Summary
This chapter considers the microbiology of cheese ripening and complements the next chapter which considers the biochemistry of cheese ripening and the development of cheese flavour. The important parameters controlling the shelf-life of cheese, viz., water activity, NaCl level, oxidation-reduction level, pH, nitrate and temperature are examined in some detail as is the growth of non-starter lactic acid bacteria, mainly lactobacilli, which grow in all cheeses during ripening. The role of the secondary cultures, e.g., brevibacteria, propionibacteria and moulds, which grow only during ripening, are considered within descriptions of the microbiology of the individual cheese varieties. The cheeses examined in detail from a microbiological view include Cheddar, Swiss-type cheese, Parmigiano Reggiano, Gouda and Edam, bacterial-, e.g., Limburger, Livarot and Tilsit, Reblochon and Gubbeen, and mould surface-ripened cheeses, e.g., Camembert and Brie, and blue cheeses. Microbial spoilage of cheese, e.g., early and late gas formation, open texture, growth of lactobacilli and propionibacteria in Dutch-type cheese, and yeast and moulds, is considered. Finally descriptions of the various genera other than starter and non-starter bacteria found in cheese, e.g., Agrococcus, Arthrobacter, Brachybacterium, Brevibacterium, Corynebacterium, Microbacterium, Propionibacterium, Micrococcus, Kokuria, Kytococcus, Staphylococcus and the various yeasts and moulds are given.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
- Control of microbial growth
- Non-starter lactic acid bacteria
- Secondary cultures
- Cheese spoilage
- Microbiology of different cheeses
11.1 Introduction
The quality of cheese is determined mainly by its flavour and texture and hence considerable effort has been devoted to elucidating the principal microbiological and biochemical changes that occur in cheese during ripening. The appearance of many varieties of cheese changes during ripening, e.g., the formation of holes, called eyes, in Swiss-type and, to a lesser extent, in Dutch-type cheese, growth of mould on the surface (e.g., Brie and Camembert) or interior (blue varieties), or the growth of microorganisms on the surface (smear-ripened cheeses). All these changes are caused by growth of microorganisms in or on the cheese. Therefore, an understanding of the factors involved in controlling their growth in cheese is important in trying to understand the development of cheese texture and flavour.
High numbers (at least 109 cfu/g) of microorganisms are present in all cheeses early in ripening and it is their lysis and the release of intracellular enzymes which mainly determine the development of flavour in the cheese (Chaps. 12 and 13).
11.2 Microbial Activity During Ripening
The factors controlling the growth of microorganisms in cheese include: water activity, concentration of salt, oxidation-reduction potential, pH, the presence of NO3, a relatively low ripening temperature and the production of bacteriocins by some starter strains. Individually, the effect of these factors may not be very great, but the interaction of all of them, acting in concert, as so-called ‘hurdles’, is the real controlling factor. Other compounds produced during curd manufacture and ripening, e.g., H2O2 and fatty acids, also inhibit microbial growth but the concentrations of these produced in cheese are not sufficiently high to have a significant effect on the microorganisms.
11.3 Water and Water Activity
All microorganisms require water for growth but it is the availability of the water, rather than the total amount present, that is the important controlling factor. Water availability is expressed by the concept of water activity (a w) which is defined as the ratio of the vapour pressure over the cheese, P, to the vapour pressure over pure water, Po, at that temperature:
The value of a w ranges from 0 to 1.0.
A reduction in moisture occurs during the manufacture of all cheeses; the lower it becomes, the harder the cheese is and the longer its keeping quality, e.g., Parmigiano Reggiano with a moisture content of ~30 % may be held for 2 years before being marketed. Cheese, unless vacuum-packed, loses moisture by evaporation during ripening. The proteins in cheese are hydrated and this ‘bound’ water is not available for bacterial growth. Any components dissolved in the moisture of the cheese, e.g., amino acids, peptides, short-chain fatty acids, salt and organic acids (lactate, acetate and propionate) reduce its vapour pressure and hence its a w. Of these, the most important in practice are salt and lactate.
Most bacteria require a minimum a w of 0.92 for growth. Yeasts grow at a lower value of a w than bacteria, and moulds at a still lower value. The limit for most yeast is ~0.83 but osmophilic yeast grow at a w values <0.60, while moulds have a lower a w limit of ~0.75. Growth of microorganisms at low a w is characterised by a long lag phase, a slow rate of growth (i.e., long generation time) and a reduction in the maximum number of cells produced, each of which helps to limit the growth of the microorganisms. Starter bacteria generally have a higher minimum a w value than other bacteria. The minimum a w for the growth of Lc. lactis, Sc. thermophilus, Lb. helveticus and P. freudenreichii are 0.93, >0.98, >0.96 and 0.96, respectively. The influence of a w on the growth of some other microorganisms associated with cheese is shown in Table 11.1. Penicillium camemberti is the mould responsible for the white coating on Camembert and Brie cheese while Brevibacterium linens and Debaryomyces hansenii are important microorganisms in the surface flora of smear-ripened cheeses. P. camemberti, B. linens and D. hansenii can grow slowly in the presence of 10, 12 and 15 % NaCl, respectively. Staphylococcus aureus and micrococci can grow quite well in the presence of 6.5 % NaCl, which is equivalent to an a w of 0.96. Compared with other fungi, Geotrichum candidum is very sensitive to a w while B. linens is quite resistant. Propionibacteria are also particularly sensitive to a w. Facultative anaerobes have different minimum a w values depending on whether the organisms are growing aerobically or anaerobically, e.g., in the presence of O2, S. aureus has a minimum a w of 0.86 but in the absence of O2, the minimum is 0.91.
Evaporation of water from the cheese surface during ripening also contributes to the reduction of the a w of cheese; examples for Emmental and Gruyère are shown in Fig. 11.1. The reason for the faster rate of decrease in the a w of Gruyère is probably due to the surface salting of Gruyère during the early stages of ripening. In addition, the a w of cheese can vary throughout its mass (Fig. 11.2). Variations are much greater in large cheeses, like Emmental (50–60 kg), than in a small cheese, like Appenzeller (6–8 kg). This is due to several factors, including the temperature gradient in the cheese during the early stages of the fermentation, the loss of moisture during ripening, the NaCl gradient in the cheese and microbial activity on the rind. These factors must be taken into account in determining the significance of a w, especially in large cheeses. Typical a w values for cheese are listed in Table 11.2. As a comparison, the a w of milk is 0.995. Since the a w of cheese decreases during ripening, some of these values must be interpreted with care; however, they are useful as a guide. Except for the soft cheeses like Brie and Camembert, most of these values are close to the minima for starter growth.
Decrease in the a w of Emmental and Gruyère cheese during ripening. The a w at time zero, 0.995, corresponds to that of milk (From Ruegg and Blanc 1981)
Typical variations in the a w of slices, from the centre to the surface, of (a) Emmental (b) Sbrinz, (c) Gruyère and (d) Appenzeller cheese. The cheeses were ~5 months old and the a w of the rinds was (a) 0.90–0.95, (b) 0.80–0.90, (c) and (d) 0.92–0.98 (From Ruegg and Blanc 1981)
11.4 Salt
The use of NaCl to prevent microbial spoilage of food is probably as old as food production itself. The concentration required depends on the nature of the food, its pH and moisture content but, generally, less than 10 % is sufficient. Salt and a w are intimately associated and the major inhibitory factor is the reduction in a w which occurs when salt (or any solute) is dissolved in water. The relationship between salt concentration and a w is shown in Fig. 11.3 and is almost but not quite linear. The linear equation is:
The relationship between salt concentration and a w (Redrawn from Hardy 1986)
where x is the amount of salt, in g/1000 g of water, and describes the relationship very well since the r2 value is 0.997. It is generally considered that an a w of <0.92 is necessary to prevent bacterial growth; this is equivalent to a salt concentration of ~12 %. In cheese, the salt concentration varies from 0.7 to 7 %. The type of ion is also important, e.g., Na+ is a much more effective inhibitor than K+. In calculating the inhibitory effect of salt in cheese, the concentration of salt dissolved in the water of the cheese, rather than the actual concentration of salt, is the important parameter, e.g., in a Cheddar cheese with 38 g moisture/100 g and 1.9 g salt/100 g, the salt-in-water (S:M) is 5 %. Generally, the S:M in Cheddar cheese varies from 4 to 6 %. Most starter bacteria grow in the presence of 3 % but not 4 % NaCl (Chap. 6) and one of the reasons why most cheeses are brine-salted may be allow starter growth and the consequent reduction in lactose level to continue unimpeded in the early days of ripening.
Cheese is either dry-salted (e.g., Cheddar) or brine-salted (most cheeses). In brine-salted cheeses, the salt concentration is influenced directly by the size of the cheese, the concentration of salt in the brine, the temperature of the brine and the length of time for which the cheese is immersed in the brine (see Chap. 9). This will also affect the a w of the cheese. Data for the effect of brining time on the salt concentration and the a w of Camembert cheese are shown in Fig. 11.4. The brine normally used contains ~20 % NaCl, has a pH of ~5.2 (adjusted with lactic acid) and a Ca2+ content of 0.2 % (adjusted with CaCl2). The pH and Ca concentration simulate the levels in cheese and help to prevent the efflux of lactate and Ca2+ from the cheese. The tolerance of starter lactic acid bacteria (SLAB) and non-starter lactic acid bacteria (NSLAB) to salt are discussed in Chap. 6 and below, respectively.
Influence of the duration of brining at 14 ° C in a 20 % NaCl brine on the a w of Camembert cheese. The NaCl concentration and a w level were determined 15 days after manufacture (From Ruegg and Blanc 1981)
11.5 Oxidation-Reduction Potential
Oxidation-reduction potential (Eh) is a measure of the ability of chemical/biochemical systems to oxidise (lose electrons) or reduce (gain electrons). Eh is generally measured using a platinum electrode coupled with a calomel reference electrode and is expressed in mV. It can also be estimated using indicator dyes which change colour at different redox potentials. A positive value indicates an oxidised state while a negative value indicates a reduced state.
The Eh of milk is about +150 mV, while that of cheese is about −250 mV. The reduction in the Eh of cheese is directly related to the fermentation of lactose to lactic acid by the starter during growth. The exact mechanism by which the Eh is reduced is unclear but is probably connected to the reduction of the small amount of O2 in the milk to H2O (or H2O2 and then to H2O) and the reduction of NAD+ to NADH. Because of these reactions, cheese is essentially an anaerobic system, in which only facultatively or obligately anaerobic microorganisms can grow. Obligate aerobes, like Pseudomonas, Brevibacterium and Micrococcus spp., will not grow within the cheese, even when other conditions for growth are favourable. The bacteria which develop on the surface of cheese are mainly obligate aerobes and are unable to grow within the anaerobic cheese environment.
11.6 pH and Organic Acids
Most bacteria require a neutral pH value for optimum growth and grow poorly at pH values <5.0. The pH of cheese curd after manufacture generally lies within the range 4.5 to 5.3 so that pH is also a significant factor in controlling bacterial growth in cheese. Lactic acid bacteria, especially lactobacilli, generally have pH optima below 7 and Lactobacillus spp. can grow at pH 4.0; most yeast and moulds can grow at a pH <3.0, although their optimum ranges from 5 to 7. B. linens, which is found on the surface of smear-ripened cheese, cannot grow below pH 6.0. Micrococcus sp., which is also commonly found on the surface of soft cheeses, cannot grow at pH 5 and only slowly at pH 5.5.
The efficacy of organic acids as inhibitors of microbial growth is thought to depend on the amount of undissociated acid present and therefore on the dissociation constant (pKa) and pH. The pKa values for propionic, acetic and lactic acids, the principal acids found in cheese, are 4.87, 4.75 and 3.08, respectively. The undissociated form of the acid is more inhibitory than the ionised form, so that, at the same pH, lactic acid is the least and propionic the most effective inhibitor. However, the concentration of the acid is also important and, in cheese, lactate is invariably present in cheese curd at much greater concentrations than either of the other two acids. Sometimes, it is thought that the difference between pH 5.2, the pH of a well-made Cheddar cheese, and pH 5.4, the pH of a poorly made Cheddar, is not very great. However, this is not so; pH is a log scale and a difference of 1 pH unit is equivalent to a tenfold difference in the H+ concentration. The difference in [H+] between 5.2 and 5.4 is twofold.
11.7 Nitrate
NO3 − , as KNO3 (saltpetre) or NaNO3, is added to the milk (20 g/100 L) for some cheeses, especially Dutch-type cheeses, like Gouda and Edam, to prevent the production of early and late gas by coliforms and Clostridium tyrobutyricum, respectively. Much of the NO3 − is lost in the whey. The maximum amount of NO3 − permitted in cheese is 50 mg/kg, calculated as NaNO3. The real inhibitor is NO2 − which is formed from NO3 − by the xanthine oxidase present in the milk or curd. How NO2 − acts in preventing microbial growth is not clear. NO2 − can also react with aromatic amino acids in cheese to produce nitrosamines, many of which are carcinogenic (Chap. 20).
Nitrate does not inhibit the growth of coliforms but changes their metabolism so that less H2 is produced from formate, a product of sugar metabolism, by the formate dehydrogenase/hydrogenase enzyme system. Nitrate represses the formation of this enzyme system and induces a formate dehydrogenase/nitrate reductase which reduces NO3 to NO2, without the formation of H2.
11.8 Temperature
Generally, the optimum temperature for the growth of bacteria is ~35 °C for mesophiles and ~55 °C for thermophiles. However, thermophilic starters have an optimum temperature of ~42 °C. Psychrophilic bacteria have an optimum temperature below 20 °C but true psychrophiles are not found in cheese. At temperatures below the optimum, growth is retarded.
The temperature of cooking varies for different cheeses and the time the temperature is maintained at higher values will affect the survival of different organisms in the cheese. A comparison of the temperature profiles of Cheddar, Emmental, and a soft cheese is shown in Fig. 11.5. Emmental is heated to 54 °C during manufacture and the temperature is retained above 40 °C for a considerable time, while Cheddar is cooked to 39 °C and soft cheeses are heated at ~35 °C. Other cheeses cooked at a high temperature include Comte, Parmigiano Reggiano and Grana. Little acid production occurs at the maximum cooking temperature for cheeses cooked to 54 °C but the thermophilic starters withstand the temperature and begin to produce acid when the temperature falls below 48 °C. Traditional Emmental cheese is made from raw milk and because of the relatively high temperature of ripening of this cheese [18–24 °C for several weeks to promote the growth of propionic acid bacteria (PAB) ], great attention must be paid to the microbial quality of the raw milk.
The temperature of ripening of cheese is also important and is dictated by two opposing forces—on the one hand, the need to control the growth of potential spoilage and pathogenic bacteria and, on the other, the need to promote the ripening reactions and the growth of the secondary microflora in the case of soft and Swiss-type cheeses. Higher temperatures promote faster ripening by the starter and non-starter microorganisms but also allow the growth of spoilage and pathogenic bacteria. Generally, Cheddar cheese is ripened at 6–8 °C while surface-ripened cheeses, like Camembert and bacterial smear-ripened cheeses, are ripened at 10–15 °C. Emmental cheese is ripened initially for 2–3 weeks at a low temperature (~12 °C), after which the temperature is increased to 20–24 °C for 2–4 weeks to promote the growth of propionic acid bacteria and the fermentation of lactate to propionate and acetate; the temperature is then reduced to ~4 °C. For soft cheeses, the humidity of the environment is also controlled to prevent excess evaporation of moisture from the cheese surface.
Increasing the temperature of ripening is probably the simplest and most cost-effective way of accelerating the ripening of cheese (Chap. 12). This will also increase the rate of growth of other bacteria which may be present.
