Abstract
Accurate risk management and disease prediction are vital in intensive care units to channel prompt care to patients in critical conditions and aid medical personnel in effective decision making. Clinical nursing notes document subjective assessments and crucial information of a patient’s state, which is mostly lost when transcribed into Electronic Medical Records (EMRs). The Clinical Decision Support Systems (CDSSs) in the existing body of literature are heavily dependent on the structured nature of EMRs. Moreover, works which aim at benchmarking deep learning models are limited. In this paper, we aim at leveraging the underutilized treasure-trove of patient-specific information present in the unstructured clinical nursing notes towards the development of CDSSs. We present a fuzzy token-based similarity approach to aggregate voluminous clinical documentations of a patient. To structure the free-text in the unstructured notes, vector space and coherence-based topic modeling approaches that capture the syntactic and latent semantic information are presented. Furthermore, we utilize the predictive capabilities of deep neural architectures for disease prediction as ICD-9 code group. Experimental validation revealed that the proposed Term weighting of nursing notes AGgregated using Similarity (TAGS) model outperformed the state-of-the-art model by 5% in AUPRC and 1.55% in AUROC.
This work is funded by the Government of India’s DST-SERB Early Career Research Grant (ECR/2017/001056) to Sowmya Kamath S.
T. Gangavarapu and A. Jayasimha—contributed equally to this work.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Healthcare analytics
- Disease group prediction
- Natural Language Processing
- Risk assessment systems
- Deep learning
1 Introduction
Risk assessment and disease prediction in Intensive Care Units (ICUs) have had a prominent impact on clinical care and management [13]. As per US healthcare reports, more than 30 million patients visit hospitals annually, and 83% of these hospitals have adopted the Electronic Medical Record (EMR) systems [6]. In the recent years, a rapid increase in the adoption of EMRs in the hospitals of developed countries is also observed, which has prompted significant research towards modeling the patient data for diverse clinical tasks like mortality, length of stay, and hospital readmission prediction using various machine and deep learning approaches [15]. Such works have further been employed towards determining diagnostic measures needed to design and implement effective healthcare policies [8]. Despite these trends in western countries, hospitals in developing countries are yet to gain momentum in the implementation of EMRs.
Caregivers in developing countries most often resort to a human evaluation of available clinical notes for decision making and cause-effect inference [10]. Clinicians and nurses document subjective assessments and crucial information about a patient’s state, which is often lost when transcribed into EMRs [4]. Clinical nursing notes remain largely unexplored for mining and modeling the rich and valuable patient-specific information. It is challenging to utilize unstructured clinical nursing notes to predict the clinical outcomes and events primarily due to their sparsity, rawness, complex linguistic and temporal structure, high-dimensionality, rich medical jargons, and abundant abbreviations [7]. How effectively the rich information embedded in unstructured clinical text is extracted and consolidated, determines the efficacy of their usage [14]. Due to the diverse manifold nature of prevalent disease symptoms, there is often a need for assigning multiple labels to a patient record in the database [1]. Risk assessment as ICD-9Footnote 1 code group prediction using clinician’s notes can help in recognition of the onset and severity of the disease. Such assessments, when preceded by a timely response and effective communication by interdisciplinary care team members have been reported to result in a reduction in the hospital mortality rate [3].
Most state-of-the-art works [8, 12] present machine learning models built on structured EMR data to facilitate various clinical prediction tasks. The few works that adopt deep learning models [5, 13] neglect the rich patient-specific information present in the clinical nursing notes. In this paper, we utilize term weighting, word embedding (Doc2Vec), and coherence-based topic modeling (Latent Dirichlet Allocation (LDA)) approaches to structure the clinical nursing notes for capturing both the syntactic and semantic relationships between the textual features of the nursing notes, to aid in the accurate prediction of the ICD-9 code group. Deriving optimal data representations and eliminating redundant data from the nursing notes is achieved using a fuzzy similarity based data cleansing approach. Furthermore, we report the results of our exhaustive experimentation with three deep architectures including Multi-Layer Perceptron (MLP), Long Short Term Memory (LSTM), and Convolutional Neural Network (CNN).
