Abstract
Activation of NF-κB is regulated via numerous ubiquitin- and proteasome-mediated steps – an important one is processing of the precursor p105 to the p50 active subunit. The mechanisms involved are largely unknown, as this is an exceptional case where the ubiquitin system does not destroy its substrate completely. Here we demonstrate that proteasomal processing of p105 requires ubiquitin, but not generation of polyubiquitin chains. In vitro, ubiquitin species that cannot polymerize mediate processing. In yeasts that express non-polymerizable ubiquitins, processing proceeds normally, whereas degradation of substrates that are dependent on polyubiquitination is inhibited. Similar results were obtained in mammalian cells. Interestingly, processing requires multiple monoubiquitinations, as progressive elimination of lysines in p105 is accompanied by gradual inhibition of p50 generation. Last, the proteasome recognizes the multiply monoubiquitinated p105. These findings suggest that a proteolytic signal can be comprised of a cluster of single ubiquitins, and not necessarily of a chain.
This chapter is based on the authors’ work: Kravtsova-Ivantsiv, Y., Cohen, S., Ciechanover, A. (2009) Modification by single ubiquitin moieties rather than polyubiquitination is sufficient for proteasomal processing of the p105 NF-κB precursor. Mol. Cell 33, 496–504, which also features description of the methods and the list of references.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
Introduction
NF-κB proteins are transcription factors playing an important role in regulation of the inflammatory response and apoptosis. Some authors link them to the pathogenesis of cancer (Naugler and Karin, 2008). The NF-κB transcriptional complexes are dimeric combinations of subunits p50, p52, and/or members of the Rel family of proteins. p50 and p52 are produced from two precursors, NF-κB1 (p105) and NF-κB2 (p100), by limited proteolytic processing at the proteasome, and represent the N-terminal domains of their precursors, whereas the C-terminal segments are degraded.
Both signal-induced and constitutive types of processing are observed. Following stimulation, p105 undergoes phosphorylation on specific serine residues by IκB kinase (IKK) with subsequent recruitment of the SCFβ-TrCP ubiquitin ligase. This results in both complete degradation and enhanced processing of the protein (Cohen et al., 2004; MacKichan et al., 1996; Orian et al., 2000). The mechanisms and enzymes involved in a constitutive processing are not studied well. A glycine-alanine rich repeat (GRR) is essential for processing (Lin and Ghosh, 1996) and probably serves as a “stop signal” for the 26S proteasome (Orian et al., 1999). Lin and colleagues (Lin et al., 1998, 2000) suggested that p50 is not a product of p105 processing, but rather is generated co-translationally by the proteasome via a mechanism involving ribosome halting. Moorthy and colleagues (Moorthy et al., 2006) claim that the processing is mediated by the 20S rather than the 26S proteasome, and the endoproteolytic cleavage is not dependent on prior ubiquitination of the precursor. Other studies have shown that ubiquitination of p105 is required for its proteasomal processing (Coux and Goldberg, 1998; Orian et al., 1995; Palombella et al., 1994; Sears et al., 1998).
Ubiquitin-mediated proteolysis is a major pathway involved in regulating numerous biological processes (Mayer, 2005, 2006a, b, 2008). The proteins destined for degradation are signaled for recognition by the 26S proteasome by generation of a polyubiquitin chain. This chain is anchored in most cases to an internal lysine residue in the substrate via an isopeptide bond between the C-terminal glycine residue of the first ubiquitin moiety and an ɛ-NH2 group of an internal lysine in the target substrate. Within the chain, each ubiquitin moiety is bound to the previously conjugated molecule via a similar bond with the ɛ-NH2 group of internal lysine 48 (Chau et al., 1989). Recent studies of protein degradation revealed the chains based on lysine 63 (Hofmann and Pickart, 2001; Kim et al., 2007), chains lacking lysine 48-based linkages, and heterogeneous ones based on lysines 11, 48, and 63 (Kirkpatrick et al., 2006). It is also accepted that the shortest signal that is recognized by the proteasome is a tetraubiquitin chain (Thrower et al., 2000). Monoubiquitination has been shown to control non-proteolytic processes such as epigenetic control mediated via histone modification (Robzyk et al., 2000), receptor routing (Terrell et al., 1998), and viral budding (Patnaik