11.9 Growth of Starter Bacteria in Cheese
The initial number of SLAB in cheese milk ranges from about 105 to 107 cfu/ml, depending on the level of inoculation of the starter. Growth of the starter during cheese manufacture results in final numbers of ~109 cfu/g of cheese within one day. During ripening, starter organisms dominate the microflora of cheese but most die off and lyse relatively rapidly (Fig. 11.6). In the case of Cheddar cheese, the rate of lysis depends on the strain, and in the case of Comté cheese, the rate of lysis of Sc. thermophilus is faster than that of Lb. helevticus. Many artisanal cheeses, especially Spanish varieties, are made without the intentional addition of a starter. In these cheeses, lactococci also comprise the major part of the microflora and, except for La Serena, also show significant rates of lysis during ripening (Fig. 11.7). The reason for the slow rate of lysis in La Serena cheese may be due to the relatively low salt level in that cheese during the early weeks of ripening.
Changes in the numbers of (a) Streptococcus thermophilus and Lactobacillus helveticus during the ripening of Comté cheese and (b) different strains of Lactococcus lactis during the ripening of Cheddar cheese (From Beuvier, personal communication and Martley and Lawrence 1972)
Once lysis occurs, intracellular enzymes, particularly peptidases, are released, which hydrolyse the caseins and fat to amino acids and fatty acids, which are the precursors of the flavour compounds in cheese (see Chaps. 12 and 13). Starters vary in their ability to lyse—some strains lyse relatively quickly while others lyse slowly. Lysis is caused by an intracellular muraminidase which hydrolyses the bacterial cell wall peptidoglycan. This enzyme is under stringent regulation, otherwise the cells would not grow. Generally, Lc. lactis subsp. cremoris strains lyse faster than Lc. lactis subsp. lactis strains which may partly explain why the former is thought to produce a better flavoured cheese than the latter. Lysis is influenced by several factors, including the level of salt and the presence of prophage, which can be induced by cooking. The presence of small numbers of lytic phage may also have a role in lysis. Cheese made with a fast-lysing starter will ripen more rapidly than one made with a slowly-lysing strain.
There is some evidence (Ganesan et al. 2006, 2007) that lactococci enter a metabolically active but non-culturable state for long periods (up to 3.5 years), after the carbohydrate in the medium is used up. During this period, they can metabolise amino acids to fatty acids, some of which, e.g., 2-methyl butyric acid production from leucine, are important in cheese flavour formation. A population of non-culturable but metabolically active cells also occurs in Emmental cheese during ripening (Falentin 2012).The contribution that such cells make to cheese ripening is not clear and should be investigated.
11.10 Growth of Non-Starter Lactic Acid Bacteria in Cheese
Most, if not all, cheeses, whether made from raw or pasteurised milk, contain adventitious, NSLAB. These are mainly facultatively heterofermentative lactobacilli (FHL) , especially Lb. casei, Lb. curvatus, Lb. paracasei, Lb. plantarum and Lb. rhamnosus, which ferment hexoses homofermentatively to lactic acid and pentoses heterofermentatively to lactate and acetate. NSLAB are also called mesophilic lactobacilli to distinguish them from the thermophilic lactobacilli used as starters. Obligately heterofermentative lactobacilli, e.g., Lb. brevis and Lb. fermentum, and obligately homofermentative pediococci, e.g., Pediococcus pentosaceus and P. acidilactici are found occasionally as NSLAB in cheese. Many of these species are also present in natural whey cultures (see Chap. 6). The dominant species of FHL found in most cheeses are Lb. paracasei and Lb. plantarum and generally several strains of each species are present. In Cheddar cheese, an average of 7 strains are present, and, in addition, there is some evidence that a succession of strains occurs during ripening (Fitzsimons et al. 2001). Lb. rhamnosus is an important component of NSLAB in New Zealand Cheddar cheese (Crow et al. 2001).
The taxonomy of Lb. casei and Lb. paracasei is controversial. The type strain of Lb. casei, ATCC 393, is actually a strain of Lb. zeae and many strains identified as Lb. casei do not hybridise with it, The Judicial Commission of the International Committee on Systematics of Bacteria (Anon 2008) ruled that this was still correct from a nomenclature viewpoint and rejected the proposal of Dellaglio et al. (2002) that Lb. casei ATCC 334 be considered as the new type strain of Lb. casei. Lb. paracasei was created by Collins et al. (1989) for strains of Lb. casei which did not hybridise with Lb. casei ATCC 393 but did hybridise with Lb. casei NCDO 151, a strain which is closely related to Lb. casei ATCC 334. The Judicial Commission also found that the name, Lb. parcasei, was legitimately published. This is quite confusing and many cheese isolates identified as Lb. casei are probably strains of Lb. paracasei. The genomes of 34 dairy, plant, human and animal strains of Lb. paracasei have been determined (Smokvina et al. 2013). The core genome consists of the cell envelope proteinase, the capacity to produce branched chain fatty acids and factors associated with host-microbe interactions, e.g., pili, while the variome consists of hypothetical proteins, phages, plasmids, cell-surface proteins, transporters and enzymes involved in EPS biosynthesis .
SLAB are found in high numbers (>108 cfu/g) in all freshly made cheese. In contrast, the initial number of NSLAB varies considerably from about 100 cfu/g in Cheddar cheese to 106 cfu/g in Casar de Cáceres (Fig. 11.8) and, within the first few week of ripening, they grow relatively quickly to high numbers (~108 cfu/g) in all cheeses at a rate which depends on the ripening temperature, the moisture content and the availability of a suitable energy source. Cheddar is the only cheese in Fig. 11.8 which is made from pasteurised milk, which partly explains the low initial number of NSLAB in this cheese, since NSLAB are partially but not completely inactivated by pasteurisation. In raw milk cheese, the number of NSLAB in the curd is higher, their growth is faster and the population is more heterogeneous. NSLAB grow much more rapidly in Casar de Cáceres and La Serena cheese than in Comté or Cheddar cheese due to the higher moisture content of the first two cheeses compared to Comté and Cheddar cheese. The higher rate of growth of NSLAB in Comté cheese compared with Cheddar is due to the higher ripening temperature of Comté (3 weeks at 14 °C, followed by 9 weeks at 18 °C, before the temperature is reduced to 7 °C) compared to Cheddar (6–8 °C throughout ripening). The higher temperature used in ripening Comté cheese is to promote the growth of PAB, which are responsible for eye formation in this cheese.
The ultimate source of NSLAB in cheese is not clear. Small numbers survive pasteurisation and the high cooking temperature (52–54 °C) used in producing hard cheeses, like Emmental, which is traditionally made from raw milk, suggesting that raw milk is the source. There is also some evidence that biofilms formed on processing equipment, e.g., raw milk silos, ultrafiltration units, if present, cheddaring belts and cheese towers, can be potent sources, with numbers ranging from 100 to 10,000 cfu/cm2 (Agarwal et al. 2006).
NSLAB are acid-tolerant and most of them are also salt tolerant, e.g., 90 % of the strains of Lb. casei, Lb. plantarum and Lb. curvatus isolated from Cheddar cheese grow in the presence of 6 % NaCl (Jordan and Cogan 1993). In contrast, SLAB are sensitive to this level of salt. The tolerance of NSLAB to salt and acid and their ability to grow in the absence of oxygen imply that they should grow well in cheese, provided an energy source is present. The energy source used by them in cheese is thought not to be lactose, since at the time of exponential growth of NSLAB, lactose is not present, unless the salt level is very high. However, the amount of lactose required to sustain 106 cfu/g is small (~1 mg/g) and so trace amounts of lactose in the cheese could be a potential source of energy. Other possible sources have also been suggested, including citrate, amino acids, and the sugars present in the glycoproteins of the milk fat globule membrane or in RNA (ribose) or DNA (deoxyribose), produced from lysis of the SLAB. Diaz-Muniz and Steele (2006) have shown that Lb. casei ATCC 334, which was isolated originally from cheese, can use citrate as an energy source in the presence of limiting, but not excessive, concentrations of galactose in a chemically defined medium and in the presence of both limiting and excess concentrations in a cheese extract medium. Neither lactose nor glucose could replace galactose. However, free galactose is unlikely to be present in cheese unless Sc. thermophilus is used as a starter. In another study, extracts of 2-, 4- and 6-month-old Cheddar cheese supported the growth of this organism to final cell densities of 107–108 cfu/ml implying that adequate energy source(s) are present in the ripening cheese (Budinich et al. 2011). Recently, Lazzi et al. (2014) showed that Lb. rhamnosus, growing in a cheese broth, could oxidise pyruvate, which can be produced from lactate, citrate or amino acids, to acetate with the concomitant production of ATP and therefore growth.
Despite extensive study, the role of NSLAB in the development of cheese flavour is unclear; some studies have shown positive effects, others negative effects and others no effect. The catabolism of amino acids, particularly the aromatic amino acids (phenylalanine, tyrosine and tryptophan), the branched chain amino acids (leucine, isoleucine and valine), and the S-containing amino acid, methionine, is important in the development of cheese flavour (see Chaps. 12 and 13). A prerequisite for this is lysis of the cell to release the intracellular enzymes responsible for flavour development. In contrast to SLAB, NSLAB lyse very slowly in cheese (Fig. 11.8) and so their intracellular enzymes are released only slowly into the cheese matrix. Evidence for the lysis of NSLAB can be inferred from the finding that a progression of different strains occurs during cheese ripening (Fitzsimons et al. 2001). It should also be remembered that the high numbers of NSLAB found in cheese would have considerable metabolic activity in their own right without having to lyse.
In Cheddar cheese, NSLAB transform the l-lactate, produced by the SLAB, to d-lactate. This is also likely to occur in other cheeses. A racemic mixture of both isomers is eventually formed. Some NSLAB can also transform lactate to acetate on the cheese surface in the presence of O2. This will result in a sharper taste of the cut surfaces of the cheese, especially if the cut surfaces remain uncovered for several hours. Pediococci are much more active than lactobacilli in forming acetate from lactate but are found only in small numbers in cheese.
Bacteriocins (see Chap. 6) can prevent the growth of NSLAB, e.g., Lacticin 3147, a bacteriocin produced by a strain of Lc. lactis, isolated from a kefir grain, prevented the growth of NSLAB in Cheddar cheese ripened for 6 months at 8 °C (Ryan et al. 1996, 2001). The bacteriocin-producing strain was not very useful as a starter culture as it produced an off-flavour in milk. Although the cheeses were not evaluated for flavour, there were no differences in the usual indices of protein breakdown in the cheese, viz., water-soluble N, phosphotunstic acid-soluble N and free amino acids. Lacticin 3147 is a two peptide bacteriocin, containing the unusual amino acids, d-alanine and lanthionine; its production is encoded on a conjugative plasmid, which also encodes phage resistance. The plasmid was transferred by conjugation to several Lc. lactis strains. Cheese made with the transconjugants had 100-times (2 log cycles) less NSLAB than the control cheese over a 6 month ripening period (Ryan et al. 2001). This was correlated with the presence of the bacteriocin in the cheese and the cheese graded slightly better than the control cheese, which was described as somewhat bitter, inferring that some NSLAB produce off-flavours in cheese. In another study, cheese was made using Lc. lactis CNRZ 481, which produces the bacteriocin , pediocin PA-1, as an adjunct to the starter culture Lc. lactis HP. The adjunct did not affect acid production by the starter strain; instead it increased lysis of the starter and inhibited the growth of NSLAB (O’Sullivan et al. 2003). Both the experimental (made with strain CNRZ 481 and HP) and control cheeses (made with HP only) graded well but the experimental cheese had a nicer flavour than the control. For more information on NSLAB in cheese see the reviews of Beresford et al. (2001), Broadbent et al. (2003), Beresford and Williams (2004) and Steele et al. (2006).
11.11 Spatial Development of Bacteria in Cheese
Generally, about 90 % of the bacteria present in the milk are retained in the curd during cheesemaking; the remaining 10 % are lost in the whey. These bacteria are immobilised or entrapped in the curd when it coagulates and grow as colonies in the three-dimensional cheese matrix. Until recently, little study of colony formation in cheese has been undertaken because of the lack of suitable techniques. Jeanson et al. (2011), using confocal microscopy, a starter, which produced a green fluorescent protein, and a model cheese in gel cassettes, studied colony formation in cheese and showed that larger colonies were produced at lower inoculation levels. Colonies were also shown to have a Poisson distribution and microgradients in pH did not occur around them. These workers also showed that, in a cube of cheese, there will be only a short distance between colonies at high inoculation rates and greater distances at lower inoculation levels, and that final cell numbers were not affected by the inoculation level. The normal inoculation rate is about 1 × 107 cfu/ml milk which gives a theoretical inter-colonial distance of 26 μm, which compares quite well with the experimental value of 34 μm found at an inoculation rate of 9.6 × 106 cfu/ml.
Cheese ripening is the result of the action of enzymes on fat and protein, ultimately forming the compounds which cause flavour to develop in the cheese (see Chap. 12). As the starter bacteria are immobilised in the cheese, it follows that diffusion of substrates to (and products from) the colonies must occur for flavour to develop. Diffusion of dextrans of different molecular masses (4.4, 70 and 155 kDa) in cheese and agar has been studied in model systems and the results showed that the larger molecular mass dextrans were able to penetrate colonies immobilised in both systems (Floury et al. 2013). Diffusion was faster in cheese than in agar. The shape of the colony was also different in cheese (spherical) than in agar (lenticular) indicating that the physical pressure exerted on the colony in cheese was similar in all directions (isotropous).
11.12 Non-Starter Lactic Acid Bacteria as Adjunct Cultures
Considerable effort has been expended in New Zealand in selecting and developing NSLAB, particularly Lb. paracasei and Lb. rhamnosus, which improve the flavour of Cheddar cheese (Crow et al. 2001; Coolbear et al. 2008). These are added to the cheese milk as a mixture of 2–4 strains at an initial level of 300–1000 cfu/ml of milk. At the same time, factory hygiene was improved so that the initial count of NSLAB in the cheese was <10 cfu/g. The idea behind this development is that the deliberately added NSLAB would dominate the ‘wild’ NSLAB microflora during ripening. Selection of strains that would dominate the ‘wild’ NSLAB may be the key to using NSLAB to improve cheese flavour, since some studies have shown that added NSLAB do not dominate the ‘wild’ NSLAB during ripening (Broadbent et al. 2003). Another point worth considering is the ability of the added NSLAB to lyse.
Nowadays, it is also common to add strains of Lb. helveticus or Sc. thermophilus as adjuncts to mesophilic starters to improve the flavour of Cheddar cheese. They give a more “rounded” flavour to the cheese and are also able to continue to produce acid if the mesophilic culture is infected with phage. There is one caveat: galactose may be present at significant levels in the cheese since most strains of Sc. thermophilus are unable to utilise galactose and excrete it into the curd during growth.
The addition of yeasts, particularly Debaryomyces hansenii and Yarrowia lipolytica, with known proteolytic and lipolytic activities, has also improved the flavour of South African Cheddar cheese (Ferreira and Viljoen 2003); this finding does not appear to have been studied in other countries.
11.13 Enterococci
Enterococci can be found at levels in excess of 107/g in many cheeses, particularly those made around the Mediterranean, and are considered to be essential for flavour development. Many of these are artisanal, raw milk cheeses, made at farm-house level, without the deliberate use of starters. Enterococci can metabolise lactose and their tolerance to salt and heat make them ideal candidates as starters.
There is considerable debate on whether enterococci should be considered to be pathogens (Franz et al. 2003; Foulquie Moreno et al. 2006; Fisher and Phillips 2009). During the past few decades, they have been incriminated as the cause of several diseases, including bacteremia, urinary tract infections and endocarditis. Many strains are promiscuous and easily pick up plasmids encoding antibiotic resistance, e.g., vancomycin. Many of these plasmids are also conjugative and are easily transferred naturally from cell to cell. Vancomycin is a glycopeptide antibiotic which acts by inhibiting cell wall biosynthesis, the incidence of vancomycin-resistant enterococci (VRE) in hospitals has increased dramatically. The use of avoparcin, which is also a glycopeptide antibiotic, as a growth promoter in animal feed has been incriminated in the increased occurrence of VREs in farm animals, including pigs and poultry. Because of this, the use of avoparcin has been banned in several European countries. Many VREs are difficult to deal with because they are also resistant to other therapeutic antibiotics, implying that alternative antibiotic therapy may not be available. In this context it is interesting that some SLAB, e.g., Leuconostoc spp. and NSLAB, e.g., Lactobacillus and Pedicoccus spp., are intrinsically resistant to vancomycin.
