biochemical characterization of mutants in chaperonin proteins CCT4 and CCT5 associated with hereditary sensory neuropathy. J Biol Chem 289(40):27470–27480. https://doi.org/10.1074/jbc.M114.576033
Shahmoradian SH, Galaz-Montoya JG, Schmid MF, Cong Y, Ma B, Spiess C, Frydman J, Ludtke SJ, Chiu W (2013) TRiC’s tricks inhibit huntingtin aggregation. Elife 2:e00710. https://doi.org/10.7554/eLife.00710
Siegers K, Bolter B, Schwarz JP, Bottcher UMK, Guha S, Hartl FU (2003) TRiC/CCT cooperates with different upstream chaperones in the folding of distinct protein classes (Retracted Article. See vol 27, 301 p, 2008). EMBO J 22(19):5230–5240. https://doi.org/10.1093/emboj/cdg483
Sinha S, Belcastro M, Datta P, Seo S, Sokolov M (2014) Essential role of the chaperonin CCT in rod outer segment biogenesis. Invest Ophth Vis Sci 55(6):3775–3784. https://doi.org/10.1167/iovs.14-13889
Skjaerven L, Cuellar J, Martinez A, Valpuesta JM (2015) Dynamics, flexibility, and allostery in molecular chaperonins. FEBS Lett 589(19 Pt A):2522–2532. https://doi.org/10.1016/j.febslet.2015.06.019
Sontag EM, Joachimiak LA, Tan Z, Tomlinson A, Housman DE, Glabe CG, Potkin SG, Frydman J, Thompson LM (2013) Exogenous delivery of chaperonin subunit fragment ApiCCT1 modulates mutant Huntingtin cellular phenotypes. Proc Natl Acad Sci USA 110(8):3077–3082. https://doi.org/10.1073/pnas.1222663110
Sot B, Rubio-Munoz A, Leal-Quintero A, Martinez-Sabando J, Marcilla M, Roodveldt C, Valpuesta JM (2017) The chaperonin CCT inhibits assembly of alpha-synuclein amyloid fibrils by a specific, conformation-dependent interaction. Sci Rep 7:40859. https://doi.org/10.1038/srep40859
Soues S, Kann M-L, Fouquet J-P, Melki R (2003) The cytosolic chaperonin CCT associates to cytoplasmic microtubular structures during mammalian spermiogenesis and to heterochromatin in germline and somatic cells. Exp Cell Res 288(2):363–373. https://doi.org/10.1016/s0014-4827(03)00248-9
Spiess C, Meyer AS, Reissmann S, Frydman J (2004) Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol 14(11):598–604. https://doi.org/10.1016/j.tcb.2004.09.015
Srikakulam R, Winkelmann DA (1999) Myosin II folding is mediated by a molecular chaperonin. J Biol Chem 274(38):27265–27273. https://doi.org/10.1074/jbc.274.38.27265
Stemp MJ, Guha S, Hartl FU, Barral JM (2005) Efficient production of native actin upon translation in a bacterial lysate supplemented with the eukaryotic chaperonin TRiC. Biol Chem 386(8):753–757. https://doi.org/10.1515/BC.2005.088
Stirling PC, Cuellar J, Alfaro GA, El Khadali F, Beh CT, Valpuesta JM, Melki R, Leroux MR (2006) PhLP3 modulates CCT-mediated actin and tubulin folding via ternary complexes with substrates. J Biol Chem 281(11):7012–7021. https://doi.org/10.1074/jbc.M513235200
Tam S, Geller R, Spiess C, Frydman J (2006) The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat Cell Biol 8(10):1155–1162. https://doi.org/10.1038/ncb1477
Tam S, Spiess C, Auyeung W, Joachimiak L, Chen B, Poirier MA, Frydman J (2009) The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat Struct Mol Biol 16(12):1279–1285. https://doi.org/10.1038/nsmb.1700
Tracy CM, Kolesnikov AV, Blake DR, Chen CK, Baehr W, Kefalov VJ, Willardson BM (2015) Retinal cone photoreceptors require phosducin-like protein 1 for G protein complex assembly and signaling. PLoS ONE 10(2):e0117129. https://doi.org/10.1371/journal.pone.0117129
Trinidad AG, Muller PA, Cuellar J, Klejnot M, Nobis M, Valpuesta JM, Vousden KH (2013) Interaction of p53 with the CCT complex promotes protein folding and wild-type p53 activity. Mol Cell 50(6):805–817. https://doi.org/10.1016/j.molcel.2013.05.002