The rest of this paper is structured as follows: Sect. 2 discusses relevant work in the area of our work. Section 3 presents our detailed methodology for deriving optimal data representations. Data modeling and deep architectures used in ICD-9 code group prediction are discussed in Sect. 4. The experiments, evaluation, and results are discussed in detail in Sect. 5. Finally, we conclude with a summary and future research possibilities in Sect. 6.
2 Related Work
The prediction of prominent clinical outcomes and benchmarking the performance of the proposed machine and deep learning models is greatly facilitated by the availability of sizeable public patient datasets such as MIMIC-III [9]. In 2016, Pirracchio [12] presented a super learner algorithm which was an ensemble of various machine learning models. For the task of ICU mortality prediction, the super learner algorithm outperformed various severity scores including SAPS-II, SOFA, and APACHE-II. The preponderance of machine learning approaches over traditional prognostic scoring systems was emphasized. However, the obtained results were not benchmarked against the latest machine and deep learning models. The clinical task of mortality prediction was presented as a case study by Johnson et al. [8], who highlighted the challenges in replicating the results reported by 28 related and recent prominent publications on publicly available MIMIC-III database. They used an extracted set of features from the database and compared the reported performance against gradient boosting and logistic regression models. In order to take into consideration the significant heterogeneity in the studies and ensure a fairer comparison between approximate approaches, Johnson et al. [8] emphasized the need to improve the way of reporting the performance of clinical prediction tasks.
Harutyunyan et al. [5] used multitask recurrent neural networks to develop a comprehensive deep learning model and benchmarked their outcomes on four disparate clinical prediction tasks on the MIMIC-III database. Their work showed encouraging results in clinical prediction tasks with the use of deep learning approaches. However, their obtained performance was only benchmarked against a standard logistic regression model and LSTM model, and did not benchmark against machine learning models (including super learner) or severity scoring systems. An extensive set of benchmarking results on various clinical tasks such as the prediction of the ICD-9 code group, length of stay, and several versions of in-hospital mortality was presented by Purushotham et al. [13] on MIMIC-III, against various severity scoring systems and machine learning models. More recently, Krishnan and Kamath [10] benchmarked their performance for the ICU mortality prediction task. They used Word2Vec embeddings of the electrocardiogram reports in the MIMIC-III database and an unsupervised data cleaning approach using K-means clustering, followed by an extreme learning machine classifier for the prediction task.
Our work advances the efforts of these previous state-of-the-art approaches by exploring the potential use and availability of unstructured clinical notes, an under-tapped resource of rich patient-specific information. The EMR coding process often decimates the treasure-trove of information present in the clinician’s notes. Our work addresses this issue by designing a clinical processing and representation generation methodology based on clinical concept extraction and topic modeling using deep learning models. Furthermore, we present an exhaustive comparative study to evaluate the performance of the proposed fuzzy similarity based data cleansing approach across a variety of deep learning models in the clinical task of multi-label ICD-9 code group prediction.
3 Materials and Methods
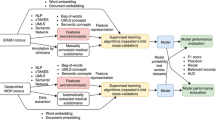
In this section, we present a brief description of the Natural Language Processing (NLP) pipeline depicted in Fig. 1. We also discuss the preprocessing steps employed to derive optimal data representations for the multi-label classification task of ICD-9 code group prediction.