There is little information on how rapidly Enterococcus spp. grow in milk but in Cheddar cheese they grow very well during manufacture and remain fairly constant during ripening (Fig. 11.9). These trials involved the separate evaluation of three strains of Ec. faecalis and one strain each of Ec. faecium, Ec. durans and Ec. casseliflavus in duplicate trials. There was little difference in the rate of growth of either the strains or the species and the data for all strains tested was amalgamated. In addition, there was no statistical difference in the grading of control and enterococci-containing cheese. In Trial 2, a small number of enterococci were present in the control but the levels were too low to have any effect on flavour development.
Growth of a 6-strain cocktail of enterococci (3 strains of Ec. faecalis and 1 strain each of Ec. faecium, Ec. durans and Ec. casseliflavus) during the manufacture and ripening of Cheddar cheese. Two trials were conducted and the data are plotted as the average ±s.d. No enterococci were found in the control of Trial 1 at any stage during manufacture or ripening; small numbers were found in the control of Trial 2 (From Rea et al. 2004)
Enterococci also remain fairly constant in other cheeses, e.g., artisanal Spanish and Italian cheeses (Fig. 11.10). Casar de Cáceres and La Serena are made from raw ewes’ milk and Afuega’l Pitu from raw cows’ milk; no deliberate inoculation with starters occurs in any of these cheeses. Pecorino Umbro is made from pasteurised ewes’ milk and a mesophilic starter is deliberately added. A surface microflora develops on some of these cheeses but the counts in Fig. 11.10 are from the internal part of each cheese. The first point on each line in the graph is the number of enterococci present in the milk at the beginning of manufacture. The data show that considerable growth occurs during manufacture and during the first days of ripening, after which they remain constant, except for Afuega’l Pitu cheese, in which the numbers decreased. The numbers of Enterococcus in Casar de Cácares and La Serena cheese were well in excess of 106/g and may have contributed to flavour development. It is sometimes difficult to distinguish between lactococci and enterococci. However, the numbers of enterococci shown in Fig. 11.10 are reliable as media selective for enterococci were used to enumerate them.
11.14 Secondary Microorganisms in Ripening Cheese
Many cheese varieties contain a secondary, non-lactic microflora, the function of which is to produce some specific characteristic change in the cheese, e.g., surface growth in the case of surface-ripened cheeses or the production of CO2, propionate and acetate in the case of some Swiss varieties, e.g., Emmental and Comté. CO2 is responsible for eye formation in the latter cheeses and propionate gives them a sweet flavour. In all of these cheeses, flavour development is dominated by the metabolic activity of the secondary flora during ripening.
Several microorganisms are involved as secondary starters, including bacteria (Agrococcus, Arthrobacter, Brevibacterium, Brachybacterium, Corynebacterium, Microbacterium, Propionibacterium, Staphyloccocus and Micrococcus spp.), yeast (Kluyveromyces marxianus and Debaryomyces hansenii) and moulds (Geotrichum candidum, P. camemberti and P. roqueforti). All of these microorganisms are not present in every cheese and, except for Propionibacterium spp. and P. roqueforti, all of them develop only on the cheese surface. All the bacteria are Gram-positive although small numbers of Gram-negative organisms are isolated occasionally (see below). These are considered more fully in the description of the microbiology of the various cheeses below.
11.15 Molecular Methods of Identification
Cheese is a very dynamic environment for the growth of microorganisms and up to 109 LAB/g can be found in them during ripening. In many cheeses, particularly surface-ripened ones, numerous different species are involved, some of which are very difficult to identify. Considerable efforts have been made to understand the microbiology of cheese and categorically identify all the microorganisms present. Traditionally, selective and non-selective media were used to isolate the different microorganisms in cheese and the colonies were then purified and identified. In recent times, molecular methods have been developed and applied to either organisms isolated from the cheese, in culture-dependent methods or to DNA (and RNA) extracted directly from the cheese, in culture-independent methods. Most workers agree that the results obtained from culture-dependent and culture-independent methods complement each other. The common culture-dependent methods are pulsed field gel electrophoresis (PFGE) and randomly amplified polymorphic DNA (RAPD) and the common culture independent methods involve direct extraction of DNA and RNA from various cheeses, amplification of 16S rRNA genes by PCR and their separation by denaturing gradient gel electrophoresis (DGGE), temperature gradient gel electrophoresis (TGGE) or single strand conformational polymorphism (SSCP). These techniques have been applied to the milk for cheesemaking, whey starters and cheese during ripening, including NSLAB and the surface microflora, and have been reviewed by Ogier et al. (2004), Randozzo et al. (2009) and Quigley et al. (2011).
More recently, so-called next generation, high-throughput, sequencing analysis has been used. This technique can give useful information on the microbial composition of the cheese, on strain composition, or on the distribution of particular genes within the cheese, depending on the amplicons used as primers for the DNA or RNA extracted from the cheese (for reviews see Bokulich and Mills 2012; Ercolini 2013). These techniques have resulted in the identification of genera not previously found in cheese, e.g., Marinilactibacillus and Stenotrophomonas (Delbès et al 2007), Prevotella and Faecalibacteria (Quigley et al. 2012) and the existence of “house”-specific microbes on washed-rind cheeses, which may have a role in determining site-specific products (Bokulich and Mills 2013). Marinilactibacilli are slightly halophilic, alkaliphilic LAB, which are found in marine environments and have also been isolated from spoiled dry-cured hams while Stenotrophomonas maltophilia is an emerging multidrug-resistant global opportunistic pathogen. Both Prevotella and Faecalibacteria are strict anaerobes; Prevotella are commensals of the rumen and hind gut of animals while Faecalibacterium are the dominant organisms in the human gut. In addition, they also form butyrate, d-lactate and formate, which are commonly found in cheese. The influence of such bacteria on the flavour of cheese has not been studied. This technique has also been used to study the growth of various pathogens, starters and the indigenous microflora in raw milk cheese (Masoud et al. 2012).
11.16 Development of Microorganisms in Different Cheeses
Cheeses are commonly divided into hard, semi-hard and soft cheeses, which primarily reflects the moisture content of the cheese, with hard cheeses containing ~38 % moisture, semi-hard containing 42 % moisture and soft cheeses containing 50 %. The higher the moisture content, the faster is the growth of different microorganisms in the cheese and consequently the quicker the flavour develops. The development of different microorganisms in several, well-known examples of these cheeses is examined below.
11.16.1 Cheddar Cheese
Cheddar is a hard, dry-salted cheese made with a mesophilic starters, which grows rapidly in the cheese from an initial level of ~107 to 108 or 109/g at salting (about 5.5 h after inoculation). Nowadays, thermophilic cultures are also added at low levels (see Chap. 6). The cheese evolved in the village of Cheddar in Somerset, England, but is now made all over the world. In the past, Cheddar was generally ripened at 6–8 °C but the tendency now is to ripen at a slightly higher temperature, which results in faster proteolysis and lipolysis and consequently faster flavour production. The manufacture of Cheddar cheese was a very labour intensive process but nowadays the process is highly mechanized and automated. Vacuum packing in rectangular blocks and rapid cooling of the blocks before the beginning of ripening is a common feature of commercial Cheddar production today.
Normally, the fermentation of lactose by starter LAB in cheese is rapid and is complete within 1 day of manufacture. However, in dry-salted cheeses, like Cheddar, a relatively large amount of lactose (~10 g/kg of cheese) is present in the curd after overnight pressing. This is due to inhibition of the metabolism of the starter cultures by the salt and the relatively low pH. The S:M in Cheddar cheese determines the subsequent rate of lactose fermentation by the starters; a high S:M level reduces the rate while a low level increases it, e.g., at a S:M of 4.1, the fermentation is virtually complete in 7 days while at a S:M of 6, it takes > 50 days (Fig. 11.11). There is little difference in the rate of lactose utilisation at intermediate S:M levels, around 5. These values are only indicative and vary depending on the sensitivity of the particular culture to salt. Lc. lactis subsp. cremoris is much more sensitive to salt than Lc. lactis subsp. lactis strains. The former cannot grow in the presence of 4 % salt while the latter can. Salt may also uncouple acid production from growth.
Effect of salt-in-moisture (S/M%) on lactose metabolism in Cheddar cheese, made with Lc. lactis ssp. cremoris C13 and 266, ripened at 12 °C (From Turner and Thomas 1980)
The development of starter and NSLAB in a Cheddar cheese during ripening at 6 °C is shown in Fig. 11.12. After 2 months, the number of lactococci was not measured because NSLAB grow on the medium (LM17) used to estimate the numbers of lactococci. Mesophilic starters generally die out relatively rapidly during the first few weeks of ripening (Fig. 11.6) but the rate is strain-dependent and probably reflects the ability of the strain to withstand the cooking temperature of the cheese and its ability to lyse. Phage may also be involved in reducing the number of cells. NSLAB grow relatively rapidly in Cheddar cheese during ripening from a low initial number (~102/g) to a final number of 107–108/g after 15 weeks, with a generation time of 8.3 days at 6 °C.
NSLAB also transform l-lactate to d-lactate, eventually producing a racemic mixture. This transformation has no effect on the flavour of the cheese but Ca d-lactate is insoluble and can precipitate as small, white crystals throughout the cheese late in ripening. Some consumers consider this a defect in the cheese. Strains of Lb. curvatus and Lb. fermentum are important in forming crystals of Ca d-lactate in ripening cheese (Somers et al. 2001). An example of the growth of NSLAB and the racemisation of L- to D-lactate during Cheddar cheese ripening is shown in Fig. 11.13. Metabolism of residual lactose by the starter lactococci is also shown in Fig. 11.13.
Relationships between non-starter lactic acid bacteria (NSLAB), production of l- and d-lactate and metabolism of lactose in Cheddar cheese during ripening at 12 ° C. The S:M% was 4.1 (From Turner and Thomas 1980)
11.16.2 Swiss-Type Cheeses
Swiss-type cheeses, of which Emmental and Comté are important examples, are hard cheeses that are characterised by the development of eyes in the cheese during ripening due to the fermentation of lactate to propionate, acetate and CO2 by propionic acid bacteria (PAB) . In the past, milk itself was the source of the PAB but nowadays carefully selected strains of PAB are deliberately added to the milk with the starter culture. The inoculum is usually small (only a few hundred cells per ml of milk) but this number is sufficient for the development of a more regular propionic fermentation. The most common species of PAB used is Propionbacterium freudenreichii and the strains used are commonly grown in sodium lactate broth at 30 °C; the grown culture survives for several weeks when stored at 4 °C.
The eyes are larger in Emmental than in Comté because the former is ripened at a higher temperature. Traditionally, both of these cheeses are made from raw milk using thermophilic cultures consisting of Sc. thermophilus and Lb. helveticus and little acid is produced in the vat during their manufacture. Nowadays, much Swiss-type cheese is made from pasteurised or thermised milk and Lb. delbrueckii subsp. lactis has replaced Lb. helveticus as the rod starter in Emmental cheese because of its lower peptidolytic activity and less propensity of the resulting cheese to late fermentation. The latter is thought to be due to a more intense propionic fermentation and additional production of CO2 from decarboxylation of amino acids late in ripening. This is seen mainly as cracks or splits in the cheese because the body of the cheese has become short and crumbly and cannot retain the excessive CO2 production.
During the initial hours in the press, the lactic acid is produced by Sc. thermophilus but, as the temperature and pH decrease, Lb. helveticus begins to grow, reaching maximum numbers 12–20 h after the addition of starter. Counts of both Sc. thermophilus and Lb. helveticus in Comté cheese, and presumably Emmental also, are higher at the periphery than at the centre (Fig. 11.14). In both cheeses, growth of the starter is limited by the high cooking temperature (52–54°C) but growth begins again as soon as the temperature decreases. The temperature falls more rapidly at the periphery than in the centre of the cheese and hence greater bacterial growth (and acid production) occurs at the periphery.
Growth of Streptococcus thermophilus (open triangle, filled triangle) and Lactobacillus helveticus (open square, filled square) at the centre (open symbols) and periphery (closed symbols) of Gruyerè cheese during manufacture (From Accolas et al. 1978)
Sc. thermophilus, Lb. delbrueckii subsp. bulgaricus and some strains of Lb. delbrueckii subsp. lactis metabolise only the glucose moiety of lactose and excrete galactose, which, along with any residual lactose, can then be metabolised by Lb. helveticus and galactose-utilising strains of Lb. delbrueckii subsp. lactis, if present. All the lactose is fermented during the first 10 or 12 h of manufacture. The L isomer of lactate is produced by both Sc. thermophilus and Lb. helveticus , while the D isomer is produced only by the latter organism.
After several weeks ripening at a low temperature (4–14 °C), the cheese is placed in a ‘warm room’ at 18–24 °C, during which P. freundreichii grows and transforms the lactate to propionate, acetate and CO2, which is responsible for eye formation. Traditionally, natural contamination of the milk was relied upon as the source of the PAB in traditionally made Comté and Emmental but nowadays, selected strains of P. freundreichii are deliberately added to the milk with the starter cultures to give an initial count of a few hundred cells per ml of milk. The eyes in Emmental cheese are much larger than in Comté cheese because Emmental is ripened at 22 °C and Comté at 18 °C. PAB are also stimulated by unidentified low molecular mass products of growth of some strains of Sc. thermophilus and Lb. helevticus (Piveteau et al. 1995).
The pathway of lactate fermentation by PAB is complicated (Fig. 11.15). The classical fermentation involves two separate pathways, in one of which propionate is produced and the other in which acetate is produced. The lactate is first oxidised to pyruvate, two moles of which are reduced to propionate and one mole oxidised to acetate and CO2. ATP is generated only in the production of acetate. The overall stoichiometry is:
Transcarboxylase is the key enzyme in the production of propionate and requires biotin for activity. Generally, PAB are able to metabolise both isomers of lactate but, in a mixture of the two, preferentially metabolise the L rather than the D isomer.
The theoretical ratio of propionate:acetate is 2:1 but the ratio in cheese is often less, averaging 1.4:1 (Crow 1986). PAB co-metabolise lactate and aspartate, which can be produced from casein by proteolysis. Aspartate is metabolised to fumarate and NH3 by aspartase activity, and the fumarate is then reduced to succinate. In the presence of aspartate, more lactate is metabolised to acetate than to propionate to maintain the redox balance in the cells; the overall effect is to reduce the ratio of propionate:acetate. Strains of PAB with a high level of aspartase produce higher levels of propionate, acetate and CO2 in cheese than those with a low level. PAB also have a prominent role in lipolysis but not proteolysis in Swiss cheese. For more information on the physiology and metabolism of PAB, see Thierry et al. (2011).
The complex interrelationships between lactose and lactate utilisation and production of propionate and acetate by the PAB in Emmental cheese are shown in Fig. 11.16—lactate production occurs early and is due mainly to the growth of Sc. thermophilus; essentially, d-lactate production does not begin until the cheese is in the press and propionate and acetate are not detected until the numbers of PAB have increased significantly. Galactose accumulates early in manufacture, during the growth of Sc. thermophilus, but is subsequently used when Lb. helveticus begins to grow.