Ursic D, Culbertson MR (1991) The yeast homolog to mouse Tcp-1 affects microtubule-mediated processes. Mol Cell Biol 11(5):2629–2640. https://doi.org/10.1128/mcb.11.5.2629
Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ (1998) Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell 93(5):863–873. https://doi.org/10.1016/S0092-8674(00)81446-4
van den Brink DM, Brites P, Haasjes J, Wierzbicki AS, Mitchell J, Lambert-Hamill M, de Belleroche J, Jansen GA, Waterham HR, Ronald Wanders JA (2003) Identification of PEX7 as the Second Gene Involved in Refsum Disease. Am J Human Genet 72(2):471–477. https://doi.org/10.1086/346093
Vinh DBN, Drubin DG (1994) A yeast Tcp-1-like protein is required for actin function in-vivo. P Natl Acad Sci USA 91(19):9116–9120. https://doi.org/10.1073/pnas.91.19.9116
Voisine C, Pedersen JS, Morimoto RI (2010) Chaperone networks: tipping the balance in protein folding diseases. Neurobiol Dis 40(1):12–20. https://doi.org/10.1016/j.nbd.2010.05.007
Waldmann T, Lupas A, Kellermann J, Peters J, Baumeister W (1995) Primary structure of the thermosome from thermoplasma-acidophilum. Biol Chem H-S 376(2):119–126. https://doi.org/10.1515/bchm3.1995.376.2.119
Wang H, Han W, Takagi J, Cong Y (2018) Yeast inner-subunit PA–NZ-1 labeling strategy for accurate subunit identification in a macromolecular complex through Cryo-EM analysis. J Mol Biol 430(10):1417–1425. https://doi.org/10.1016/j.jmb.2018.03.026
Wei PL, Huang CY, Tai CJ, Batzorig U, Cheng WL, Hunag MT, Chang YJ (2016) Glucose-regulated protein 94 mediates metastasis by CCT8 and the JNK pathway in hepatocellular carcinoma. Tumour Biol 37(6):8219–8227. https://doi.org/10.1007/s13277-015-4669-3
Willardson BM, Howlett AC (2007) Function of phosducin-like proteins in G protein signaling and chaperone-assisted protein folding. Cell Signal 19(12):2417–2427. https://doi.org/10.1016/j.cellsig.2007.06.013
Willison KR (2018a) The structure and evolution of eukaryotic chaperonin-containing TCP-1 and its mechanism that folds actin into a protein spring. Biochem J 475(19):3009–3034. https://doi.org/10.1042/bcj20170378
Willison KR (2018b) The substrate specificity of eukaryotic cytosolic chaperonin CCT. Philos Trans R Soc Lond B Biol Sci 373(1749). https://doi.org/10.1098/rstb.2017.0192
Wu CZ, Chang LC, Lin YF, Hung YJ, Pei D, Chen JS (2015a) Chaperonin-containing t-complex protein-1 subunit beta as a possible biomarker for the phase of glomerular hyperfiltration of diabetic nephropathy. Dis Markers 2015:548101. https://doi.org/10.1155/2015/548101
Wu X, Zhang H, Chen D, Song Y, Qian R, Chen C, Mao X, Chen X, Zhang W, Shao B, Shen J, Yan Y, Wu X, Liu Y (2015b) Up-Regulation of CCT8 related to neuronal apoptosis after traumatic brain injury in adult rats. Neurochem Res 40(9):1882–1891. https://doi.org/10.1007/s11064-015-1683-1
Xu XM, Wang J, Xuan ZY, Goldshmidt A, Borrill PGM, Hariharan N, Kim JY, Jackson D (2011) Chaperonins facilitate KNOTTED1 cell-to-cell trafficking and stem cell function. Science 333(6046):1141–1144. https://doi.org/10.1126/science.1205727
Yam AY, Xia Y, Lin HT, Burlingame A, Gerstein M, Frydman J (2008) Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat Struct Mol Biol 15(12):1255–1262. https://doi.org/10.1038/nsmb.1515
Yamamoto YY, Uno Y, Sha E, Ikegami K, Ishii N, Dohmae N, Sekiguchi H, Sasaki YC, Yohda M (2017) Asymmetry in the function and dynamics of the cytosolic group II chaperonin CCT/TRiC. PLoS ONE 12(5):e0176054. https://doi.org/10.1371/journal.pone.0176054