et al., 2000). As for degradation, the methylated ubiquitin (MeUb), in which all the amino groups in the internal lysines were modified – resulting in its inability to polymerize – could stimulate in vitro degradation of lysozyme, although at a lower efficiency compared with WT ubiquitin (Hershko and Heller, 1985). There exists a single and specific monoubiquitination of Pax3 – a key regulator of myogenesis – which targets it for proteasomal degradation (Boutet et al., 2007). Interestingly, the degradation of certain proteins by the proteasome appears to be ubiquitin independent (reviewed recently in Jariel-Encontre et al. (2008)), though the only firmly established case is ornithine decarboxylase (ODC) (Murakami et al., 1992). Experimental evidence strongly suggests that the 19S regulatory particle of the proteasome recognizes the ubiquitinated substrates. Subunit Rpn10/S5a/Mcb1 was shown to bind ubiquitin chains through ubiquitin-interacting motifs (UIMs) with a preference to polymeric ubiquitin (Deveraux et al., 1994). Since deletion of this subunit had only a mild effect on protein degradation in yeast (van Nocker et al., 1996), it is clear that additional ubiquitin-binding subunit(s) exist. Indeed, it was shown that the 19S ATPase subunit S6’/Rpt5 binds polyubiquitin chains in an interaction that is modulated by ATP hydrolysis (Lam et al., 2002); however the biological significance of this association has not been further unraveled. It was reported recently that Rpn13/ARM1 functions as a novel ubiquitin receptor (Husnjak et al., 2008). Rpn13 carries a conserved amino-terminal domain termed the pleckstrin-like receptor for ubiquitin (Pru). This motif binds monoubiquitin and K48-linked diubiquitin with high affinity compared to other ubiquitin receptors (Husnjak et al., 2008; Schreiner et al., 2008). Present chapter demonstrates that proteasomal processing of p105 can proceed following multiple monoubiquitinations, suggesting that a cluster of single moieties and not necessarily a polyubiquitin chain can serve as a proteasomal recognition proteolytic signal.
Results
Involvement of the Ubiquitin System in Processing of p105
As described in the Introduction, certain basic aspects related to the mechanism of p105 processing are still controversial. These controversies prompted us to re-examine the role of the ubiquitin–proteasome system (UPS) in the process. Processing of 35S-labeled p105 in crude reticulocyte fraction II (that lacks ubiquitin) is strongly dependent on the addition of ubiquitin (Fig. 10.1a). It requires the presence of E1 and E2 (Fig. 10.1b, compare lane 8 to lane 4) and probably also of an E3 present in fraction II (Orian et al., 1995). The reaction is ATP dependent (Fan and Maniatis, 1991; Orian et al., 1995), suggesting again that the 26S proteasome catalyzes p105 processing. The involvement of the 20S proteasome in the process was confirmed by the finding that MG132, clasto-lactacystin β-lactone, and epoxomicin inhibited processing significantly (Fig. 10.1b, lane 5 and Fig. 10.1c).
Ubiquitin system-mediated processing of WT and S927A p105 s to p50 in a reconstituted cell-free system. (a) Processing of p105 requires ubiquitin. In vitro-translated and 35S-labeled p105 was processed in the presence or absence of ubiquitin as indicated. Processing of WT (b, c) or S927A (c, d) p105 s is ATP-, proteasome-, E1-, E2-, and fraction II-dependent. 35S-labeled p105 s were processed in the presence or absence of ATP and ATP-regenerating system, MG132, clasto-lactacystin β-lactone, epoxomicin, DMSO, and E1 and E2, as indicated. Ub denotes ubiquitin and FrII denotes fraction II; % of processing is the ratio of radioactivity of p50/radioactivity of p50 + p105 multiplied by 100. All values relate to 100% which is the set point for processing of p105 in a complete system
The experiment shown in Fig. 10.1b was carried out using WT p105. The same results (Fig. 10.1d) were obtained in an identical experiment in which we used p105S927A, a mutant that cannot be specifically phosphorylated at this site by IKK, and therefore cannot recruit the βTrCP ubiquitin ligase and undergo signal-induced processing and/or degradation (Heissmeyer et al., 2001; Lang et al., 2003; Salmeron et al., 2001). It should be noted that p105S927A can be phosphorylated on other Thr/Ser/Tyr residues, yet these modifications are probably irrelevant to βTrCP recruitment (Heissmeyer et al., 2001; Lang et al., 2003).
These experiments clearly demonstrate that processing of p105 in a cell-free system is dependent on ubiquitination and cleavage by the 26S proteasome.