3.1 Dataset and Cohort Selection
The Massachusetts Institute of Technology Lab for Computational Physiology developed MIMIC-III, a freely accessible large healthcare database. It comprises comprehensive and diverse de-identified healthcare data of more than 40, 000 intensive care patients. Also provided are 223, 556 nursing notes corresponding to 7, 704 distinct ICU patients (diagnoses_icd table) extracted from 2, 083, 180 note events (noteevents table). For the preparation of our datasets, two criteria were employed in selecting the MIMIC-III subjects. First, the age at the time of a subject’s admission to the ICU was used to identify subjects with age less than 15 (patients and admissions tables), and their records were removed. Second, only the first admission of each MIMIC-III subject to the hospital was considered, and all later admissions were discarded. Such selection was made to assure the prediction with the earliest detected symptoms aiding in faster risk assessment. Both steps were carried out in accordance with the existing literature [8, 10, 13]. Overall, the dataset elicited from the selected tables of the database encompassed nursing notes corresponding to 7, 638 subjects with a median age of 66 years (Quartile Q\(_1\)–Q\(_3\): 52–78 years).
3.2 Data Cleaning and Aggregation
It was observed that the data extracted from the MIMIC-III database had erroneous entries due to various factors including outliers, noise, duplicate or incorrect records, and missing values. First, we identified and removed the erroneous entries in these nursing notes by using the iserror attribute of the noteevents table set to 1. Second, the MIMIC-III subjects with duplicate records were identified and such records were deduplicated. After handling these erroneous entries, the resulting data corresponded to 6, 532 patients.
A significant challenge in modeling the voluminous nursing notes and facilitating multi-label ICD-9 code group classification is the aggregation of multiple such notes of a specific MIMIC-III subject. Such notes may have many similar terms which can affect the vector representations significantly. Monge-Elkan (ME) token-based fuzzy similarity scoring approach is coalesced with Jaro internal scoring scheme to enable decision-making while handling multiple near-duplicate nursing notes of a subject. ME similarity handles alternate names, clinical abbreviations, and medical jargons. Jaro similarity, as an internal scoring strategy effectively handles spelling errors and produces a normalized similarity score between 0 and 1. Given two nursing notes \(\eta _p\) and \(\eta _q\) with \(|\eta _p|\) and \(|\eta _q|\) tokens (\(\mathcal {C}_i^{(p)}\)s and \(\mathcal {C}_j^{(q)}\)s) respectively, their ME similarity score with Jaro internal scoring is computed using,
where, the Jaro similarity score of two given tokens \(\mathcal {C}_m\) of length \(|\mathcal {C}_m|\) and \(\mathcal {C}_n\) of length \(|\mathcal {C}_n|\) with c matching characters and t transpositions, is computed using,
A pair of nursing notes are merged only if the ME score of that pair is lower than a preset threshold. Only the first record is retained and the second is purged when the ME score of a pair of nursing notes is greater than the threshold. Note that only the nursing notes and not the ICD-9 code groups are merged or purged based on similarity. To facilitate multi-label classification, we merge the corresponding ICD-9 codes across multiple nursing notes of a patient. We refer to the resultant nursing note for a patient obtained as a result of merging as the aggregate nursing note of that patient. In this study, the fuzzy-similarity threshold (\(\theta \)) was empirically determined to be 0.825.
3.3 Data Preprocessing
The next step in the NLP pipeline is preprocessing the nursing notes to achieve data normalization. Preprocessing includes tokenization, stop-word removal, and stemming/lemmatization. Firstly, multiple spaces, punctuation marks, and special characters are removed. During tokenization, we split the text in each clinical nursing note into numerous smaller words (tokens). Using the NLTK English stopword corpus, stopwords among the generated tokens are removed. Furthermore, any references to images (file names such as ‘MRI_Scan.jpg’) are removed, and character case folding is performed. Token removal based on its length based was not performed to mitigate any loss of vital medical information (such as ‘MRI’ in ‘MRI Scan’). Finally, suffix stripping was facilitated by stemming, followed by lemmatization which aimed at converting the stripped words to their base forms. The words appearing in less than ten nursing notes were removed to lower the computational complexity and avoid overfitting.
