Abstract
Liver diseases, especially liver cancer, are a major threat to human health. In order to assist doctors in efficiently diagnosing the condition and developing treatment plans, automatic segmentation of the liver from CT images has a strong clinical need. However, it is difficult to design an accurate segmentation algorithm because of the blurred boundary of CT images and the great difference of pathological changes. In this paper, we propose a cascade model for liver segmentation from CT images, which uses cascade U-nets with adversarial learning to obtain more accurate segmentation results. The experimental results show that the proposed algorithm is competitive and its dice value reaches 0.955.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
1.1 Motivation
Liver is the largest abdominal organ in human body, which is often threatened by diseases and drug damage. According to the Global Hepatitis Report 2017 released by the World Health Organization, about 325 million people worldwide are infected with chronic hepatitis B virus or hepatitis C virus, which can lead to chronic infection for life and eventually lead to progressive liver damage. CT image is a very convenient way to detect abdominal organs. Radiologists often need to observe the shape and texture of patients’ liver through CT images to find visible lesions. However, it is tedious and inefficient to analyze abdominal CT images manually by radiologists. Therefore, it is very important to study the automatic liver segmentation technique in CT images. However, as shown in Fig. 1, this task faces many challenges, such as great variation of size and shape, unclear boundaries and different degrees of lesions, so automatic segmentation technology has not yet entered the clinical stage.
1.2 Related Work
Traditional manual segmentation requires extensive radiology experience and is very time consuming. In order to help radiologists improve their work efficiency, researchers have turned their attention to automatic liver segmentation technology. In recent years, many automatic segmentation methods have been proposed. Pham et al. [16] proposed a method based on grayscale and texture. This method includes global threshold, region growth, voxel classification and edge detection, but it is easy to segment along the blurred boundary, resulting in boundary leakage. Chan et al. [1] proposed a variational semi-automated liver segmentation method, which employs prior knowledge and morphological features into account. Zheng et al. [22] and Zhang et al. [21] proposed a point-based statistical shape model (SSM), which can obtain higher accuracy in the case of small amount of data.
2 Background
2.1 Image Segmentation Network
The fully convolutional network (FCN) [13] is the basic architecture for many semantic segmentation tasks, which consists of cascaded downsampling path and upsampling path. The U-net [18] extends FCN by introducing skip connection between downsampling path and corresponding upsampling path. The skip connection improves the network performance by facilitating the transmission of information. The DeepLab architecture [12] involved atrous convolutions and poolings into the CNN architecture. Based on the DeepLab [2], Chen et al. proposed the latest DeepLabV3 [3]. The upsampling path of this model only consists of very few convolution layers, which is different from the upsampling path used in the FCN and the U-net. Jegou et al. [10] introduces the Dense connection [8] into FCN for segmentation tasks.
In recent years, fully convolutional network has developed rapidly in the field of semantics segmentation, and has achieved remarkable results in the competition of image segmentation. Exploration of this new segmentation method has begun for medical image segmentation. Christ et al. [4] and Vorontsov et al. [19] proposed a cascaded fully convolutional network, which combines two fully convolutional networks. The first network is used to segment liver, and the second network is used to segment liver lesions on the basis of the first network. The segmentation results are processed by 3D conditional random field to make results more accurate. The authors of [5] proposed a deeply supervised network for liver segmentation. The input of the network is part of the 3D bounding box, which has to be slid on a target scan during the test time. In order to alleviate the problem of vanishing gradient, the network uses additional deconvolution layers to generate feature maps from two intermediate layers, and the gradients of loss are calculated from several branches. A three-dimensional convolutional neural network is proposed in [7]. This method contains two steps. First, a deep 3D CNN is trained to learn prior map of the liver. Then, in order to optimize segmentation result, both global and local appearance information from the prior segmentation are adaptively incorporated into a segmentation model. Rafiei et al. [17] proposed a 3D-2D fully convolutional network, which makes full use of the spatial information in CT volume to segment the liver while computation and memory consuming are moderate.