NSLAB, particularly, Lb. casei and Lb. rhamnosus , slow down the propionic acid fermentation in Emmental cheese (Froehlich-Wyder and Bachmann 2004). This is thought to be due to citrate metabolism since Cit− mutants of the NSLAB showed much less inhibition. The exact mechanism of this reaction remains unclear. NSLAB are also important in the production of flavour compounds, particularly esters and alcohols, in Swiss-type cheese (Bouton et al. 2009). This was shown by adding Lb. paracasei and Lb. rhamnosus to microfiltered milk and following their subsequent development and the production of various flavour compounds during ripening. Microfiltration is a membrane filtration technique which can be applied to raw skim milk. It retains the bacteria but allows the casein micelles and the rest of the milk constituents to pass through. Pasteurised cream is added subsequently to the filtrate to bring the fat up to the normal level before the cheese is made. This technique was also useful in studying the effect of the raw milk flora on cheese flavour. Swiss-type cheese was made from raw, microfiltered, pasteurised or pasteurised milk to which the microfiltered retenate was added, the latter effectively simulating a raw milk cheese. Cheese made from milk containing the raw milk flora, i.e., the raw milk cheese and the milk containing the retentate, had the better overall aroma, which correlated with higher levels of NSLAB, PAB (which were not added deliberately to the milk), and enterococci, implying that the raw milk microflora is important in determining the flavour of this cheese (Beauvier et al. 1997).
Comté cheese is covered by an orange-coloured smear, called the morge, composed mainly of corynebacteria, micrococci and yeast. Levels of ~1010/cm2 are present in the ripened cheese and it has been calculated that the total number of bacteria in the smear of Comté cheese is the same as the total number in the cheese mass; the species involved do not appear to have been identified.
11.16.3 Parmigiano Reggiano Cheese
This is a very hard PDO (Protected Designation of Origin ) Italian cheese made from partly skimmed raw cows’ milk using natural whey starters, and is made in the Emilia-Romagna region of Northern Italy. Grana Padano is a similar PDO protected cheese made over a wider area of Northern Italy. The cooking temperature is 55–56 °C and the curd is left in the whey for 40–60 min before being placed in a circular mould. The cheese is brine-salted (27 % NaCl at 16 °C for 20–24 h), before ripening it at 16–18 °C and 85 %RH for a minimum of 2 years. It should not be confused with Parmesan cheese which is a similar cheese made mainly in North America.
The microbiology of Parmigiano Reggiano is shown in Fig. 11.17. The natural whey starter used contained Lb. helveticus, small numbers of Sc. thermophilus (<100/ml) and, unusually, Lb. delbrueckii subsp. bulgaricus, which is a starter for yoghurt production, but no lactococci or enterococci (Coppola et al. 2000). The LAB isolated during the first days of ripening were similar to those in the starter; however, after 8 days of ripening they were then supplanted by Lb. paracasei subsp. paracasei, Lb. rhamnosus and Pd. acidilactici. Small numbers of lactococci were identified but only in the early days of ripening. Relatively high numbers (~104 cfu/g) of enterococci, identified as Ec. faecalis and Ec. faecium, Kocuria kristinae, K. rosea, Kytococcus sedentarius and Arthrobacter agilis, were present but all of these decreased throughout ripening. Kocuria and Kytococcus spp. are essentially aerobic organisms and why they were present in an essentially anaerobic system is not clear.
Changes in the numbers of lactic acid bacteria, enterococci and micrococci in Parmigiano Reggiano cheese during ripening. (From Coppola et al. 2000)
11.16.4 Gouda and Edam Cheeses
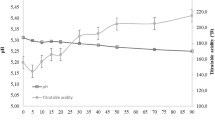
Gouda and Edam are the most important semi-hard cheeses and are of Dutch origin. Both are made in similar ways using DL mixed-starter cultures and are brine-salted. The shapes are very different with Gouda being flat and cylindrical and weighing upto 14 kg, usually covered in a yellow wax while Edam is spherical and covered in red wax and weighs 1–2 kg. Both have small eyes due to CO2 production from citrate by the Cit+ lactococci and leuconostocs in the DL starter culture. Whey is removed during manufacture and replaced with warm water to reduce the level of lactose in the curd. This means that lactose disappears from the cheese very early in ripening. The milk is usually bactofuged to eliminate spores of Clostridium tyrobutyricum and Cl. butyricum which cause late gas formation and off-flavour production in the ripening cheese, due to CO2, H2 and butyric acid production from lactate. The sediment or sludge from the bactofugation is sterilised and added back to the milk to increase the cheese yield. If the milk is not bactofuged, 0.015 % NaNO3 is added to the milk to prevent the growth of clostridia. NO3 − is not the actual inhibitor but NO2 − produced from it by xanthine oxidase, naturally present in the milk. The NO2 − slows down the germination of the spores until the NaCl has diffused sufficiently into the cheese to serve the same purpose. The changes in pH and the levels of lactate and lactose occurring during manufacture and the early stages of ripening of Gouda cheese are shown in Fig. 11.18. The decrease in pH and the increase in lactate are rapid and the lactose has been completely utilised in 9–10 h. The pH in the ripening cheese continues to rise due to the utilisation of citrate and the increased level of proteolysis by the rennet and starters.
Changes in the levels of lactose and lactate and the decrease in pH in Gouda cheese during manufacture. Note the large decrease in lactose coinciding with washing the curd (From van den Berg et al. 2004)
A high-resolution AFLP technique has been used to determine the complex composition of a DL mixed culture, commonly used in Gouda cheese manufacture in the Netherlands (see Chap. 6). One leuconostoc and seven lactococcal lineages were found. This technique was also used to follow what happened to the different lactococcal lineages during cheese manufacture and ripening. The Prt− Lc. lactis subsp. cremoris lineage dominated the microbial community during the first phase of manufacture in which fast acid production is the key process. This is in agreement with the notion that 10 % or less Prt+ strains are sufficient to supply peptides and free amino acids to the Prt− component in an actively growing culture (Erkus et al. 2013; Smid et al. 2014). Brining the cheese triggered a sixfold decrease in the Prt− lineage during the first 2 weeks of ripening. Assuming that loss of viability coincides with cell lysis, the Prt− lineage is likely to be the primary supplier of intracellular lipases and proteinases for the initial production of fatty acids, peptides and amino acids in the cheese matrix. The two citrate-utilising Lc. lactis subsp. lactis lineages displayed much better survival characteristics then the non-citrate utilising components of the culture. This was linked to the capacity of these cells to metabilise arginine by the arginine deiminase pathway and to cheese flavour production. The Leuc. mesenteroides lineage was responsible for 37 % araT transcription, encoding aromatic amino tranferase, during the first 24 h of cheese manufacture, even though it comprised only 1.8 % of the culture.
11.16.5 Surface-Ripened Cheeses
Surface-ripened cheeses are subdivided into bacterial- and mould-ripened cheeses, depending on the major microorganisms involved. Bacterial surface-ripened cheeses include Comté, Livarot, Reblochon, Limburger and Tilsit, and are characterised by the development of a red to orange-coloured, smear on the surface. Mould surface-ripened cheeses include the well-known French varieties, Brie, Camembert, Coulommier and Carre de l’Est and are characterised by a white, felt-like growth on the cheese surface. Bacterial surface-ripened cheeses are also called smear-ripened cheeses, because of the glistening appearance of the cheese surface; they are also called washed-rind cheeses, because their rind is washed several times with brine during ripening, or red-smear cheeses, because of the red-orange colour which characteristically develops on the surface of these cheeses. The ripened cheeses generally have a strong, pungent smell, reminiscent of smelly socks.
Typically, hard, surface-ripened cheeses, e.g., Comté, are made with thermophilic starter cultures and semi-hard, e.g., Tilsit and Pont l’Evéque, and soft surface-ripened cheeses, e.g., Reblochon , are made with mesophilic cultures. Cheeses made with thermophilic cultures are cooked to temperatures around 54 °C whereas only limited cooking (~35 °C) is given to washed-rind cheeses made with mesophilic cultures which consequently have a relatively high moisture content. After light pressing, sometimes overnight, the cheeses are brined (usually saturated brine, pH 5.2; 0.2 % Ca) for 4–18 h depending on their size, small cheeses are brined for shorter times than larger ones. Sometimes, the only pressing received is that of the weight of the curd itself. The cheeses are then drained for a few hours after which they are smeared. Both bacterial- and mould-ripened cheese are ripened at 10–15 °C at a high relative humidity to prevent loss of moisture and consequent drying out of the cheese surface.
11.16.5.1 Deacidification
Environmental factors, particularly the temperature of ripening, and the composition of the cheese, e.g., high moisture (in most cheeses of this type), low initial pH, and high lactate levels, determine the succession of microorganisms that grow on the surface of surface-ripened cheese. The pH of a young cheese after acidification of the cheese curd by the starter lactic acid bacteria is ~5.0. This low pH selects microorganisms (yeast and moulds) which grow on lactate metabolising it to CO2 and H2O and deaminating amino acids producing NH3 and keto acids, causing the pH on the surface to rise and permit the development of bacteria, particularly coryneforms. This is called deacidification and occurs in both mould- and smear-ripened cheeses. The presence of moulds and yeast on the surface of cheese is to be expected since the cheese has a relatively low pH (both can grow at a pH value <3), a ready substrate, lactate, for energy production and a relatively low a w. Once the surface pH increases to >5.8, salt-tolerant bacteria (STB) begin to grow. The interrelationship between the increase in pH and the numbers of STB and yeast in Tilsit cheese is shown in Fig. 11.19. The STB were counted on Plate Count Agar containing 8 % salt. The pH increases steadily from about 5.5 to 7.5 in the first 2 weeks of ripening after which it remains more or less constant. Simultaneously, the numbers of yeast and STB also increase with the STB increasing more rapidly than the yeast.
Deacidification also enhances the action of enzymes, including lipases, proteinases and peptidases, many of which have optima close to neutrality. The lipases and proteinases hydrolyse the fat and protein to fatty acids and peptides and amino acids, while the peptidases hydrolyse the smaller peptides to amino acids. Both the fatty acids and amino acids are the precursors of many of the flavour compounds in surface-ripened cheese (see Chaps. 12 and 13).
11.16.5.2 Bacterial Surface-Ripened Cheeses
Bacterial surface-ripened cheeses can be classified as hard, e.g., Gruyère and Comté, semi-hard, e.g., Tilsit, Brick and Limburger or soft, e.g., Münster, Livarot and Reblochon. Most bacterial surface-ripened cheese is brine-salted. However, Comté is an exception and is dry-salted by rubbing salt and smear on to its surface several times a week during the first 3 weeks of ripening.
Two types of smearing are used, either the “old-young” method, which is traditionally practiced in Germany, or dipping or washing the surface of the cheese with a solution containing various combinations of yeast and bacteria, traditionally different combinations of Geotrichum candidum , Debaryomyces hansenii or Brevibacterium linens , obtained from commercial sources (used in most other countries). In the “old-young” method, a smear from ripened (old) cheese is washed off the surface of the cheese and is then used to inoculate the surface of the young cheese. This ensures that the surface microorganisms that contributed to the ripening of the old, ripened cheese are transferred to the young, fresh cheese. Then, the cheese is ripened at 10–15 °C at an RH > 90 % for several weeks to allow the surface microflora to develop and produce the red or orange colour. The cheese is smeared at the beginning of ripening and then once or twice at 2–4 days intervals to give a uniform distribution of the organisms on the cheese surface. This reduces the risk of unwanted contaminants like moulds colonizing the cheese surface. Generally, visible growth on the surface is apparent within a few days of the beginning of ripening. After 2–3 weeks, the desired microflora has developed and soft and semi-soft cheese are then wrapped or transferred to another ripening room at a lower temperature for further maturation.
Growth of Listeria monocytogenes is a considerable problem on smear cheeses. This organism can grow at 0 °C, pH 4.4 and in 12 % salt and causes listeriosis in humans (Chap. 19). The “old-young” method of smearing can also result in contamination of the young cheese by this and other pathogenic bacteria, which is totally undesirable in a cheese.
Until recently, the bacteria involved were not clear and many were misnamed. The major reason for this is that coryneforms, the dominant bacteria in the smear, are quite difficult to identify accurately unless molecular techniques are used. For a long time, Brevibacterium linens was thought to be the major bacterium on the surface of smear-ripened cheese. Nowadays, it is known to constitute only a minor portion of the flora of a mature cheese. B. linens does not grow below pH 5.5 or 6 and, in fact, has been shown to be a mixture of two different species, B. linens and, a new species, B. aurantiacum (Gavrish et al. 2004).
The microflora of five smear-ripened cheeses, viz. Limburger from Germany, Reblochon and Livarot from France, Tilsit from Austria and Gubbeen from Ireland, has been examined in detail recently, using both traditional and molecular techniques to identify the microorganisms (Cogan et al. 2014). Limburger cheese had the simplest microflora, containing two yeasts, D. hansenii and G. candidum, and two bacteria, Arthrobacter arilaitensis and B. aurantiacum. Livarot was the most complicated, comprising 10 yeasts and 38 bacteria, including many Gram-negatives. Reblochon also had a very diverse microflora containing 8 yeast and 13 bacteria (excluding Gram-negatives which were not identified) while Gubbeen comprised 7 yeast and 18 bacteria and Tilsit 5 yeasts and 9 bacteria. D. hansenii was by far the dominant yeast and was found in all cheeses, followed in order by G. candidum, which was found in all cheeses except Gubbeen, Candida catenulata, which was found only in Livarot and Gubbeen, K. lactis, which was found only in Reblochon and Livarot and C. lusitaniae which was found only in Tilsit and Gubbeen.
B. aurantiacum was the dominant bacterium and was found in each batch of cheese. The next most common bacteria in order were Staphylococcus saprophyticus , which was found in all cheese except Limburger, Arthrobacter arilaitensis, which was found in all cheeses, Corynebacterium casei, which was found only in Reblochon, Tilsit and Gubbeen, C. variabile, which was found only in Reblochon, Tilsit and Gubbeen and Microbacterium gubbeenense, which was found in all cheeses except Limburger. All of these are coryneform bacteria, except S. saprophyticus. Micrococci and staphylococci dominated the bacterial flora early in ripening but later they were outgrown by corynebacteria. Other bacteria were isolated in low numbers, suggesting that each of the five cheeses has a unique microflora. The smear bacteria are all Gram-positive, salt-tolerant (the surface layer of surface-ripened cheese can contain up to 15 % NaCl), aerobic or facultatively anaerobic microorganisms and hence grow easily at the high salt level in the surface layer of these brine-salted cheeses. Gram-negative bacteria were isolated from the two French cheeses, Reblochon and Livarot, but only those from Livarot were identified and included Hafnia alvei, Proteus vulgaris, Alcaligenes faecalis and Psychrobacter spp. (Cogan et al. 2014). Halomonas venusta, H. variabile and an unidentified Halomonas sp. have been found in several other smear cheeses (Maoz et al. 2003; Mounier et al. 2005). Halomonas and Vibrio are salt tolerant. Halomonas are considered to indicate hygiene problems and are normally associated with seawater and salterns and, therefore, it is likely that the salt used in the brining process is the source of this organism. Hafnia, Proteus, Alcaligenes, Psychrobacter, Halomonas and Vibrio spp. are Gram-negative and the effect of such bacteria on the flavour of smear cheeses is unclear.