Processing of p105 Does Not Require Polyubiquitination
Once it was established that processing requires ubiquitination, it was important to determine whether it also requires generation of a polyubiquitin chain(s). As can be seen in Fig. 10.2a, processing proceeds even in the presence of ubiquitin species that cannot polymerize – UbK48R (lane 4), MeUb (lane 5), and UbK0 (lane 6). We hypothesized that p105 is subjected to processing mediated via two independent mechanisms: (i) signal-mediated (which can occur also in the crude extract) that leads to phosphorylation and cleavage of p105 that requires polyubiquitination and is mediated by the βTrCP ubiquitin ligase and (ii) basal/signal-independent processing that requires monoubiquitination and is mediated by an as yet to be identified ligase. WT ubiquitin can promote both processes, and therefore the cleavage in its presence is more efficient than in the presence of the non-polymerizable ubiquitin species. Supporting this hypothesis is our finding that p105 is partially phosphorylated in crude extract (Orian et al., 2000), and that this modification can result in increased processing (Cohen et al., 2004). To demonstrate that this is indeed the case and p105 can undergo two types of processing that are apparently dependent on two different modes of ubiquitination, we utilized p105S927A (Heissmeyer et al., 2001; Lang et al., 2003; Salmeron et al., 2001). Indeed, efficiency of processing of this p105 mutant is similar using either WT or non-polymerizable ubiquitin species (Fig. 10.2b, lanes 5 and 6, and Fig. 10.2c).
Processing of p105 in a cell-free system does not require generation of polyubiquitin chains. (a–c) Processing of 35S-labeled WT (a, b) and p105S927A (b, c) was monitored in a cell-free system in the presence of WT, K48R, methylated, and K0 ubiquitins as indicated. (d) Processing of p105S927A mediated by WT ubiquitin is not affected by competition with MeUb. 35S-labeled p105S927A was processed in the presence of the indicated ubiquitin species. (e) Generation of high MW ubiquitin adducts of p105S927A is inhibited by MeUb and the inhibition is alleviated by WT ubiquitin. 35S-labeled p105S927A was ubiquitinated in a cell-free system using WT and methylated ubiquitins as indicated. (f) Degradation of Ring1BI53Sin a cell-free system requires WT Ub. In vitro-translated and 35S-labeled Ring1BI53Swas degraded in a cell-free system in the presence of WT or methylated ubiquitins as indicated. Degradation was calculated based on the radioactivity remained in the lane along time relative to time 0. MeUb denotes methylated ubiquitin and Ub conj. denotes ubiquitin conjugates. Processing (panels a–d) was calculated as described under Fig. 10.1, except that the 100% set point reflects processing in a system that contains WT ubiquitin
To further demonstrate that p105 can be processed following monoubiquitination, we monitored the generation of p50 from p105S927A in the presence of WT ubiquitin and increasing concentrations of MeUb. This ubiquitin species can be activated by E1 and conjugated to the substrate, and therefore can be used to inhibit degradation of substrates that require polyubiquitination for their recognition by the proteasome (Ben-Saadon et al., 2004; Breitschopf et al., 1998; Herman-Bachinsky et al., 2007; Hershko and Heller, 1985). As can be seen in Fig. 10.2d, MeUb did not have any inhibitory effect on processing of p105. To demonstrate that the modified ubiquitin was indeed active, we showed that it inhibits generation of high molecular mass ubiquitin adducts of p105 (Fig. 10.2e, compare lane 4 to lane 3), and this inhibition can be alleviated by competition with WT ubiquitin (Fig. 10.2e, compare lane 5 to lane 4).
As a control, we tested the requirement for WT ubiquitin for in vitro degradation of Ring1B (Ben-Saadon et al., 2006). Since Ring1B is an ubiquitin ligase that catalyzes also self-ubiquitination, we used a RING finger mutant of this enzyme (Ring1BI53S) in which this activity is abrogated. As is shown in Fig. 10.2f, WT ubiquitin catalyzes initially formation of high MW conjugates (lane 5) that disappear along time (lane 6). In striking contrast, the methylated species of ubiquitin leads to accumulation of non-degradable, lower MW adducts (lanes 8 and 9), representing multiply monoubiquitinated species of Ring1B that cannot be digested by the proteasome.