2.2 GAN
GAN [6] has achieved great success in the field of image generation. It is inspired by the theory of zero-sum game. It regards generation problem as conflict and cooperation between two players: Discriminator and generator. First, images will be generated from noise by the generator, and then, with the help of discriminator, quality of the image will be improved step by step. CGAN [14] has gained impressive results in various image-to-image conversion problems, such as image super-resolution [11], image inpainting [15], and style transfer [12]. CGAN regards original image as a constraint, feeding it into the network along with noise as input, and adding conditional variables into loss functions of both generator and discriminator. Under this circumstance, the generated image is of higher quality and closer to the real image. Zhang et al. [20] firstly proposed a stacking model of GAN called StackGAN. The model can be divided into two stages: The first stage generates a coarse image mainly containing primitive shapes and colors based on the given text description. Then, the second stage takes the coarse image and text description as input, to generate high-resolution images gradually. Huang et al. [9] designed a stacked GAN, which is trained to invert the hierarchical representations of a bottom-up discriminative network. Each GAN in this stacked model learns to generate lower-level representations that are conditional on higher level representations in order to generate more qualified images.
3 Method
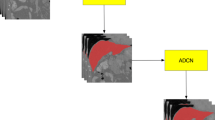
In this paper, we propose a cascade model with adversarial learning for liver segmentation. The algorithm is able to segment the liver accurately and efficiently from CT slices. It consists of two stages: In the first stage, the 2D slice images are fed into the first stage U-net segmentation network, and the segmentation results of the first stage are obtained. In the second stage, CT slices are concatenated with the results of the first U-net, and then input into the second stage U-net segmentation network to obtain more accurate segmentation results. The training of the above two stages utilizes the adversarial loss. The model is illustrated in the Fig. 2.
3.1 Model
Generative Adversarial Networks (GANs) consists of two components: an generator G and a discriminator D. The two components are competing in a zero-sum game, in which the generator G aims to produce a realistic image given an input z, that conforms to a certain distribution. The discriminator D is forced to distinguish if a given image is generated by G or it is indeed a real one from the dataset. The adversarial competition enables the generator and the discriminator to achieve better performance, whilst making it hard for D to differentiate generation of G from the real data. Conditional Generative Adversarial Networks (CGANs) extends GANs by introducing an additional observed information, namely conditioning variable, to both the generator G and the discriminator D.
The architecture of the proposed model is shown in the Fig. 3. It consists of two stacked CGANs: CGAN1(G1, D1), CGAN2(G2, D2), with the second stacked on top of the first. CGAN is an extension of GAN. It introduces additional conditional variables for both generator and discriminator. In the proposed model, CT slice is the conditional variable. The architectures of the generator G and discriminator D (ignoring BN and ReLU) can be seen in Tables 1 and 2, respectively.
Both the generator G1 and the discriminator D1 of the first CGAN are added with condition, the CT image x. G1 is trained to produce mask G1 (z, x) corresponding to CT image. y denotes the ground truth corresponding x.
The objective function of the CGAN1 is:
In order to obtain deterministic results from G1, we eliminate random noise and simplify the formula as follows:
In addition to adversarial loss, L1 loss is also applied to obtain more accurate pixel-level classification results:
So, the final objective function of CGAN1 is:
λ is L1 loss weighted coefficient. For CGAN2, it consists of G2 and D2. We use similar objective functions. The adversarial loss of CGAN2 is:
The difference is that CGAN2 combines CT slices and the output of CGAN1 as input. Finally, the objective function of the whole model is:
The output of CGAN1 is fed into CGAN2 as a prior knowledge to help it get more accurate segmentation results.