3.4 Feature Modeling of Clinical Concepts
Let \(\mathbb {S}\) denote the set of all aggregate nursing notes. Each aggregate nursing note \(\eta _i\) in \(\mathbb {S}\) constitutes a variable length of tokens from a sizeable vocabulary \(\mathbb {V}\), thus making \(\mathbb {S}\) very complex. Therefore, a transformation of unstructured clinical text into an easier-to-use machine processable form is critically important. The performance and efficacy of the utilized deep architectures are heavily reliant on the optimal vector representations of the underlying corpus. We used vector space modeling and coherence-based topic modeling for feature modeling to enable an optimal representation of the patient cohort.
Vector Space Modeling. Vector space modeling of clinical concepts aims at representing each nursing note as a point in a multidimensional vector space of d dimensions (usually, \(d \ll |\mathbb {V}|\)). The term weighting scheme is a transformation of the Bag of Words (BoW) that assigns weight to tokens in an unsupervised manner. This scheme captures both the importance (occurrence frequency) and rarity (specificity) of a token in the given vocabulary. The weight (\(W_{i}^{(p)}\)) assigned to a term \( w _i^{(p)}\) (of total \(|w^{(p)}|\) terms) in a nursing note \(\eta _p\) (of total N nursing notes) occurring \( f _{i}^{(p)}\) times is given by,
The term weights of all the tokens in an aggregate nursing note are computed to obtain a vector (\(\{W_{i}^{(p)}\}_{i=1}^{|\mathbb {V}|}\)) in the machine processable form.
Although the term weighting scheme effectively captures the syntactic relation between the textual features, it often suffers from the curse of high dimensionality and sparsity. Furthermore, it does not capture the intuition that semantically similar nursing notes have similar representations (e.g., ‘bone’ and ‘fracture’). Doc2Vec or Paragraph Vectors (PVs) overcome these shortcomings by efficiently learning the term representations in a data-driven manner. Doc2Vec numerically represents variable length documents as low dimensional, fixed length document embeddings. It is a simple neural network with one shallow hidden layer that learns the distributed representations and provides content-related measurement. It captures both semantic and syntactic textual features obtained from the nursing notes text corpus. The implementations in the Python Scikit-learn and Gensim packages were used on the transcribed clinical words, to extract the features modeled using vector space models. In this study, we used the PV distributed memory variant of Doc2Vec with a dimension size of 500 (trained for 25 epochs) due to its ability to preserve the word order in the nursing notes.
Coherence-Based Topic Modeling. Topic modeling aims at finding a set of topics (collection of terms) from a collection of documents (nursing notes) that best represents the documents in the corpus. LDA, a popular cluster analysis approach is a generative topic model based on the Bayesian framework of a three-layer structure including documents, topics, and terms. A soft probabilistic and flat clustering of terms into topics and documents into topics is facilitated by LDA. It posits that each nursing note and each term, belong, with a certain probability, to a set of topics. This topic modeling approach can capture the context of occurrence which is essential for accurate predictability by the underlying deep architectures.
Similar to other clustering methods, the challenge is to determine the correct number of LDA clusters. To cope with this issue, the Topic Coherence (TC) between topics is used to derive the optimal number of topics. TC is a way to evaluate the topic models with a much higher guarantee on human interpretability. In our work, we adopt coherence-based LDA as it accounts for the semantic similarity between the higher scoring terms. The implementation available in Python Gensim package was used implement LDA with TC. A normalized pointwise mutual information score was used as a confirmation measure due to its high correlation with human interpretability [2]. The number of topics for LDA models was set to 100 and the LDA matrix was built on a BoW representation of the nursing notes. Furthermore, the number of topics was determined to be 100, by comparing the coherence scores of several LDA models obtained by varying the number of topics from 2 to 500 (increments of 100).
4 ICD-9 Code Group Prediction
ICD-9 codes are a taxonomy of diagnostic codes used by medical personnel including doctors and public health agencies to classify diseases and a wide variety of symptoms, infections, disorders, causes of injury etc. Researchers have stressed the need to differentiate between full-code predictions and category-level (group) predictions due to the high granularity in the diagnostic code hierarchy [11]. Each code groupFootnote 2 includes a set of similar diseases, and almost every health condition can be classified into a unique code group. In this research, we focus on ICD-9 code group predictions as a multi-label classification problem, with each patient’s nursing note mapped to more than one code group. All the ICD-9 codes for a given admission are mapped into 19 distinct diagnostic classes. Note that there are no records in the MIMIC-III database within the ICD-9 code range of 760–779. Furthermore, this study classifies all the Ref and V codes into the same code group.