Several new species were identified during the above study including Agrococcus casei (Bora et al. 2007), C. casei (Brennan et al. 2001a) Mb. gubbeenense (Brennan et al. 2001b) and Mycetocola reblochoni (Bora et al. 2008). New species have also been isolated from other smear cheeses, e.g., S. succinus subsp. casei and S. equorum subsp. linens from a Swiss smear-ripened cheese (Place et al. 2003a, b), Brachybacterium tyrofermentans and Brach. alimentarius (Schubert et al. 1996) from the smear of hard cheese and Arthrobacter bergerii and Arthrobacter arilaitensis from Camembert and Reblochon cheese, respectively (Irlinger et al. 2005). The genomes of Arth. arilaitensis Re117 (Monnet et al. 2010), C. casei UCMA 3821 and LMG S-19264 (Monnet et al. 2012b; Walter et al. 2014) and C. variabile DSM 44702, which was isolated from Gubbeen cheese as C. mooreparkensis , (Schroder et al. 2011), have been sequenced and ranged in size from 3.11 to 3.85 Mb. In addition, strains Re117 and LMG S-19264 contained two plasmids and strain DSM 44702 a putative phage. Genes of importance for the growth of these bacteria and their putative role in cheese ripening were detected, including the uptake of iron, osmoprotection and catabolism of lactate, citrate, protein and fat, but whether these genes are transcribed in the cheese during ripening has not been studied, except for those involved in the metabolism of iron (Monnet et al. 2012a). Iron is essential in the respiration of lactose, lactate and citrate, where it functions as part of the cytochrome system. The cheese surface is surrounded by oxygen implying that the organisms on the surface can respire, if they have the necessary enzymes. Bovine milk is low in iron (0.2–0.4 mg/L) and the main mechanisms used by these bacteria to transport iron are siderophores which are strong chelators of Fe3+. Thirty genes involved in chelation and transport of iron by siderophores were identified in strain Re117 (Monnet et al. 2010) and 29 in strain DSM 44702 (Schröder et al. 2011). Further studies (Monnet et al. 2012a) showed that the availability of iron on the cheese surface is limiting for the growth of these bacteria on the cheese surface. The osmoprotectant genes detected included the transport of ectoine, a derivative of proline, proline itself, and glycine betaine, a derivative of glycine where the H atoms on the amino group are replaced by methyl groups. These are all highly soluble compounds which allow these organisms to tolerate and grow at high concentrations of salt. C. variabile DSM 44702, contained the genes for transport of lactate and citrate and the conversion of lactate to pyruvate, which can then be oxidised through the TCA cycle. The genes encoding enzymes involved in proteolysis included a proteinase, and a proline iminopeptidase, which releases proline from the amino terminal end of peptides, while the genes encoding enzymes involved in lipolysis include esterases, lipases and the enzymes involved in β-oxidation of fatty acids. The different species have also been shown to develop different colours on the cheese surface.
Arthrobacter, Brevibacterium, Brachybacterium, Corynebacterium and Microbacterium are often called coryneform bacteria. All of them are Gram-positive, catalase-positive, non-sporeforming and are generally non-motile. A major feature of their growth is that exponential phase cells are pleomorphic, showing the presence of irregularly shaped rods, including wedge, club, V and curved shapes. In addition, Arthrobacter, Brevibacterium and Brachybacterium spp. go through a marked rod/coccus cycle during growth, with rod forms dominating the exponential phase of growth (1–2 days) and coccal forms dominating the stationary phase (5–7 days). All belong to the Actinomycete (high GC) branch of the Gram-positive bacteria.
A study of the surface microflora of five Italian washed-rind cheeses, Taleggio, Gorgonzola, Casera, Scimudin and Formaggio di Fossa, has also been conducted using molecular techniques (Fontana et al. 2010). Most of the bacteria were cocci including S. saprophyticus, S. equorum, S. vitulinus, S. caprae, Micrococcus luteus and M. caseolyticus and only two coryneforms, B. linens or more likely B. aurantiacum, since the reference strain used was actually the type strain of B. aurantiacum, and C. flavescens. These data suggest that the microflora of Italian smear-ripened cheeses differ significantly from others similar European cheeses.
Bokulich and Mills (2013) used high-throughput sequencing technology to study the microbial ecosystems in two US artisanal cheesemaking plants, producing fresh and smear- and mould-ripened cheeses, in great detail. Fermentation-associated microorganisms , especially Lactococcus and Debaryomyces dominated most surfaces, suggesting that these microorganisms establish biofilms on equipment surfaces and may play an important role in transferring microorganisms to the cheese. In addition, environmental microorganisms from the processing environment dominated the surface microflora of smear cheeses in both plants, demonstrating the importance of the environment in populating the cheese surface, even when it is deliberately inoculated with smear bacteria. Gram-positive bacteria dominated the cheese surface. However, Gram-negative bacteria were also found on the surfaces of mature cheeses, many of which were halotolerant, e.g., Pseudoalteromonas spp., Halomonas spp. and Vibrio casei and which may have originated in the salt used in cheesemaking.More recently, Wolfe et al. (2014) examined the ecology of the rinds of 137 cheeses, including washed-rind, mould-ripened and cheeses with natural, undisturbed rinds, from 10 countries (England, France, Germany, Ireland, Italy, Portugal, Spain, Sweden, Switzerland and the US) using molecular techniques. Bacteria from 14 genera (Arthrobacter, Brachybacterium, Brevibacterium, Corynebacterium, Nocardiopsis, Yaniella, Staphylococcus, Halomonas, Pseudomonas, Psychrobacter, Pseudoalteromonas,Vibrio, Hafnia/Serratia and Sphingobacterium) and fungi from 10 genera (Debaryomyces, Galactomyces, Candida, Scopulariopsis, Fusarium, Acremonium, Peniillium, Aspergillus, Chrysosporium and Sporendoema) were found at more than 1 % abundance but not in all cheeses. The average number of bacterial and fungal genera were 6.5 (range 1–13) and 3.2 (range 1–7), respectively. More than 60 % of the bacteria and 25 % of the fungi were environmental contaminants. This is the first report of the presence of the actinobacteria, Nocardiopsis and Yaniella, on cheese rinds. The data also showed that Pseualteromonas spp contain methionine-γ-lyase which converts methionine to methanthiol, a key component of the flavour of washed-rind cheeses and which to date has only been found in B. linens.
The finding of staphylococci in cheese raises issues regarding their pathogenicity even though the strains isolated were coagulase negative. A French study (Coton et al. 2010) has shown that S. equorum S. xylosus, S. saprophyticus and S. epidermidis were the dominant species in numerous French cheeses examined over a 16 year period from 1990; 11 other coagulase-negative species were also identified. S. epidermidis and S. saprophyticu s were also found in clinical samples but PFGE analysis showed no relationship between the clinical and the food strains.
11.16.5.3 Sources of the Bacteria on Surface-Ripened Cheese
Commercially, only B. linens , D. hansenii and G. candidum are used as secondary cultures to deliberately inoculate the cheese surface. However, in several studies, very few of these cultures were re-isolated from the cheese and, when they were, it was only from the initial stages of ripening. These results imply that smear cheese production units must have an adventitious ‘house’ flora and that the use of commercial secondary cultures in the production of smear-ripened cheeses is questionable. One way around this problem is to identify the dominant organism present in a particular cheese and then give them back to the cheesemaker and this has been shown to be effective in practice. Brines, many of which can be several years old, were shown to be an important source of S. saprophyticus and D. hansenii and the skin of the arms and hands of workers were important sources of C. casei and C. variabile (Mounier et al. 2006). This raises interesting questions concerning the ecology of surface-ripened cheese and human skin since the dominant genera on both skin and the cheese surface are Staphylococcus and Corynebacterium species. In addition, micrococci, coryneforms, yeast and moulds are present as a biofilm on the shelves (Mariani et al. 2007) and wooden vats (‘gerles’ in French) (Didienne et al. 2012) used in ripening the French smear-ripened cheeses, Reblochon and Salers, respectively. An example from Reblochon shelves is shown in Fig. 11.20. Therefore, other wooden tools used in cheesemaking should also be considered likely sources of microorganisms.
Scanning electron microscopy of the surface of a ripening shelf. A (×300), ripening biofilm does not cover all the surface of the shelf. B (×1500), bacteria are located in the cracks. C (×4000) and D (×6000), details of Geotrichum candidum (cylinders) and bacteria (From Mariani et al. 2007)
Defined-strain secondary cultures for bacterial surface-ripened cheese are also being developed and the successful use of a defined-strain culture containing, D. hansenii, B. linens, A. nicotianae (probably Mb. gubbeenense), C. ammoniagenes (probably C. casei) and S. sciuri has been shown on a pilot scale; such cultures are not yet available commercially (Bockelmann et al. 2005) The fact that commercial cultures are not subsequently recovered from cheese may militate against the use of defined cultures but a better understanding of the microbiology, ecology and interactions which occur between bacteria on the cheese surface will help considerably in developing them.
11.16.5.4 Mould Surface-Ripened Cheeses
Camembert is a mould-ripened cheese with a relatively high moisture content (~50 %) and short ripening time. It is probably the most common mould-ripened cheese and is produced in two forms, traditional and non-traditional Camembert. Traditional Camembert, more commonly known as Camembert de Normandie, is made from raw milk and is dry-salted whereas non-traditional Camembert is made worldwide, generally from pasteurised milk and is brine-salted. Traditional Camembert is also a PDO (Protected Designation of Origin) cheese, made only in designated areas of Normandy in France. DL mixed cultures are used in production of both types and no pressing of the curd occurs. Instead the moulds, containing the curd, are inverted a few times to help syneresis of whey from the curd. Generally, P. camemberti, K. lactis, G. candidum and B. linens are necessary to develop the optimum flavour of Camembert cheese. These microorganisms are obligate aerobes and grow only on the cheese surface and are either added to the milk with the starter cultures (in the case of non-traditional Camembert) or sprayed on the cheese surface after manufacture (traditional Camembert). A new type of Camembert is also made with a mixture of mesophilic and thermophilic cultures. Part of the whey is drawn off during manufacture, ensuring that a less acidic cheese is produced. These are called stabilised or solubilised cheeses and are often sold under trade names. The manufacture and ripening of Brie, another very common mould surface-ripened, is similar to that of Camembert except that Brie has a larger diameter and a thinner wheel.
A detailed study (Leclercq-Perlat et al. 2004) of some of the microbial changes which occur on the surface of experimental Camembert cheese made under aseptic conditions but with the deliberate addition of both starter and the common ripening organisms, P. camemberti, K. lactis, G. candidium and B. linens, is shown in Fig. 11.21 and gives some understanding of the complex microbiological and biochemical changes occurring in the cheese during ripening. The ripening conditions for the cheese were as follows: 24 h after moulding, the cheeses were brined at 14 °C for 25 min: they were then transferred to a sterile ripening chamber, held at 14 °C, 85 % RH for 24 h, after which they were ripened at 12 °C and 95 % RH for 14 days. On day 6 the cheeses were turned, and on day 14, the temperature and RH were changed to 14 °C and 85 % RH for 1 day, after which the cheeses were wrapped and stored at 4 °C until day 45.
Top figure, changes in the numbers of K. lactis (open circles), G. candidum (open triangles) and Brevibacterium linens (open squares) on the rind; middle figure, lactose in the rind (closed triangles) and core (open triangles) and lactate in the rind (closed circles) and core (open circles) and bottom figure, pH in the rind (solid triangles) and core (open triangles) in Camembert cheeses during ripening for 45 days. The temperature and RH maintained during the ripening are also indicated (From Leclercq-Perlat et al. 2004)
Deacidification was much more rapid at the cheese surface than in the core. The surface pH increased slowly initially from ~4.6 on day 1 to 5.1 on day 6 (Fig. 11.21). This slow change in pH reflects two opposing conditions, consumption of lactate by the rapidly growing K. lactis and G. candidum and continuing production of lactic acid from the residual lactose, by the starter, which occurred, even though a significant amount of salt was present on the surface, from the brining. The rind was completely devoid of lactose by day 7 and the pH reached 7.7 on the surface on day 11. The number of lactococci probably reached ~109/g at the beginning of ripening and remained at this level in both the interior and the surface throughout ripening.
Numbers of K. lactis and G. candidum increased rapidly during the first 2 days of ripening and then, more slowly, until day 7 after which growth of G. candidum continued and that of D. hansenii ceased; both organisms reached final cell densities of ~8 log cfu/g DM. In contrast, numbers of B. linens remained static at 2 log cfu/g DM for the first 5 days of ripening, after which they increased to 4 log cfu/g DM from day 5 to day 9 and then more slowly reaching 9 log cfu/g DM by day 40. P. camemberti is an obligate aerobe and grows only on the cheese surface. Its numbers were also estimated but gave no real idea of how rapid growth was as the mycelium was destroyed during the analysis. However, growth of P. camemberti was visible from day 7 and completely covered the cheese surface on day 12.
11.16.6 Blue Cheeses
Blue cheeses are characterised by blue veins of the mould, Penicillium roqueforti, running through the cheese and include some famous varieties, e.g., Gorgonzola, Stilton, Danablu and Roquefort. Gorgonzola and Stilton are made from pasteurised bovine milk, Danablu from thermised bovine milk while Roquefort is made from raw ovine milk. The colour can vary from brown to green to blue depending on the age of the cheese. In many blue cheeses no starters are used but mesophilic cultures are used for Stilton, Danablau and Roquefort while a mixture of both mesophilic and thermophilic starters are used for Gorgonzola. Generally, spores of P. roqueforti are added with the starter. Salting is either by brining or dry salting and the cheese is pierced to allow limited entry of O2 to promote the growth of P. roqueforti. Essentially cheese is an anaerobic system but P. roqueforti is unique among moulds in being able to grow at low levels of O2 which the piercing process allows to diffuse into the cheese from the air.
There is little published information on the microbial changes which occur in blue cheese during ripening except for Cabrales, a Spanish blue cheese, made from raw milk without the deliberate addition of starters or P. roqueforti (Nunez 1978; Nunez et al. 1981). Adventitious LAB in the milk are responsible for acid production during manufacture and ripening. The coagulum is cut 2 h after addition of farm-made goat rennet and is then scooped into moulds which are held at 16–18 °C for 48 h to allow whey drainage to occur. Then, the curd is removed from the mould, covered with coarse salt, held for a further 48 h at 16–18 °C and then ripened at 10–12 °C for 10–15 days and transferred to caves for further ripening at 9–12 °C at 90–95 % RH.
The microbiological changes which occur in Cabrales cheese during ripening are shown in Fig. 11.22. In each graph, the first and second points refer to the counts in the milk and the curd on days 1 and 2, respectively. Growth of lactococci is relatively rapid during the first few days after which their numbers decrease but more rapidly on the surface than in the interior of the cheese. Numbers of mesophilic lactobacilli remain more or less constant at 106 cfu/g on the surface but increase to 108 cfu/g in the interior, after which they decrease. Micrococcus spp. show the opposite trend, being higher on the surface than in the interior while yeast show significant growth on the surface of the cheese early in ripening after which they decrease. Coliforms grow during cheesemaking but their numbers decrease rapidly over the next 2 weeks. This is probably due to the very rapid decrease in pH which reaches ~5.0 in 48 h due to the growth of the lactococci after which it increases to 6.5 in the interior and to 7 on the surface due to metabolism of lactate to CO2 by the yeast and moulds.
Growth of different organisms and changes in the pH at the surface (S) and in the interior (I) of Cabrales cheese during ripening (From Nunez 1978)
Pronounced pH, NaCl and a w gradients occur in blue cheese. Deacidification also occurs since P. roqueforti utilizes the lactate in the cheese and produces NH3 from deamination of amino acids, causing the pH to increase. Blue cheeses also contain large numbers of yeasts which also metabolise lactate and produce NH3. However, their role has not been studied extensively.
The most important organisms in flavour development of Blue cheese are the SLAB and P. roqueforti and G. candidum but yeast, particularly D. hansenii, Kluveromyces marxianus and Yarrowia lipolytica, and NSLAB, particularly Lb. paracasei, Lb. casei and Lb. plantarum, also probably play a role in flavour development. Interactions can occur between yeast and P. roqueforti; certain strains of D. hansenii have been shown to stimulate and strains of Y. lipolytica to inhibit the growth of P. roqueforti. Brines are an important source of yeast and their composition and the conditions of ripening vary from country to country. In France, the brine used for Blue cheese production contains 19–20 % NaCl, has a pH of 4–6 and a temperature of 13–16 °C while in Denmark, the concentration of salt and the temperature are higher (22–23 % and 19 °C, respectively), and the pH somewhat lower (4.5) (Cantor et al. 2004). The higher salt and temperature will significantly increase the rate of salt diffusion into the cheese. Yeast numbers in brine can reach 106 cfu/ml with D. hansenii being the predominant one while numbers on the surface of Roquefort cheese reach ~109 cfu/g of dry matter (Besancon et al. 1992).
11.17 Microbial Spoilage of Cheese
The most common microbial defects of cheese are early and late gas. They are relatively uncommon in cheese to-day, due to improved hygienic quality of the milk and better quality control in cheese plants.