Monoubiquitination(s) Is Sufficient for Processing of p105 In Vivo
To test the role of a certain species of ubiquitin in any process, a most useful tool appears to be a yeast strain in which all the endogenous ubiquitin genes were deleted (Spence et al., 1995), and only the studied ubiquitin species is expressed. The yeast cells are kept alive by expression of WT ubiquitin under a galactose-regulated promoter. This promoter can be turned off by the addition of glucose with subsequent rapid depletion of cellular ubiquitin (Hanna et al., 2003; Hanna et al., 2007). Following depletion of endogenous cellular ubiquitin, any species of ubiquitin (such as WT, K48R, or K0 ubiquitins) can be expressed under the CUP1 promoter, for example, along with a proteolytic target substrate. Since it was shown that p105 is faithfully processed to p50 in the yeast Saccharomyces cerevisiae (Sears et al., 1998), we chose the yeast cell as our initial experimental platform.
As can be seen in Fig. 10.3a, processing of p105S927A proceeded unaffected regardless of whether WT or K48R ubiquitins were expressed. Yet, K48R ubiquitin can still be involved in generation of other than K48-linked chains recognized by the proteasome. To ascertain that this does not occur, we expressed in yeast UbK0. As can be seen in Fig. 10.3b, processing of p105S927A proceeds in a similar rate to that observed when WT Ub is expressed, further strengthening the notion that it occurs following monoubiquitination(s). In contrast, degradation of short-lived proteins that apparently require generation of lysine 48-based polyubiquitin chains, such as Cln2 (Fig. 10.3c, d), Gcn4 (Fig. 10.3e), and MyoD (Fig. 10.3f; Sadeh et al., 2008), was significantly inhibited following expression of UbK48R or UbK0.
Monoubiquitination is sufficient for processing of p105 in yeast cells. (a) and (b) (i) Generation of p50 in Saccharomyces cerevisiae does not require synthesis of a polyubiquitin chain(s). Processing of HA-p105S927A was monitored in a yeast cell that expresses either UbK48R or UbK0 as indicated. (ii) Quantitative analysis of p105 processing. Processing was calculated as described under Fig. 10.1. For each time point, the respective value of processing at time 0 was subtracted, and the 100% set point represents processing in a system that contains WT ubiquitin that was incubated for 4 h following addition of CHX. Hollow and solid circles represent processing in the presence of WT ubiquitin and UbK48R (or UbK0), respectively. (c) and (d) Stabilization of HA-Cln2 in a yeast cell that expresses K48R (c) or K0 (d) ubiquitins. Degradation of HA-Cln2 was monitored following addition of cycloheximide (i panels). (ii) Quantitative analysis of the degradation experiments. Hollow and solid circles represent degradation in the presence of WT ubiquitin and UbK48R (or UbK0), respectively. (e) and (f) Stabilization of HA-Gcn4 (e) and MyoD (f) in a yeast cell that expresses UbK48R. Degradation of the indicated proteins was monitored following addition of cycloheximide (i panels). (ii) Quantitative analysis of the degradation experiments. Chx denotes cycloheximide and WB denotes Western Blot
Similar results were obtained in mammalian cells, where processing of p50 proceeded unaffected in the presence of K0 ubiquitin (Fig. 10.4ai). Two lines of experimental evidence demonstrate that the expressed UbK0 affected conjugate formation and degradation in cells: (i) the polyubiquitin chains conjugated to the expressed p105 are of lower MW compared to those generated in the presence of WT Ub (Fig. 10.4aii, aiii) and (ii) degradation of c-Myc, a bona fide short-lived substrate of the ubiquitin system (Gross-Mesilaty et al., 1998), was significantly inhibited following expression of UbK0 (Fig. 10.4b).
Processing of p105S927A in mammalian cells does not require generation of a polyubiquitin chain(s). (a) HEK 293 cells were transfected with cDNAs coding for Flag-p105 (lanes 1–2), and HA-WT Ub (lanes 1 and 3) or HA-UbK0 (lane 2). (i) Analysis of expression and processing of p105. 10% of the cell lysates were resolved via SDS-PAGE, blotted onto nitrocellulose membrane, and p105 and p50 were detected using anti-Flag. Processing was calculated as described under Fig. 10.2. p105 and its conjugates were immunoprecipitated from the remaining portion of the lysate (90%) using anti-Flag, and detected by either anti-Flag (ii) or anti-HA (iii). (b) Stabilization of HA-c-Myc in cells that express UbK0. HEK 293 cells were transfected with cDNAs coding for HA-c-Myc (lanes 1–9), along with an empty vector (lanes 1–3), cDNAs coding for Flag-WT Ub (lanes 4–6), or Flag-UbK0 (lanes 7–9). Degradation of HA-c-Myc was monitored following the addition of cycloheximide. Tubulin was used to assess equal protein loading. IP denotes immunoprecipitation
Taken together, these findings strongly suggest that processing of p105 in cells can be promoted by monoubiquitination(s).