Yin H, Miao X, Wu Y, Wei Y, Zong G, Yang S, Chen X, Zheng G, Zhu X, Guo Y, Li C, Chen Y, Wang Y, He S (2016) The role of the Chaperonin containing t-complex polypeptide 1, subunit 8 (CCT8) in B-cell non-Hodgkin’s lymphoma. Leuk Res 45:59–67. https://doi.org/10.1016/j.leukres.2016.04.010
Ying Z, Tian H, Li Y, Lian R, Li W, Wu S, Zhang HZ, Wu J, Liu L, Song J, Guan H, Cai J, Zhu X, Li J, Li M (2017) CCT6A suppresses SMAD2 and promotes prometastatic TGF-beta signaling. J Clin Invest 127(5):1725–1740. https://doi.org/10.1172/JCI90439
Yokota S, Yamamoto Y, Shimizu K, Momoi H, Kamikawa T, Yamaoka Y, Yanagi H, Yura T, Kubota H (2001) Increased expression of cytosolic chaperonin CCT in human hepatocellular and colonic carcinoma. Cell Stress Chaperon 6(4):345–350. https://doi.org/10.1379/1466-1268(2001)006<0345:Ieoccc>2.0.Co;2
Yokota S, Yanagi H, Yura T, Kubota H (1999) Cytosolic chaperonin is up-regulated during cell growth - preferential, expression and binding to tubulin at G(1)/S transition through early S phase. J Biol Chem 274(52):37070–37078. https://doi.org/10.1074/jbc.274.52.37070
Zang Y, Jin M, Wang H, Cui Z, Kong L, Liu C, Cong Y (2016) Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM. Nat Struct Mol Biol 23(12):1083–1091. https://doi.org/10.1038/nsmb.3309
Zang Y, Wang H, Cui Z, Jin M, Liu C, Han W, Wang Y, Cong Y (2018) Development of a yeast internal-subunit eGFP labeling strategy and its application in subunit identification in eukaryotic group II chaperonin TRiC/CCT. Sci Rep 8(1):2374. https://doi.org/10.1038/s41598-017-18962-y
Zhang J, Baker ML, Schroder GF, Douglas NR, Reissmann S, Jakana J, Dougherty M, Fu CJ, Levitt M, Ludtke SJ, Frydman J, Chiu W (2010) Mechanism of folding chamber closure in a group II chaperonin. Nature 463(7279):379–383. https://doi.org/10.1038/nature08701
Zhang Q, Yu D, Seo S, Stone EM, Sheffield VC (2012) Intrinsic protein-protein interaction-mediated and chaperonin-assisted sequential assembly of stable bardet-biedl syndrome protein complex, the BBSome. J Biol Chem 287(24):20625–20635. https://doi.org/10.1074/jbc.M112.341487
Zhang Y, Wang Y, Wei Y, Wu J, Zhang P, Shen S, Saiyin H, Wumaier R, Yang X, Wang C, Yu L (2016) Molecular chaperone CCT3 supports proper mitotic progression and cell proliferation in hepatocellular carcinoma cells. Cancer Lett 372(1):101–109. https://doi.org/10.1016/j.canlet.2015.12.029
Zhao X, Chen XQ, Han E, Hu Y, Paik P, Ding Z, Overman J, Lau AL, Shahmoradian SH, Chiu W, Thompson LM, Wu C, Mobley WC (2016) TRiC subunits enhance BDNF axonal transport and rescue striatal atrophy in Huntington’s disease. Proc Natl Acad Sci USA 113(38):E5655–E5664. https://doi.org/10.1073/pnas.1603020113
Zou Q, Yang ZL, Yuan Y, Li JH, Liang LF, Zeng GX, Chen SL (2013) Clinicopathological features and CCT2 and PDIA2 expression in gallbladder squamous/adenosquamous carcinoma and gallbladder adenocarcinoma. World J Surg Oncol 11:143. https://doi.org/10.1186/1477-7819-11-143
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Jin, M., Liu, C., Han, W., Cong, Y. (2019). TRiC/CCT Chaperonin: Structure and Function. In: Harris, J., Marles-Wright, J. (eds) Macromolecular Protein Complexes II: Structure and Function . Subcellular Biochemistry, vol 93. Springer, Cham. https://doi.org/10.1007/978-3-030-28151-9_19
Download citation
DOI: https://doi.org/10.1007/978-3-030-28151-9_19
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-28150-2
Online ISBN: 978-3-030-28151-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)