Early gas generally occurs within 1 or 2 days after manufacture. It is characterised by the appearance of many small holes throughout the cheese and is caused by coliform bacteria . The gas is mainly H2 which is produced from formate, a product of lactose metabolism, by formic hydrogenylase . It is more problematic in soft and semi-hard cheese than in hard cheese because of the higher a w in the former cheeses. An effective way of controlling early gas is to add KNO3 or NaNO3 at a low level (0.2 %) to the milk. NO3 − does not prevent the growth of coliform but acts as an alternative electron acceptor, allowing complete oxidation of lactose to CO2 and H2O, rather than fermentation to formate, thus effectively reducing the production of H2 from formate. Early gas production can occasionally be caused by yeast due to CO2 production from lactose or lactate.
Late gas formation, or late blowing, does not occur until late in ripening and is due to the fermentation of lactate to butyrate and H2 by Clostridium tyrobutyricum and Cl. butyricum . The butyrate gives the cheese a pronounced off-flavour and the H2 is responsible for large, deformed holes or eyes. Late gas can be particularly prevalent in Dutch- and Swiss-type cheese; in the latter cheese clostridia grow with the PAB, during the “hot” room ripening period. Silage is a potent source of these bacteria and, for this reason, it is forbidden to feed it to cows, whose milk is intended for cheesemaking, in Switzerland. Clostridial spores survive passage through the cows’ digestive system and appear in the dung. The degree of contamination of the milk with dung depends on the hygienic conditions during milking. The spores are not inactivated to any extent by pasteurisation of the milk but germinate in the cheese during ripening. Late gas production can also be controlled by removal of the spores from the milk by bactofugation but this often results in inferior quality cheese, or by the addition of 0.15 % KNO3 to the milk before renneting. In addition, many thermophilic cultures are thought to stimulate the growth of clostridia, through the production of peptides and amino acids. Strains of Cl. tyrobutyricum vary widely in their abilities to grow under different conditions of pH, salt concentration and temperature; 7 out of 10 strains grew at pH 5 (at 37 oC), 5 strains grew in the presence of 3 % NaCl (at pH 7 and 37 oC) and no strain grew in 3.5 % NaCl or at 10 oC (at pH 7 and 37 oC) (Ruusunen et al. 2012). This data suggests that the low ripening temperatures, low pH and relatively high concentrations of most hard cheeses during ripening prevent the growth of many strains of Cl. tyrobuyricum.
The bacteriocin, nisin, produced by some strains of Lc. lactis subsp. lactis, is effective in controlling the growth of clostridia and is used for this purpose in processed cheese. However, it is not suitable for use in natural cheese because many strains of starter are sensitive to it. Defined-strain starters that produce nisin are now being marketed to a limited extent for use in Dutch-type cheeses. Other bacteriocin-producing starters , e.g., lacticin 3147, have also been shown to control late gas formation in cheese (Martinez-Cuesta et al. 2010). Increasing the level of salt, lowering the pH of the cheese rapidly through the use of an active starter and addition of lysozyme can also be effective in preventing late gas production.
Lysozyme , which is found in milk, saliva, tears and other body fluids, hydrolyses the cell walls of sensitive bacteria, like Cl. tyrobutyricum, causing them to lyse. It is commonly used in Italy and is added to the milk with the starter at a level of 25 mg/L. It is generally considered to have no effect on the growth of starters, although some strains in Italian natural whey cultures are inhibited by it.
Other microorganisms have occasionally been implicated as spoilage organisms. Citrate-metabolising lactobacilli have been incriminated as the cause of open texture or slit openness in Cheddar cheese, due to the production of CO2 from citrate. The optimum pH for uptake of citrate ranges from 4 to 5 and significant metabolism of citrate occurs in the absence of an energy source at pH 5.2, the pH of many semi-hard and hard cheeses. Rapid refrigeration of the cheese blocks after manufacture and good hygiene reduce this defect. If the permeability of the wrapping material allows O2 into the cheese during storage, some NSLAB, particularly pediococci, can oxidise lactate to acetate and CO2 on the cheese surface. Open texture in cheese has been reviewed by Martley and Crow (1996).
Growth of NSLAB (Lb. casei, Lb. plantarum and Lb. brevis) in Dutch-type cheese to more than 107 cfu/g in 4–6 weeks causes the production of gas and putrid off-flavour development. Brines are considered to be a major source of these organisms and it is recommended that they should contain <103 lactocbacilli/ml (van den Berg et al. 2004). The increased levels of NSLAB in brines is due to growth of yeast on deposits which naturally occur just above the brine surface, and on racks and other equipment. Growth of the yeast decreases the local pH in the curd allowing the NSLAB to grow. Entry of the NSLAB from the brine is facilitated by poor pressing of the cheese and consequent lack of closing of the rind. Weak brines also support the growth of these NSLAB, and it is important that the brine strength should be maintained at >16 % NaCl, pH < 4.5 and 13 °C.
Ec. malodoratus , as its name implies, causes the production of bad flavours, and has been found in Gouda cheese.
Growth of thermo-resistant streptococci , particularly strains of Sc. thermophilus , occurs in the regeneration section of pasteurisers during long pasteurisation runs and reaches high numbers (106/ml) in the pasteurised milk. Their number may increase to 108/g of cheese early in ripening and cause unclean yeasty flavours to develop in the cheese. Thus, in many cheese plants, pasteurisers are routinely cleaned after continuous runs of 8 h.
PAB are also prone to grow in cheese, particularly Dutch cheese, during prolonged ripening, causing development of a sweet taste and very open texture, due to excessive gas production. Under normal conditions their growth is significantly slowed down by the relatively high salt level ( PAB are very sensitive to salt) and the ripening temperature of <12 °C; some cheeses may be ripened at 5–6 °C to prevent growth of PAB.
Yeasts and moulds are occasionally incriminated as spoilage organisms in cheese. The surface of cheese, especially when it is moist, e.g., an unwrapped soft or semi-soft cheese, is an ideal environment for their growth. These cause little damage to the cheese but are unsightly. They can be washed off the cheese surface with a dilute brine solution. Sometimes hard cheeses are dipped in a dilute solution of the antifungal antibiotic,natamycin (pimaricin), to prevent the growth of yeast and moulds on their surfaces Sorbic acid is allowed in Italy as a preservative for hard, fresh and processed cheese. Sorbate-resistant moulds, Paecilomyces variotti, and yeast, D. hansenii from Crescenza and Provolone cheese are able to grow in the presence of 3 mg of sorbic acid/g and are also able to transform sorbic acid to trans-1,3-pentadiene, which has a taste and odour like kerosene (Sensidini et al. 1994).
Cladosporium cladosporioides, Penicillium commune, C. herbarum, P. glabrum and a Phoma spp. are responsible for the “thread mould” defect of Cheddar cheese (Hocking and Faedo 1992). This defect occurs as a black, dark brown, or dark green spot or thread (hence the term ‘thread mould’) in the folds, creases and gusset ends of the plastic bags used to wrap Cheddar cheese during ripening. It can occur on the cheese surface but is more often associated with free whey drawn from the fresh cheese block during vacuum packaging. These moulds are obviously able to grow in the presence of low levels of O2.
Growth of P. commune , which is closely related to P. camemberti, can result in discoloration of cheese surfaces and the production of off-flavours (Lund et al. 1995).
11.18 Probiotics
These are live cultures, generally of Lactobacillus or Bifidobacterium spp., which, when administered in adequate amounts, confer a health benefit on the host. They have been exploited extensively in the dairy industry in the development of novel functional foods. More than 40 different strains are being used but, with the exception of Lb. casei Shirota, Lb, rhamnosus GG and B. animalis Bb12, very few of them have been clinically proven to have a positive effect on human health. To realize their health benefits, the probiotic strain must be viable at the point of sale and present at concentrations of ~106 cfu/g. The health benefits include alleviation of lactose intolerance, prevention and reduction of diarrhoea, prevention of allergies, reduction of the risk associated with mutagenecity and carcinogenicity, reduction in cholesterol, inhibition of Helicobacter pylori, the cause of some stomach ulcers, and other intestinal pathogens, prevention of inflammatory bowel disease and modulation of the immune system. Yoghurt and fermented milks are the main vehicles for the delivery of probiotics but cheese is also useful since it has a higher pH, more solid consistency and relatively higher fat, which help to protect probiotic bacteria. When cheese is used it is important to determine what effect the survival of the probiotic culture has on the various ripening parameters and on the development of cheese flavour. In addition, particularly where probiotic lactobacilli are added, suitable media to distinguish between the probiotic and NSLAB strains must be developed. For a review of probiotics see Vasiljevic and Shah (2008).
11.19 Non-Lactic Genera of Bacteria Found in Cheese
A brief description of the genera of bacteria other than starter and NSLAB found in cheese is given below. Many, especially the corynebacteria, are difficult to identify by simple tests and generally chemical analysis of cells for the types of polar lipids, menaquinones, fatty acids, the presence or absence of mycolic acid and amino acid composition of the cell wall, are required. More recently, molecular methods, e.g., rep-PCR with different primers, PFGE or 16S rRNA sequencing are being used but these techniques require sophisticated equipment.
11.19.1 Agrococcus
These are Gram-positive, catalase-positive, non-motile, aerobic ovoid to short rods which contain diamino butyric acid in their cell wall. The cells occur singly, in pairs, in short flexible chains, or in small irregular clusters. The phospholipids are phosphatidylglycerol, diphosphatidylglycerol. The main menaquinones are MK-12 and MK-11. No mycolic acids are present. The type species is Agrococcus jenensis. They do not grow at 42 °C and have been isolated from air, soil and medieval wall paintings as well as smear cheese.
11.19.2 Arthrobacter
These are Gram-positive, catalase-positive, non-motile, strictly aerobic rods which go through a marked rod/coccus cycle during growth, with the rod forms dominating in exponentially growing cultures and the coccus forms dominating stationary phase cultures. Their cell wall contains lysine. Little or no acid is produced from glucose or other sugars. They have non-exacting nutritional requirements; generally, biotin is the only vitamin required. Their habitat is soil and they do not withstand pasteurisation. They can be confused with micrococci (see below). A. agilis has been isolated from cheese (Coppola et al. 2000).
11.19.3 Brachybacterium
These are Gram-positive, facultatively anaerobic short rods which exhibit a rod-coccus growth cycle. Their optimum temperature is ~30 °C. Their cell wall contains meso-diaminopimelic acid and glucose, galactose and rhamnose but not mycolic acids. Five species are recognised and two of these, Br. alimentarium and Br. tyrofermentans have been isolated from Comté and Beaufort cheese, respectively. Their nutritional characteristics do not appear to have been studied but Br. alimentarium and Br. tyrofermentans can grow in the presence of 14 and 16 % NaCl, respectively.
11.19.4 Brevibacterium
Brevibacteria are Gram-positive, catalase-positive, non-motile, strictly aerobic rods which go through a marked rod/coccus cycle during growth. Their cell wall contains meso-diaminopimelic acid and they metabolise sugars by respiration. There are five species, B. linens, B. aurantiacum, B. casei, B. iodinum and B. epidermidis. The first three species have been isolated from cheese, the fourth from milk and the fifth from skin. B. linens has recently been shown to be a mixture of two species, B. linens and B. aurantiacum both of which produce yellow- or orange-coloured colonies (Gavrish et al. 2004) while those of B. iodinum are purple, due to the production of a phenazine derivative; the other two species produce gray-white colonies. Brevibacteria grow poorly, if at all, at 5 °C, have an optimum temperature of 20–25 °C and grow in the presence of a high concentration of NaCl; B. linens and B. iodinum grow in the presence of 8–10 % NaCl and B. casei and B. epidermidis in the presence of 15 % NaCl. Nutritionally, they have not been studied very well but most strains of B. linens require amino acids and vitamins for growth. Their metabolism is respiratory and they do not produce acid from glucose. They are easily confused with Arthrobacter spp. B. linens metabolises methionine to methional, which is thought to be responsible from the characteristic “dirty sock” odour of smear-ripened cheeses. B. linens can be determined specifically through the production of a stable pink colour within 2 min on treatment of a small amount of a colony with a drop of 5 M KOH or 5 M NaOH or a salmon pink colour within 1 min on treatment with glacial acetic acid. Brevibacteria are acid-sensitive and will not grow a at pH values <6.0. Their major habitats are dairy products, especially cheese, activated sludge and human skin.
11.19.5 Corynebacterium
These are Gram-positive, catalase-positive, non-motile, slightly curved rods with tapered ends; club-shaped forms may be found also. Some species are strict aerobes and others are facultative anaerobes. A rod-coccus cycle does not occur. Methylene blue stains show the presence of deep blue coloured metachromatic granules. Meso-diaminopimelic acid and short-chain (22–36 C atoms) mycolic acids are found in their cell wall. They are nutritionally exacting, requiring several vitamins, amino acids, purines and pyrimidines for growth. Two bacteria, Microbacterium flavum and Caseobacter polymorphus, which were isolated from cheese, have been reclassified as C. flavescens and C. variabile, respectively. C. polymorphus was isolated originally from the surface of Dutch smear-ripened cheese and produces grey-white, slightly pink or slightly red colonies.
11.19.6 Microbacterium
Microbacteria are small, Gram-positive, non-motile or motile, obligately aerobic rods which do not go through a rod/coccus cycle; however, in older cultures (3–7 days), the rods are short and a proportion may be coccoid. Currently, there are 13 species only one of which, M. lacticum, has been found in milk. Their optimum temperature is 30 °C. Colonies vary in colour from gray-white to pale green or yellow. Their cell wall peptidoglycan contains lysine. Generally, their metabolism is respiratory but acid is produced from glucose and some other sugars in peptone-containing media. Most strains require biotin, pantothenic acid and thiamine for growth. The main species found in milk is M. lacticum, which is thermoduric, surviving heating at 63 °C for 30 min. The organism is not found in aseptically-drawn milk and there is strong evidence that the major source of contamination of milk with this organism is improperly cleaned dairy equipment.
11.19.7 Propionibacterium
These are Gram-positive, non-motile, pleomorphic rods, which may be coccoid, bifid or, sometimes, branched in shape. They occur singly, in pairs, in short chains or in clumps with ‘Chinese lettering’ arrangements. Colonies vary in colour and can be white, grey, pink, red, yellow or orange. Although these organisms are catalase positive, they are essentially anaerobic or microaerophilic bacteria. Propionibacteria are divided into the ‘classical’ and ‘acnes’ groups. The classical group are found mostly in dairy products, particularly cheese, and also in silage and olive fermentations while the acnes group are found mainly on human skin. The classical group is divided into four species: P. freudenreichii, the most common one, P. jensenii, P. thoenii and P. acidipropionici. The peptidoglycan of P. freudenreichii contains meso-diaminopimilic acid (DAP) while the L isomer is found in the other three species. P. freudenreichii was considered to exist as two subspecies, P. freudenreichii subsp. freudenreichii and P. freudenreichii subsp. shermanii but genetic analyses have shown that both subspecies are identical; the only phenotypic difference is that P. freudenreichii subsp. freudenreichii is able to ferment lactose while the other subspecies cannot. The colour of P. freudenreichii colonies changes during incubation from grey to tan or pink. P. freudenreichii and P. jensenii are non-hemolytic while P. thoenii and P. acidipropionici are β-haemolytic. PAB generally have relatively simple nutritional requirements although they generally require pantothenic acid, biotin or thiamine for growth and many of them can use NO3 − as the sole source of N.
11.19.8 Pediococcus
These are Gram-positive, catalase-negative cocci which occur as tetrads. Tetrad formation is due to cell division in two directions in a single plane and is characteristic of this genus. There are eight species. They are homofermentative, producing either DL- or L-lactate from sugars and most strains can grow in the presence of 6.5 % NaCl. They generally have complex nutritional requirements. Most pediococci do not ferment lactose and therefore do not grow well in milk. Those strains which metabolise lactose may also lack a proteinase to hydrolyse milk protein to the amino acids and peptides, required for their growth. Some of them can grow at pH 8.5 and 4.2. Some pediococci can metabolise citrate but the products are acetate and formate rather than diacetyl and acetoin. They are found occasionally as a minor part of the NSLAB flora in some hard cheeses. Their influence on the production of cheese flavour is not clear.