4.1 Deep Neural Architectures
We used three deep neural architectures including MLP, LSTM, and CNN to make the ICD-9 code group predictions. We used the implementations available in the Python Keras package with Tensorflow backend for this purpose. All the presented deep models were trained to minimize a binary cross entropy loss function using an Adam optimizer, with a batch size of 128, for eight epochs.
Multi-layer Perceptron. MLP is a feed-forward neural network with an input layer, one or more hidden layers, and an output layer. The first layer takes the clinical terms in an aggregate nursing note as the input and uses the output of each layer as the input to the following layer. Each node of the hidden or output layer l is associated with a bias (\(b^{(l)}\)) and each node to node connection (from layer l to \(l + 1\)) has a weight (\(W^{(l,l+1)}\)). A node in a layer l with an input \(s^{(l)}\) is activated in the layer \(l + 1\) using an activation function \(\mathbf {g}\) as \(\mathbf {g}(W^{(l,l+1)}\cdot s^{(l)} + b^{(l+1)})\). Furthermore, MLP uses the backpropagation algorithm to calculate the gradient of the loss function, allowing it to learn an optimal set of weights and biases needed to minimize a suitable loss function. The ability of MLPs to solve problems stochastically enables them to approximate solutions even for extremely complex problems. In this research, we utilize an MLP network with one hidden layer of 75 nodes with a Rectified Linear Unit (ReLU) activation function and an output layer of 19 nodes with a sigmoid activation function.
Long Short Term Memory. LSTMs are a special type of Recurrent Neural Networks (RNNs) that effectively capture the long-term dependencies. LSTMs overcome the problem of vanishing gradients, typically observed in traditional RNNs. Capturing the context and long-term dependencies in the raw clinical text would be crucial in the accurate prediction of the ICD-9 code groups. A recurrent neuron in RNNs has a simple activation structure, similar to that in MLP. In LSTM networks, however, the recurrent neuron, termed as the LSTM memory cell is equipped with a much more complex structure. More specifically, given a nursing note \(\eta _i\) at a time step t, with an embedding of \(s_t^{(i)}\), the output (\(h_t\)) and the state (\(c_t\)) of an LSTM memory cell can be given by,
where, \(\odot \) represents element-wise multiplication, \(\text {tanh}(\cdot )\) is the hyperbolic tangent function, and i, f, o, and g are the values at the input gate, forget gate, output gate, and cell state respectively and are computed as (\(\sigma (\cdot )\) denotes the sigmoid function),
Note that \(W^{(l,l+1)}\) varies between the layers but is shared through time. In this study, the dimensions of the embedding and LSTM hidden state are 289 (17 time steps with 17 features each) and 300. The multi-label prediction is achieved by a sigmoid activation of the final LSTM output.
Convolutional Neural Network. CNN is a deep feed-forward neural network architecture which uses a variation of the MLP aimed at minimal processing. Let an aggregate nursing note \(\eta _i\) be modeled to produce an embedding of \(s_{1:n}^{(i)}\), where \(t_{k:k+l}\) refers to the concatenation of the terms \(t_k, t_{k+1},\dots , t_{k+l}\). The computation of a new feature involving a convolution operation using a filter \(f \in \mathbb {R}^{wn}\) on a window of w terms and bias b is given as \(\mathbf {g}(f \cdot s^{(i)}_{k:k+w-1} + b)\). The filter f is applied to every possible window of terms in the embedding \(\{s^{(i)}_{1:w},s^{(i)}_{2:w+1},\dots ,s^{(i)}_{n-w+1:n}\}\) to produce a feature map. The same process can be extended to multiple filters (with varying window sizes) to obtain multiple feature maps. The features from the penultimate layer are passed to a fully connected layer using an activation function. CNNs drastically reduce the number of hyper-parameters (weights and biases) to be learned by the network, thus reducing the training overhead. In this research, we employed one fully connected layer of 289 nodes with ReLU activation function, one CNN layer with \(3 \times 3\) convolution window size and 19 feature map size. The multi-label prediction is achieved by a sigmoid activation of the final convolved output.