3.2 Training
Training is divided into two phases. The first phase employs an alternating training scheme. Specifically, each time a mini-batch CT slices is fed into model, firstly, update G1, D1, with G2, D2 fixed. Then, update G2, D2, and fix G1, D1. After 10 epochs training, enter the second stage. In the second phase, the entire model is end-to-end trained for several epochs updating CGAN1 and CGAN2 simultaneously.
4 Experiment
4.1 Dataset
We employ dataset provided by Liver Tumor Segmentation Challenge (LiTS) which is organized by ISBI and MICCAI in conjunction to evaluate the proposed method. The open dataset includes 131 CT volumes and corresponding expert standard labels. The dataset was collected from six different clinical sites using different scanners and scanning methods with different slice resolutions and slice spacings. We randomly divided the data set into 10 folds, each with about 13 volumes, and a fold is used for validation, while the others for training, namely 10-fold cross validation. For data preprocessing, we crop the image intensity values of all volumes to the range of [−200, 250] HU, in order to reduce irrelevant information.
4.2 Implementation
The proposed model was implemented based on tensorflow1.12.0. All experiments were performed on workstations equipped with two Intel Xeon E5-2609 processors, 64G RAM and one NVIDIA TITAN XP GPU.
We use the Adam solver to train the CGAN1 and CGAN2 with a mini-batch size of 8. The model was trained for 15 epochs and the initial learning rate was set to 0.0002. In the second phase of training, the learning rate is divided by 10. In our experiments, λ is set to 100.
5 Comparison with Other Methods
The proposed model is compared with two baseline methods. U-net is widely used in biomedical image segmentation tasks. A 3D-2D model proposed in [17] makes full use of volume’s 3D spatial information.
Dice is employed for evaluating our model. The Dice score is defined as:
where |X| and |Y| are the pixel values of predicted result and ground truth respectively, and the Dice score is in the interval [0,1]. A perfect segmentation yields a Dice score of 1. Dice is usually divided into Dice per case and Dice global where Dice per case score refers to an average Dice score per volume while Dice global score is the Dice score evaluated by combining all datasets into one. In this paper, we employ Dice per case score to evaluate liver segmentation performance.
Some quantitative results of several methods are shown in Table 3 through cross validation. It is observed that our model has achieved more accurate segmentation results. This is because we introduced adversarial loss, which can be seen as a high-level loss that provides more global optimization information for the model. On the other hand, the cascade structure can help the model to further improve performance. Using the output of the first stage as the prior knowledge of the second stage can provide more reliable information for the second stage CGAN and help it achieve more accurate segmentation. Some results are demonstrated in Fig. 4.
6 Conclusion
In this paper, we propose a cascade model for liver segmentation with adversarial. This model is able to segment the liver accurately and efficiently from CT slice images. It consists of two stages: In the first stage, the 2D CT slice image is input into the first-level CGAN segmentation network to obtain the segmentation result of the first stage. In the second stage, the CT slice image is concatenated with the corresponding segmentation results of the first stage as input of the second stage CGAN2 to obtain more accurate segmentation results. The main contributions of this work are the followings: First, the generative adversarial method is introduced into the liver segmentation task in CT images. Compared with the traditional L1 and L2 losses, adversarial loss can get a clear prediction result and reduce the ambiguity of the segmentation results. Second, due to the cascade structure, the segmentation result of the first phase can provide a prior knowledge for the second phase. Thus, more accurate segmentation results can be obtained. Further research can explore more efficient CGAN cascading methods.