11.19.9 Micrococcus
Micrococci are Gram-positive, catalase-positive, strictly aerobic, non-motile cocci (0.2–2.0 μm in diameter) which occur in pairs, clusters or tetrads. The cell wall contains l-lysine. Division occurs in several planes resulting in the formation of regular and irregular clusters. Their natural habitat is skin and currently 17 species are recognised. All grow in the presence of 5 % NaCl and many in the presence of 10–15 % NaCl. Many species produce yellow, orange or red colonies. Their nutritional requirements are variable. M. luteus, the type species, produces yellow colonies and grows on glutamate as the sole source of C and N, in the presence of thiamin and/or biotin. Some species can utilise ammonium phosphate as the N source but many species have complex nutritional requirements. They are commonly found on the surface of smear-ripened cheese but the species found are not clear; they dominate the surface microflora of Comté and blue cheese.
Based on 16S rRNA sequencing, the Micrococcus and Arthrobacter genera overlap. A major taxonomic reassessment of the Micrococcus genus has been carried out and a further four new genera have been proposed, Dermacoccus, Kocuria, Kytococcus and Nesterenkonia (Stackebrandt et al. 1995). Kocuria and Kytococcus spp. are found in cheese.
11.19.10 Kocuria
These are Gram-positive, catalase-positive, strictly aerobic cocci, although strains of K. kristinae are slightly facultative anaerobes. Their cell wall contains lysine and alanine and they can grow in 7.5 % NaCl with some species growing in the presence of 10–15 %. The GC content varies between 66 and 75 mol %. This genus now contains Micrococcus varians, Mc. roseus and Mc. kristinae which are described as K. varians, K. rosea and K. kristinae, respectively. The latter two species have been found in significant numbers in Parmigiano Reggiano cheese (Coppola et al. 2000).
11.19.11 Kytococcus
These are Gram-positive, catalase-positive, strictly aerobic cocci which can grow in the presence of 10 % NaCl. Their cell wall contains lysine and their GC content is 68–69 mol %. Ky. sedentarius has been found in significant numbers in Parmigiano Reggiano cheese (Coppola et al. 2000).
11.19.12 Staphylococcus
These are Gram-positive, catalase-positive, facultatively anaerobic, non-motile cocci (0.5–1.5 μm in diameter), which characteristically divide in more than one plane to form clusters. They can also occur in pairs and tetrads. Currently, 19 species of staphylococci are recognised and many produce yellow or orange colonies. They are facultative anaerobes and grow better aerobically than anaerobically. Most strains grow in the presence of 10 % NaCl and between 10 and 40 °C. Acid is produced anaerobically from several sugars, including glucose and lactose. They are fastidious, requiring from 5 to 12 amino acids and several B vitamins for growth. Major habitats of staphylococci include the nasal membranes, skin and the GI and genital tracts of warm-blooded animals. S. aureus causes mastitis in cows and boils and carbuncles in humans and is considered to be a pathogen. Many strains of S. aureus produce a heat-stable enterotoxin which causes food poisoning; growth to about 106 cfu/g in food is necessary to produce sufficient toxin (0.1–1.0 μg/kg) to cause food intoxication. Coagulase activity is accepted as the indicator of pathogenicity in staphylococci and S. aureus, S. intermedius and S. hyicus produce it. S. intermedius has been found in the nasal passage of horses, dogs, mink and foxes and S. hyicus on the skin of pigs and less frequently on the skin and in the milk of cows.
Micrococcus and Staphylococcus appear as regular and irregular clusters of cells when examined microscopically and traditionally both genera have been placed in the family Micrococcaceae, indicating that they are closely related. However, phylogenetic studies show that they are quite distant from each other, Staphylococcus spp. belong to the Clostridium branch of the Gram-positive bacteria and contain 30–39 mol % GC while Micrococcus belong to the Actinomycete branch and contain 63–73 mol % GC.
It is relatively easy to distinguish micrococci from staphylococci. The simplest way is to check for acid production from glucose under aerobic and anaerobic conditions. Staphylococci produce acid from glucose aerobically and anaerobically while micrococci either do not produce acid or produce it only aerobically. In addition, micrococci are resistant to lysostaphin, a cell wall degrading enzyme, and are sensitive to erythromycin (0.04 mg/ml) and bacitracin (0.04 U) while staplylococci give the opposite reactions.
11.20 Yeast and Moulds
Yeast and moulds are generally not nutritionally demanding, and are larger and grow more slowly than bacteria. Therefore, they do not compete with bacteria in environments in which bacteria grow, e.g., at pH values around 7. However, they grow quite well at pH values of 2–4 where bacteria either do not grow or grow very poorly. The low pH of freshly made cheese is therefore partially selective for their growth. Yeast and moulds are eukaryotes, i.e., they contain a clearly identifiable nucleus, and most of them also contain chitin, a β-1,4 polymer of N-acetylglucosamine, which is responsible for their rigid structure.
Colonies of yeast are generally soft in consistency while those of moulds are hard and large and often show different colourations. In addition, they look quite different under the microscope; yeast are generally round or pear-shaped while moulds show a mycelial network of filamentous hyphae . Some fungi are dimorphic producing hyphae under one set of circumstances and yeast-like cells under another. The human pathogen, Candida albicans , is the best example of dimorphism and grows like a yeast in body fluids but develops hyphae to invade tissue.
Both yeast and moulds are classified as fungi and are divided into 3 major groups, Ascomycetes , Zygomycetes and Deuteromycetes . Classification of fungi is complex and only a few important features are described here. These include determining whether cells in the mycelium are septate (showing the presence of a cross-wall) or non-septate (absence of a cross-wall), the types and ways spores are produced and whether reproduction is sexual or asexual. Ascomycetes and Zygomycetes are septate, while Deuteromycetes are non-septate. The spores produced by Ascomycetes are formed in a sac called the ascus (for this reason these spores are called ascospores) and are involved in sexual reproduction. The spores produced by Deuteromycetes and by Zygomycetes are called conidia (see below) and sporangiophores, respectively and are not involved in sexual reproduction. Yeast generally multiply by budding in which a protuberance is formed on the wall of the cell which eventually breaks off to form a new cell in which further budding occurs. Sometimes, several buds are produced by the same cell and remain attached to it. Some yeast (Schizosaccharomyces spp.) multiply by binary fission. Sexual reproduction is given the generic name, teleomorph, while asexual reproduction is called the anamorph. The same fungus has often been given different names depending on the type of reproduction and some examples of this are shown in Table 11.3. Taxonomically, the teleomorphic name is normally used but there are exceptions, e.g. the anamorphic name, Geotrichum candidum, is more commonly used than the teleomorphic one, Galactomyces candidum.
The species of yeast found on the surface of different cheeses show considerable diversity (Table 11.3). The dominant species in all cheeses, except Romadour from one plant, is D. hansenii. Kluvyeromyces spp. are also dominant in the French (Roquefort, Camembert and St. Nectaire) cheeses and in the Spanish (Cabrales) cheese but appears to be absent from the German and Austrian cheeses (Weinkase, Limburg, Romadour and Tilsit). S. cerevisiae and Pichia spp. are also important in Camembert and Cabrales cheese. All these yeasts are members of the Ascomycetes group. S. cerevisiae is also involved in wine, beer and bread-making. Very few yeast are capable of fermenting lactose but K. lactis is an exception. This may be one reason for its dominance in some surface-ripened cheese. Whether variation occurs within the same cheese has not been studied to any great extent. The evidence in Table 11.3 suggests that it does occur, at least in Limburg; both cheeses examined contained D. hansenii and G. candidum in significant numbers but, in addition, T. delbrueckii was found in one cheese and Y. lipolytica in the other.
The most important moulds in cheese are P. camemberti, P. roqueforti and G. candidum and all are members of the Deuteromyces group. P. camemberti is responsible for the white growth on the surface of Camembert and Brie while P. roqueforti is responsible for the blue veins found in Roquefort and other blue cheeses. It is generally thought that G. candidum is present on the surface of most mould and bacterial-ripened cheese. The results in Table 11.3 would suggest that it is found only in Weinkase, Romadour, Limburg and Tilsit. Scanning electron micrographs of Camembert cheese show the presence of G. candidum and it is likely that the reason it was not reported to be present in the other cheeses in Table 11.3 is that the various workers involved considered it to be a mould rather than a yeast.
Microscopic observation is very important in classifying fungi because their various structures can be seen clearly. Both P. camemberti and P. roqueforti reproduce asexually from conidia (spores) which are extruded from a flask-shaped cell called a phialide which is borne on the conidiophore or spore-bearing hyphae (Fig. 11.23). The multiplication of G. candidum is quite different. The hyphae grow to a considerable extent, then stop and septa are formed transversely, separating the hyphae into short compartments which eventually fragment into separate conidia, which start the reproductive process again. G. candidum has characteristics of both yeast and moulds and, in the past, was often called a yeast-like fungus. When it was first isolated, it was called Oidium lactis , which was later changed to Oospora lactis, and later still to its current name; it is commonly known as the dairy mould. Its natural habitat is soil where it is involved in the decay of organic matter (Boutrou and Gueguen 2005).
Major features of Penicillum roqueforti, Penicillum camemberti and Geotrichum candidum (From Samson et al. 1995)
Many moulds produce toxins, which are carcinogenic, e.g., the aflatoxins produced by Aspergillus flavus . However, the strains involved in cheese do not produce toxins. Physiological conditions for the production of toxins by microorganisms are generally much narrower than those for growth.
Most fungi grow quite well at the pH of cheese and most of those found in cheese are also quite tolerant to salt, e.g., the growth of P. camemberti is largely unaffected by 10 % NaCl (Table 11.1) and some strains of P. roqueforti can tolerate 20 % NaCl. G. candidum is an exception and is quite sensitive to salt. A slight reduction in growth occurs in the presence of 1 % NaCl and it is completely inhibited by ~6 % NaCl. Therefore, too much brining will prevent its growth on the cheese surface. Perhaps its intolerance to salt explains why it is sometimes deliberately added in the manufacture of some surface-ripened cheeses.
Generally, yeast are facultative anaerobes while moulds are considered to be obligate aerobes. However, P. roqueforti is an exception and can grow in the presence of limited levels of O2, which is exemplified by its growth throughout the mass of blue cheese. Yeast and moulds are generally heat sensitive and are killed by pasteurisation.
Occasionally yeast have been incriminated in the spoilage of cheese, either through the production of gas (CO2) or the development of off-flavours. Unripened cheeses, e.g., Cottage, Quarg, etc., especially if they contain sucrose, as a sweetener, are particularly prone to spoilage by yeast.
Suggested Reading
Beresford TP, Fitzsimons NA, Brennan NL et al (2001) Recent advances in cheese microbiology. Int Dairy J 11:259–274
Beresford T, Williams A (2004) The microbiology of cheese ripening. In: Fox PF, McSweeney PLH, Cogan TM et al (eds) Cheese: Chemistry, Physics and Microbiology, vol 1, 3rd edn. Elsevier, Amsterdam, pp 287–317
Boutrou R, Gueguen M (2005) Interests in Geotrichum candidum for cheese technology. Int J Food Microbiol 102:1–20
Cantor MD, van den Temple T, Hansen TK et al (2004) Blue cheeses. In: Fox PF, McSweeney PLH, Cogan TM et al (eds) Cheeses: Chemistry, Physics and Microbiology, vol 2, 3rd edn. Elsevier, Amsterdam, pp 175–198
Cogan TM, Goerges S, Gelsomino R et al (2014) Biodiversity of the surface microbial consortia from Limburger, Reblochon, Livarot, Tilsit and Gubbeen cheeses. Microbiol. Spectrum 2(1): CM-00010-2012. doi:10.1128/microbiolspec
Crow VL, Curry B, Hayes M (2001) The ecology of non-starter lactic acid bacteria (NSLAB) and their use as adjuncts in New Zealand Cheddar. Int Dairy J 11:275–283
Erkus O, de Jager VCL, Spus M et al (2013) Multifactorial diversity sutains microbial community stability. ISME J 7:2126–2136
Franz CMAP, Stiles M, Schleifer KH et al (2003) Enterococci in foods – a conumdrum for food safety. Int J Food Microbiol 88:105–122
Froehlich-Wyder MT, Bachmann HP (2004) Cheeses with a propionic fermentation. In: Fox PF, McSweeney PLH, Cogan TM, Guinee TP (eds) Cheeses: Chemistry, Physics and Microbiology, vol 2, 3rd edn. Elsevier, Amsterdam, pp 141–156
Ganesen B, Stuart MR, Weimer BC (2007) Carbohydrate starvation causes a metabolically active but nonculturable state in Lactococcus lactis. Appl Environ Microbiol 73:2498–2512
Jeanson S, Chadoeuf J, Madec MN et al (2011) Spatial distribution of bacterial colonies in a model cheese. Appl Environ Microbiol 77:1493–1500
Smid EJ, Erkus O, Spus M et al (2014) Functional implications of the microbial community structure of undefined mesophilic starter cultures. Microb Cell Fact 13(Suppl 1):S2
Thierry A, Deutsch SM, Falentin H et al (2011) New insights into physiology and metabolism of Propionibacterium freudenreichii. Int J Food Microbiol 149:19–27
van den Berg G, Meijer WC, Duesterhoeft EM et al (2004) Gouda and related cheeses. In: Fox PF, McSweeney PLH, Cogan TM, Guinee TP (eds) Cheeses: Chemistry, Physics and Microbiology, vol 2, 3rd edn. Elsevier, Amsterdam, pp 103–140
References
Accolas JP, Veaux M, Vassal L et al (1978) Évolution de la flore lactique thermophile au courss du pressage des fromage à pate cuite. Lait 58:118–132
Agarwal S, Sharma K, Swanson BG et al (2006) Nonstarter lactic acid bacteria biofilms and calcium lactate crystals in Cheddar cheese. J Dairy Sci 89:1452–1466
Anon (2008) The type strain of Lactobacillus casei is ATCC 393, ATCC 334 cannot serve as the type because it represents a different taxon, the name Lactobacillus paracasei and its subspecies names are not rejected and the revival of the name ‘Lactobacillus zeae’ contravenes Rules 51b (1) and (2) of the International Code of Nomenclature of Bacteria. Opinion 82. Int J Syst Evol Microbiol 58: 764–1765
Baroiller C, Schmidt J (1980) Contribution à l’étude de l’origine des levures du fromage Camembert. Lait 70:67–84