5 Experimental Results and Discussion
The experiments were performed using a server running Ubuntu OS with 56 cores of Intel Xeon processors, 128 GB RAM, 3 TB hard drive, and two NVIDIA Tesla M40 GPUs. To validate the proposed data modeling and prediction approaches, we performed an exhaustive benchmarking over the nursing notes data obtained from the MIMIC-III database. The primary challenge is the multi-label classification, where a set of ICD-9 code groups are predicted for each nursing note and a pairwise comparison of actual and predicted ICD-9 code groups for nursing note is performed. Seven standard evaluation metrics were used to evaluate the performance of each of the three deep learning models with respect to each of the four presented data modeling approaches. The evaluation metrics include Accuracy (ACC), F1 score, MCC score, Label Ranking Loss (LRL), Coverage Error (CE), Area Under the Precision-Recall Curve (AUPRC), and Area Under the ROC Curve (AUROC). Five-fold cross-validation was used to evaluate the predictability of the proposed models. Furthermore, the mean and standard errors (of the mean) of all the performance scores are presented. Table 1 shows the performance of all data modeling approaches and all deep prediction models processed using the proposed fuzzy token-based similarity approach (\(\theta = 0.825\)). We observe that the proposed Term weighting of nursing notes AGgregated using Similarity (TAGS) model, modeled with MLP outperforms more complex vector space and topic models.
AUPRC measures the number of true positives from positive predictions and is more relevant since the data extracted from the MIMIC-III database is highly imbalanced. Most previous works including the state-of-the-art model [13] are heavily reliant on the structured nature of the EMRs modeled in the form of feature sets to aid the prediction of clinical outcomes. Table 2 presents the AUPRC and AUROC performance of the proposed ICD Code group prediction models. From Fig. 2, it can be noted that the proposed TAGS model consistently outperforms the existing state-of-the-art model by 5% in AUPRC and 1.55% in AUROC. The previous works do not benchmark metrics other than AUROC and AUPRC presented in this study. We argue that the presented metrics aid in the measurement of various aspects of the predictive model’s performance including precision and recall which are vital in critical clinical tasks. The richness and abundance of information captured by the unstructured nursing notes are often lost in the structured EMRs coding process [4]. From the results, it can be noted that the TAGS model captures the discriminative features of the clinical nursing notes, eliminates redundancy, and purges anomalous data effectively aiding the deep learning classifier to learn and generalize, and such modeling results in the improvement of the clinical decision-making process.
6 Concluding Remarks
Clinical nursing notes hold a treasure trove of patient-specific information. The voluminous and heterogeneous nature of the unstructured nursing notes with complex linguistic structure makes it hard to model them. In this paper, we presented a fuzzy similarity based matching approach to eliminate redundant and anomalous data resulting in reduced cognitive burden and enhancement in the clinical decision-making process. Vector space modeling and Coherence topic modeling approaches were built on the aggregated data to capture the syntactic and latent semantic information in the nursing notes and effectively leverage it for disease prediction. It was observed that the proposed TAGS model achieved superior performance when benchmarked against the structure EMR based state-of-the-art model by 5% in AUPRC and 1.55% in AUROC. Furthermore, its performance was benchmarked using seven evaluation metrics which are vital in the assessment of the predictive capability of the proposed models, especially in clinical tasks. Our model built on unstructured text eliminates the dependency on EMRs which is extremely useful in countries with low EMR adoption rates. As part of future work, we aim at validating the TAGS model on real-time clinical records. We also intend to improve the predictive capabilities of our models, focusing on building time-aware prediction architectures in real-time.