Processing of p105 Requires Multiple Monoubiquitinations
In order to distinguish whether proteasomal processing of p105 requires multiple monoubiquitinations or whether a modification by a single moiety is sufficient, we tested the cell-free processing of a series of p105 mutants in which we eliminated progressively the number of lysine residues in the degradable C-terminal domain (Fig. 10.5a). The lysines in this domain were shown to be the targets of ubiquitination that leads to processing (Cohen et al., 2004; Cohen et al., 2006). The in vitro reaction was carried out using UbK48R. As can be seen in the experiment presented in Fig. 10.5b, processing efficiency was in direct correlation with the number of lysines in p105. A similar result was obtained using the yeast strain that lacks endogenous ubiquitin (Fig. 10.5c; see above).
Processing of p105 requires multiple monoubiquitinations. (a) (i) Schematic representation of WT p105. Numbers that follow “K” denote sequential lysine residues downstream to the GRR. K425 is denoted by K1. Numbers in bold marked by dashed lines denote the respective residue along the protein sequence. (ii) (a–f) p105 constructs in which the indicated lysines were mutated/deleted alone or in clusters. (b) Efficiency of processing of p105 in vitro is dependent on the number of lysine residues in the C-terminal domain. 35S-labeled p105 s (as described under a) were processed in a cell-free reconstituted system in the presence or absence of UbK48R as indicated. Quantitative analysis was carried out in two steps. First, for each reaction mixture to which UbK48R was not added, the amount of p50 generated was calculated as the ratio between the p50 signal to that of the summed signal of p50 and p105. Then, the same calculation was carried out for the corresponding reaction to which UbK48R was added. For each mutant, the net processing was calculated by subtracting the amount (%) of p50 generated in the presence from that generated in the absence of Ub. Arbitrarily, the net processing of WT p105 was denoted as 100%. In the second step we calculated the net processing for each mutant relative to the processing of WT p105. The numbers shown represent these values. (c) Efficiency of processing of p105 in vivo is dependent on the number of lysine residues in the C-terminal domain. Processing of HA-p105 and HA-p105K19-30R was monitored in a Saccharomyces cerevisiae strain that lacks endogenous UBI genes. Calculation of processing was carried out in a similar manner to that described under b, except that comparison was made to processing in time 0 rather than to processing in a reaction to which Ub was not added
The 26S Proteasome Binds Multiply Monoubiquitinated p105
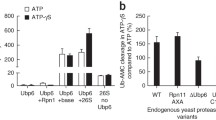
One can assume that if multiply monoubiquitinated p105 is recognized and processed by the 26S proteasome, the enzyme complex will also bind the ubiquitin-tagged substrate. To test this assumption experimentally, we carried out ubiquitination of immobilized p105 in a cell-free reconstituted system and looked for association of the ubiquitin-tagged protein with the proteasome. As can be seen in Fig. 10.6a, p105 that was ubiquitinated with either WT or MeUb (lanes 2 and 3) could bind proteasomes as is evident by the detection of the α2 and α6 subunits of the 20S complex in the precipitated complex (Fig. 10.6b, lanes 2 and 3). When the mixture did not contain ubiquitin (Fig. 10.6a, lane 1), binding did not occur either (Fig. 10.6b, lane 1).
Multiple monoubiquitinations are sufficient to mediate interaction of p105 with the proteasome. 35S-labeled p105 was immunoprecipitated (panel a, lanes 1–3) and ubiquitinated (lanes 2–3) in the presence of WT or methylated ubiquitin as indicated. Lane 4 represents a reaction mixture that contained all components except for p105 and was processed identically to the other mixtures. FrII served as the source for the 26S proteasome. Proteins were resolved via SDS-PAGE and blotted onto nitrocellulose membrane. Labeled p105 and its conjugates were detected using PhosphorImager. The membrane was incubated with antibodies to the proteasome subunits α2 and α6 that were visualized (b) using the appropriate secondary antibody. Lane 5 in (b) represents separation of 5% of the input of FrII used for the ubiquitination reaction
Discussion
Our findings further corroborate the notion that processing of p105 is mediated by the ubiquitin–proteasome system (Fig. 10.1). Surprisingly, ubiquitin species that cannot generate polyubiquitin chains could also support processing (Fig. 10.2). To demonstrate in cells the validity of the results obtained in the cell-free system, we monitored processing of p105 in a yeast strain in which all the endogenous ubiquitin genes were substituted with exogenous genes coding for either WT, K48R, or K0 ubiquitins. Efficiency of processing of p105 in the presence of UbK48R and UbK0 was not different from that monitored in the presence of WT ubiquitin (Fig. 10.3a, b). In contrast, the degradation of polyubiquitin chain-dependent substrates, such as Cln2, Gcn4, and MyoD, was severely affected (Fig. 10.3c–f). Similar findings were observed in a mammalian cell (Fig. 10.4). Using a series of p105 mutants in which we progressively substituted/eliminated all lysine residues in the C-terminal degradable domain, we found a direct relationship between the efficiency of generation of p50 and the number of ubiquitin anchors in the tail (Fig. 10.5), suggesting that a cluster rather than a single ubiquitin moiety is recognized by the proteasome. Last, we have shown that the multiply monoubiquitinated p105 can bind to the proteasome (Fig. 10.6).