Beauvier E, Berthaud K, Cegarra S et al (1997) Ripening and quality of Swiss-type cheese made from, pasteurised or microfiltered milk. Int Dairy J 7:311–323
Besancon X, Smet E, Chabalier C et al (1992) Study of surface yeast flora of Roquefort cheese. Int J Food Microbiol 17:9–18
Bockelmann W, Willems KP, Neve H et al (2005) Cultures for the ripening of smear cheeses. Int Dairy J 15:719–732
Bokulich NA, Mills DA (2012) Next generation approaches to the microal ecology of foodstuffs. BMK Rep 45:377–389
Bokulich NA, Mills DA (2013) Facility-specific “house” microbiome drives microbailmlandscapes of artisanal cheesemaking plants. Appl Environ Microbiol 79:5214–5223
Bora N, Vancanneyt M, Gelsomino R et al (2007) Agrococcus casei, sp. nov., isolated from the surfaces of smear-ripened cheeses. Int J Syst Evol Microbiol 57:92–97
Bora N, Vancanneyt M, Gelsomino R et al (2008) Mycetocola reblochonii sp. nov., isolated from the surface microbial flora of Reblochon cheese. Int J Syst Evol Microbiol 58:2687–2693
Bouton Y, Buchin S, Duboz G et al (2009) Effect of mesophilic lactobacilli and enterococci adjunct cultures on the final characteristics of a microfiltered milk Swiss-type cheese. Food Microbiol 26:183–191
Brennan NM, Brown R, Goodfellow M et al (2001a) Corynebacterium mooreparkense sp. nov., and Corynebacterium casei sp. nov. isolated from the surface of a smear-ripened cheese. Int J Syst Evol Microbiol 51:843–852
Brennan NM, Brown R, Goodfellow M et al (2001b) Microbacterium gubbeenense sp. nov., isolated from the surface of a smear-ripened cheese. Int J Syst Evol Microbiol 51:1969–1976
Broadbent JR, Houck K, Johnson E et al (2003) Influence of adjunct use and cheese microenvironment on non starter bacteria in reduced fat Cheddar type cheese. J Dairy Sci 86:2773–2782
Budinich MF, Perez-Diaz I, Cai H et al (2011) Growth of Lactobacillus paracasei ATCC 334 in a cheese model system: a biochemical approach. J Dairy Sci 94:5263–5277
Collins MD, Phillips B, Zanoni P (1989) Deoxyribonucleic acid homology studies of Lactobacillus casei, Lactobacillus paracasei sp.nov., subsp. paracasei and subsp. tolerans, and Lactobacillus rhamnosus sp. nov., comb.nov. Int J Syst Bacteriol 39:105–108
Coolbear T, Crow V, Harnett J et al (2008) Developments in cheese microbiology in New Zealand – use of starter and non-starter lactic acid and their enzymes in determining flavour. Int Dairy J 18:705–712
Coppola R, Nanni M, Jorizzo M et al (2000) Microbiological characteristics of Parmigiano Reggiano cheese during cheesmaking and the first months of ripening. Lait 80:479–490
Coton E, Desmonts MH, Leroy S et al (2010) Biodiversity of coagulase negative staphylococci in French cheesese, dry fermented sausags, processing environments and clinical samples. Int J Food Microbiol 137:221–229
Crow VL (1986) Metabolism of aspartate by Propionibacterium freudenreichii: effect on lactate fermentation. Appl Environ Microbiol 52:359–365
Cuesta P, Fernández-García E, González de Llano D et al (1996) Evaluation of the microbiological and biochemical characteristics of Afuga’l Pitu cheese during ripening. J Dairy Sci 79:1693–1698
Delbes C, Ali-Mandjee L, Montel MC (2007) Monitoring bacterial communities in raw milk and cheese by culture dependent and independent 16S RNA genus-based analyses. Appl Environ Microbiol 73:1882–1891
Del Pozo BF, Gaya P, Media M et al (1985) Changes in the microflora of La Serena ewe’s milk cheese during ripening. J Dairy Res 55:449–455
Dellaglio F, Felis GE, Torriani S (2002) The status of the species Lactobacillus casei (Orla Jensen 1916) Hansen and Lessel 1971 and Lactobacillus paracasei Collins et al. 1989. Request for an opinion. Int J Syst Evol Microbiol 52:285–287
Demarigny Y, Beuvier E, Dasen A et al (1996) Influence of raw milk microflora on the characteristics of Swiss-type cheeses. 1. Evolution of microflora during ripening and characterization of facultatively heterofermentative lactobacilli. Lait 76:371–387
Devoyod JJ, Sponem D (1970) La flore microbiennee du fromage de Roquefort 6. Les levures. Lait 50:524–543
Diaz-Muniz I, Steele JL (2006) Conditions required for citrate utilization during growth of Lactobacillus casei ATCC 334 in chemically defined medium and Cheddar cheese extract. Antonie Van Leeuwenhoek 90:233–243
Didienne R, Defargues C, Callon C et al (2012) Characteristics of microbial biofilm on wooden vats (‘gerlees’) in PDO Salers cheese. Int J Food Microbiol 156:91–101
Eliskases-Lechner F, Ginzinger W (1995a) The bacterial flora of surface-ripened cheeses with special regard to coryneforms. Lait 75:571–584
Eliskases-Lechner F, Ginzinger W (1995b) The yeast flora of surface-ripened cheese. Milchwissenschaft 50:458–462
Ercolini D (2013) High-throughput sequencing and metagenomics: moving forward in the cultureindependent analysis of food microbial ecology. Appl Environ Microbiol 79:3148–3155
Falentin H, Henaff N, LeBic P et al (2012) Reverse transcription quantitative PCR revealed persistency of thermophilic lactic acid bacteria metabolic activity until the end of ripening of Emmental cheese. Food Microbiol 29:132–160
Ferreira AD, Viljoen BC (2003) Yeasts as adjunct starters in matured Cheddar cheese. Int J Food Microbiol 86:131–140
Fisher K, Phillips C (2009) The ecology, epidemiology and virulence of Enterococcus. Microbiology 155:1749–1757
Fitzsimons NA, Cogan TM, Condon S et al (2001) Spatial and temporal distribution of non-starter lactic acid bacteria in Cheddar cheese. J Appl Microbiol 90:600–608
Floury J, Jeanson S, Madek MN et al (2013) Porosity of Lactococcus lactis subsp. lactis LD61 colonies immobilised in model cheese. Int J Food Microbiol 163:64–70
Fontana C, Cappa F, Rebecchi A et al (2010) Surface microbiota analysis of Taleggio, Gorgonzola, Casera, Scimudin and Formaggio di Fossa Italian cheeses. Int J Food Microbiol 138:205–211
Foulquie Moreno MR, Sarantinopoulos P, Tsakalidou E et al (2006) The role and application of enterococci in food and health. Int J Food Microbiol 106:1–24
Ganesen B, Dobrowski P, Weimer BC (2006) Identification of the leucine to 2 methylbutyric acid pathway of Lactococcus lactis. Appl Environ Microbiol 72:4264–4272
Gavrish EY, Krauzova VI, Potekhina NV et al (2004) Three new species of brevibacteria, Brevibacterium antiquum sp. nov., Brevibacterium aurantiacum sp. nov., and Brevibacterium permense sp. nov. Microbiology 73:176–183
Hardy J (1986) Water activity and cheese salting. In: Eck A, Gillis JC (eds) Cheesemaking, from Science to Quality Assurance, 2nd edn. Editions Tec and Doc, Paris, pp 60–81
Hocking AD, Faedo M (1992) Fungi causing thread mould spoilage of vacuum packaged Cheddar cheese during maturation. Int J Food Microbiol 16:123–130
Irlinger F, Bimet F, Delettre J et al (2005) Arthrobacter bergerei sp. nov. and Arthrobacter arilaitensis sp. nov., novel coryneform species isolated from the surfaces of cheeses. Int J Syst Evol Microbiol 55:457–462
Jordan KN, Cogan TM (1993) Identification and growth of non-starter lactic acid bacteria in Irish Cheddar cheese. Irish J Agric Food Res 32:47–55
Leclercq-Perlat MN, Buono F, Lambert D et al (2004) Controlled production of Camembert-type cheeses. Part 1: microbiological and physiochemical evolutions. J Dairy Res 71:346–354
Lazzi C, Turroni S, Mancini A et al (2014) Ranscriptomic clues to understand the growth of Lactobacillus rhamnosus in cheese. BMC Microbiol 14:28–42
Lund F, Filtenberg O, Frisvad JC (1995) Associated mycoflora of cheese. Food Microbiol 12:173–180
Mariani C, Briandet R, Chamba JF et al (2007) Biofilm ecology of wooden shelves used in ripening the French raw milk smear cheese Reblochon de Savoie. J Dairy Sci 90:1653–1661
Martinez-Cuesta MC, Benggoechea J, Bustos I et al (2010) Control of late blowing in cheese by adding lacticin 3147-producing Lactococcus lactis IFLP 3593 to the starter. Int Dairy J 20:18–24
Maoz A, Mayr R, Scherer S (2003) Temporal and stability and biodiversity of two complex antilisterial cheese ripening microbial consortia. Appl Environ Microbiol 69:4012–4018
Masoud W, Vogensen FK, Lillevang S et al (2012) The fate of indigenous microbiota, starter culture, Escherichia coli, Listeria innocua, and Staphylococcus aureus in Danish raw milk and cheeses determined by pyrosequencing and quantitative real time (qRT)-PCR. Int J Food Microbiol 153:192–202
Martley FG, Crow VL (1996) Open texture in cheese: the contribution of gas production by microorganisms and cheese manufacturing practices. J Dairy Res 63:489–507
Martley FG, Lawrence RC (1972) Cheddar cheese flavour. 2. Characteristics of single strain starters associated with good or poor flavour development. N Z J Dairy Sci Technol 7:38–44
Monnet C, Loux V, Gilbrat JF et al (2010) The Arthrobacter arilaitensis Re117 genome sequence reveals its genetic adaptation to the surface of cheese. PLoS One 5(11), e15489
Monnet C, Back A, Irlinger F (2012a) Growth of aerobic ripening bacteria at the cheese surface is limited by the availability of iron. Appl Environ Microbiol 78:2185–3192
Monnet C, Loux V, Bento P et al (2012b) Genome sequence of Corynebacterium casei UCMA 3821, isolated from a Smear-Ripened Cheese. J Bacteriol 194:728–729
Mounier J, Gelsomino R, Goerges S et al (2005) Surface flora of four smear ripened cheeses. Appl Environ Microbiol 71:6489–6500
Mounier J, Goerges S, Gelsomino R et al (2006) Sources of the adventitious microflora of a smear-ripened cheese. J Appl Microbiol 101:668–681
Nunez M (1978) Microflora of Cabrales cheese: changes during maturation. J Dairy Res 45:501–508
Nunez M, Medina M, Gaya P et al (1981) Les levures et les moissures dans le fromage bleu de Cabrales. Lait 61:62–79
Ogier JC, Lafarge V, Girard V et al (2004) Molecular fingerprinting of dairy microbial ecosystems by use of temporal temperature and denaturing gradient gel electrophoresis. Appl Environ Microbiol 70:5628–5643
O’Sullivan L, Ross RP, Hill C (2003) A lactocin 481-producing adjunct culture increases starter lysis while inhibiting non-starter lactic acid bacteria proliferation during Cheddar cheese ripening. J Appl Microbiol 95:1235–1241
Piveteau P, Condon S, Cogan TM (1995) Interactions between lactic and propionic acid bacteria. Lait 75:331–343
Place RB, Hiestand D, Burri S et al (2003a) Staphylococcus succinus subsp. casei subsp. nov., a dominant isolate from surface-ripened semi-hard cheeses. Syst Appl Microbiol 25:353–359
Place RB, Hiestand D, Gallman HR et al (2003b) Staphylococcus equorum subsp. linens, subsp. nov., a starter culture component for surface-ripened semi-hard cheeses. Syst Appl Microbiol 26:30–37
Poullet B, Huertas M, Sanchez A et al (1991) Microbial study of Casar de Cáceres cheese throughout ripening. J Dairy Res 58:231–238
Quigley L, O’Sullivan O, Beresford TP et al (2011) Molecular approaches to analysing the microbial composition of raw milk and raw milk cheese. Int J Food Microbiol 150:81–94
Quigley L, O’Sullivan O, Beresford TP et al (2012) High throughput sequencing for detection of subpopulations of bacteria not previously associated with cheese. Appl Environ Microbiol 78:5717–5724
Randozzo CL, Caggia C, Neviani E (2009) Application of molecular approaches to study lactic acid bacteria in artisanal cheeses. J Microbiol Methods 78:1–9
Rea MC, Franz CMAP, Holzapfel WH et al (2004) Development of enterococci and production of tyramine during the manufacture and ripening of Cheddar cheeses. Irish J Agric Food Res 43:247–258
Ruegg M, Blanc B (1981) Influence of water activity on the manufacture and aging of cheese. In: Rockland LB, Stewart GF (eds) Water Activity: Influences on Food Quality. Academic, New York, pp 791–811
Ruusunen M, Surakka A, Korkeala H et al (2012) Clostridium tyrobutyricum strains show wide variation in growth at different NaCl, pH and temperature conditions. J Food Prot 75:1791–1795
Ryan MP, Rea MC, Ross RP et al (1996) An application in Cheddar cheese manufacture for a strain of Lactococcus lactis producing a novel broad-spectrum bacteriocin, lacticin 3147. Appl Environ Microbiol 62:612–619
Ryan MP, Ross RP, Hill C (2001) Strategy for manipulation of cheese flora using combinations of Lacticin3147-producing and -resistant cultures. Appl Environ Microbiol 67:2699–2704
Samson RA, Hoekstra ES, Frisvad JC et al (1995) Introduction to Food-borne Fungi. Centraalbureau voor Schmmelcultures, Delft
Schröder J, Maus I, Trost E et al (2011) Complete genome sequence of Corynebacterium variabile DSM 44702 isolated from the surface of smear-ripened cheeses and insights into cheese ripening and flavour generation. BMC Genomics 12:545–574
Schubert K, Ludwig W, Springer N et al (1996) Two coryneform bacteria isolated from the surface of French Gruyère or Beaufort cheeses are new species of the genus Brachybacterium: Brachybacterium alimentarium sp. nov and Brachybacterium tyrofermentair sp. nov. Int J Syst Microbiol 46:81–87
Sensidini A, Rondinini G, Peressini D et al (1994) Presence of an offflavour associated with the use of sorbates in cheese and margarine. Italian J Food Sci 6:237–242
Somers EB, Johnson ME, Wong ACL (2001) Biofilm formation and contamination of cheese by nonstarter lactic acid bacteria in the dairy environment. J Dairy Sci 84:1926–1936
Stackebrandt E, Koch C, Gvozdiak O et al (1995) Taxonomic dissection of the genus Micrococcus: Kocuria gen. nov., Nesterenkonia gen nov., Kytococcus gen nov., Dermacoccus gen nov and Micrococcus Cohn 1872 gen. Emend. Int J Syst Microbiol 45:682–692
Stadhouders J, Langeveld LPM (1966) The microflora of the surface of cheese. Factors affecting its composition. In: 17th International Dairy Congress. Delhi, vol D, pp 577–585
Steele JL, Budinich MF, Cai H et al (2006) Diversity and metabolic activity of Lactobacillus casei in ripening Cheddar cheese. Aust J Dairy Technol 61:53–60
Smokvina T, Wels M, Polka J et al (2013) Lactobacillus paracasei comparative genomics: towards species pan-genome definition and exploitation of diversity. PLoS One 8, e68731
Turner KW, Thomas TD (1980) Lactose fermentation in Cheddar cheese and the effect of salt. N Z J Dairy Sci Technol 15:265–276
Turner KW, Morris HA, Martley FG (1983) Swiss type cheese. 2. The role of thermophilic lactobacilli in sugar fermentation. N Z J Dairy Sci Technol 18:117–124
Valdes-Stauber N, Scherer S, Seiler H (1997) Identification of yeasts and coryneform bacteria from the surface microflora of brick cheeses. Int J Food Microbiol 34:115–129
Vasiljevic T, Shah NP (2008) Probiotics – from Metchnikoff to bioactives. Int Dairy J 18:714–728
Vergeade J, Guiraud J, Larpent JP et al (1976) Etude de la flore de levure du Saint-Nectaire. Lait 56:275–285
Walter F, Albersmeierb W, Kalinowski J et al (2014) Complete genome sequence of Corynebacterium casei LMG S-19264T (=DSM 44701T), isolated from a smear-ripened cheese. J Biotechnol 189:76–77
Wolfe BE, Sutton JE, Santerelli M et al (2014) Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell 158:422–433
Author information
Authors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer New York
About this chapter
Cite this chapter
Fox, P.F., Guinee, T.P., Cogan, T.M., McSweeney, P.L.H. (2017). Microbiology of Cheese Ripening. In: Fundamentals of Cheese Science. Springer, Boston, MA. https://doi.org/10.1007/978-1-4899-7681-9_11
Download citation
DOI: https://doi.org/10.1007/978-1-4899-7681-9_11
Published:
Publisher Name: Springer, Boston, MA
Print ISBN: 978-1-4899-7679-6
Online ISBN: 978-1-4899-7681-9
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)