References
Chan, T.F., Vese, L.A.: Active contours without edges. IEEE Trans. Image Process. 10(2), 266–277 (2001)
Chen, L.C., Papandreou, G., Kokkinos, I., Murphy, K., Yuille, A.L.: Semantic image segmentation with deep convolutional nets and fully connected crfs. arXiv preprint arXiv:1412.7062 (2014)
Chen, L.C., Papandreou, G., Schroff, F., Adam, H.: Rethinking atrous convolution for semantic image segmentation. arXiv preprint arXiv:1706.05587 (2017)
Christ, P.F., et al.: Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. In: Ourselin, S., Joskowicz, L., Sabuncu, Mert R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 415–423. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_48
Dou, Q., Chen, H., Jin, Y., Yu, L., Qin, J., Heng, P.-A.: 3D deeply supervised network for automatic liver segmentation from CT volumes. In: Ourselin, S., Joskowicz, L., Sabuncu, Mert R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 149–157. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_18
Goodfellow, I., et al.: Generative adversarial nets. In: Advances in neural information processing systems, pp. 2672–2680 (2014)
Hu, P., Wu, F., Peng, J., Liang, P., Kong, D.: Automatic 3D liver segmentation based on deep learning and globally optimized surface evolution. Phys. Med. Biol. 61(24), 8676 (2016)
Huang, G., Liu, Z., Van Der Maaten, L., Weinberger, K.Q.: Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4700–4708 (2017)
Huang, X., Li, Y., Poursaeed, O., Hopcroft, J., Belongie, S.: Stacked generative adversarial networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 5077–5086 (2017)
Jégou, S., Drozdzal, M., Vazquez, D., Romero, A., Bengio, Y.: The one hundred layers tiramisu: fully convolutional densenets for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, pp. 11–19 (2017)
Ledig, C., et al.: Photo-realistic single image super-resolution using a generative adversarial network. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4681–4690 (2017)
Li, C., Wand, M.: Precomputed real-time texture synthesis with markovian generative adversarial networks. In: Leibe, B., Matas, J., Sebe, N., Welling, M. (eds.) ECCV 2016. LNCS, vol. 9907, pp. 702–716. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46487-9_43
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), June 2015
Mirza, M., Osindero, S.: Conditional generative adversarial nets. arXiv preprint arXiv:1411.1784 (2014)
Pathak, D., Krahenbuhl, P., Donahue, J., Darrell, T., Efros, A.A.: Context encoders: feature learning by inpainting. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2536–2544 (2016)
Pham, M., Susomboon, R., Disney, T., Raicu, D., Furst, J.: A comparison of texture models for automatic liver segmentation. In: Medical Imaging 2007: Image Processing, vol. 6512, p. 65124E. International Society for Optics and Photonics (2007)
Rafiei, S., Nasr-Esfahani, E., Soroushmehr, S., Karimi, N., Samavi, S., Najarian, K.: Liver segmentation in CT images using three dimensional to two dimensional fully connected network. arXiv preprint arXiv:1802.07800 (2018)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Vorontsov, E., Tang, A., Pal, C., Kadoury, S.: Liver lesion segmentation informed by joint liver segmentation. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), pp. 1332–1335. IEEE (2018)
Zhang, H., et al.: StackGAN: text to photo-realistic image synthesis with stacked generative adversarial networks. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 5907–5915 (2017)
Zhang, X., Tian, J., Deng, K., Wu, Y., Li, X.: Automatic liver segmentation using a statistical shape model with optimal surface detection. IEEE Trans. Biomed. Eng. 57(10), 2622–2626 (2010)
Zheng, S., et al.: A novel variational method for liver segmentation based on statistical shape model prior and enforced local statistical feature. In: 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), pp. 261–264. IEEE (2017)
Acknowledgement
This work is supported by Shanghai Science and Technology Commission (grant No. 17511104203) and NSFC (grant NO. 61472087).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Chen, Y., Li, S., Yang, S., Luo, W. (2019). Liver Segmentation in CT Images with Adversarial Learning. In: Huang, DS., Bevilacqua, V., Premaratne, P. (eds) Intelligent Computing Theories and Application. ICIC 2019. Lecture Notes in Computer Science(), vol 11643. Springer, Cham. https://doi.org/10.1007/978-3-030-26763-6_45
Download citation
DOI: https://doi.org/10.1007/978-3-030-26763-6_45
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-26762-9
Online ISBN: 978-3-030-26763-6
eBook Packages: Computer ScienceComputer Science (R0)