Notes
- 1.
International Statistical Classification of Diseases and Related Health Problems.
- 2.
The code ranges used for mapping can be found at http://tdrdata.com/ipd/ipd_SearchForICD9CodesAndDescriptions.aspx.
References
Baumel, T., Nassour-Kassis, J., Cohen, R., Elhadad, M., Elhadad, N.: Multi-label classification of patient notes a case study on ICD code assignment. arXiv preprint arXiv:1709.09587 (2017)
Bouma, G.: Normalized (pointwise) mutual information in collocation extraction. Proc. GSCL, 31–40 (2009)
Collins, S.A., Cato, K., Albers, D., Scott, K., et al.: Relationship between nursing documentation and patients’ mortality. Am. J. Crit. Care 22(4), 306–313 (2013)
Dubois, S., Romano, N., Kale, D.C., Shah, N., Jung, K.: Learning effective representations from clinical notes. arXiv preprint arXiv:1705.07025 (2017)
Harutyunyan, H., Khachatrian, H., Kale, D.C., Galstyan, A.: Multitask learning and benchmarking with clinical time series data. arXiv preprint arXiv:1703.07771 (2017)
Henry, J., Pylypchuk, Y., Searcy, T., Patel, V.: Adoption of electronic health record systems among us non-federal acute care hospitals: 2008-2015. ONC Data Brief 35, 1–9 (2016)
Jo, Y., Lee, L., Palaskar, S.: Combining LSTM and latent topic modeling for mortality prediction. arXiv preprint arXiv:1709.02842 (2017)
Johnson, A.E., Pollard, T.J., Mark, R.G.: Reproducibility in critical care: a mortality prediction case study. In: Machine Learning for Healthcare Conference, pp. 361–376 (2017)
Johnson, A.E., et al.: MIMIC-III, a freely accessible critical care database. Sci. Data 3, 160035 (2016)
Krishnan, G.S., Sowmya Kamath, S.: A supervised learning approach for ICU mortality prediction based on unstructured electrocardiogram text reports. In: Silberztein, M., Atigui, F., Kornyshova, E., Métais, E., Meziane, F. (eds.) NLDB 2018. LNCS, vol. 10859, pp. 126–134. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-91947-8_13
Larkey, L.S., Croft, W.B.: Automatic assignment of ICD9 codes to discharge summaries. Technical report, University of Massachusetts at Amherst, Amherst, MA (1995)
Pirracchio, R.: Mortality prediction in the ICU based on MIMIC-II results from the super ICU learner algorithm (SICULA) project. Secondary Analysis of Electronic Health Records, pp. 295–313. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-43742-2_20
Purushotham, S., Meng, C., Che, Z., Liu, Y.: Benchmarking deep learning models on large healthcare datasets. J. Biomed. Inform. 83, 112–134 (2018)
Wang, Y., et al.: MedSTS: a resource for clinical semantic textual similarity. Lang. Resour. Eval., 1–16 (2018)
Waudby-Smith, I.E., Tran, N., Dubin, J.A., Lee, J.: Sentiment in nursing notes as an indicator of out-of-hospital mortality in intensive care patients. PLoS ONE 13(6), e0198687 (2018)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Gangavarapu, T., Jayasimha, A., Krishnan, G.S., S., S.K. (2019). TAGS: Towards Automated Classification of Unstructured Clinical Nursing Notes. In: Métais, E., Meziane, F., Vadera, S., Sugumaran, V., Saraee, M. (eds) Natural Language Processing and Information Systems. NLDB 2019. Lecture Notes in Computer Science(), vol 11608. Springer, Cham. https://doi.org/10.1007/978-3-030-23281-8_16
Download citation
DOI: https://doi.org/10.1007/978-3-030-23281-8_16
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-23280-1
Online ISBN: 978-3-030-23281-8
eBook Packages: Computer ScienceComputer Science (R0)