At this stage it appeared that it will not be useful to identify precisely the lysine residues targeted. That is because processing efficiency increased with the number of ubiquitin anchors regardless of their site, suggesting that recognition is not dependent on a particular sequence of amino acids and specific lysines. Thus, deletion/mutation of lysine residues in two clusters, B and C (Fig. 10.5aii, mutants c and d), resulted in similar processing efficiency (Fig. 10.5b lanes 5, 6 and 7, 8). In addition, lack of knowledge of the structure of the multiply monoubiquitinated region and of the identity of the proteasome subunit(s) that recognizes the modified p105 would have hampered further elucidation of the mechanism involved.
Taken together, these findings strongly suggest that the proteasome can recognize multiple single ubiquitin moieties, and not only a polyubiquitin chain.
These findings raise several interesting questions. One is whether monoubiquitination that leads to proteasomal targeting occurs in cells, and if so, what restricts elongation of the chain by the ligase. The same question can be asked for other processes where monoubiquitination has been suggested to occur, such as endocytosis of membrane proteins that serves to target them to the lysosome/vacuole. In these cases – where some experimental manipulations were also used to restrict polyubiquitination – it was hypothesized that ubiquitin-binding domains of the downstream proteins that associate with the conjugated ubiquitin mask Ile44 on the surface of the ubiquitin molecule that is required for chain elongation, or Lys48 which serves as the primary ubiquitin anchor, and thus restrict further ubiquitination. Alternatively, the E2-E3 that catalyze ubiquitination may limit the extent of modification, or the polyubiquitin chain that is nevertheless formed, is trimmed by a DUB (Hicke et al., 2005). Isolation and characterization of the enzymes involved in p105 targeting will be necessary in order to shed light on this unsolved question. Another important problem will be to identify the proteasome subunit to which the multiply monoubiquitinated p105 binds, whether it is one of the already described ubiquitin-binding subunits or rather a novel one. A likely possibility is that the multiple ubiquitins are binding to multiple different ubiquitin recognition sites that may even reside on different subunits, which collectively results in a high degree of avidity. Following identification of the binding subunit(s), it will be important to identify its “ligand” – the cluster of ubiquitin moieties that bind to it. Here one would like to know how “fixed” or “flexible” is the cluster and what are the principles that govern p105 binding to the proteasome – density of the modifying ubiquitins, their distance from one another, whether they generate a specific 3D structure, etc. Last, it will be interesting to study whether monoubiquitination is confined to proteins that undergo limited processing, or it targets also substrates that are completely destroyed. In this context, studying the mechanism(s) of degradation of another protein that undergoes limited processing – the yeast Spt23 transcription factor involved in regulating the level of unsaturated fatty acids (Hoppe et al., 2000) – will be useful.
References
For the complete list of referenced studies, please see Kravtsova-Ivantsiv, Y., Cohen, S., Ciechanover, A. (2009) Modification by Single Ubiquitin Moieties Rather Than Polyubiquitination is Sufficient for Proteasomal Processing of the p105 NF-κB Precursor. Mol Cell 33, 496–504.
Acknowledgments
The detailed findings described in this manuscript were published in: Kravtsova-Ivantsiv, Y., Cohen, S., and Ciechanover, A. (2009). Modification by Single Ubiquitin Moieties Rather Than Polyubiquitination is Sufficient for Proteasomal Processing of the p105 NF-κB Precursor. Mol. Cell 33, 496–504. They are published here with permission from Cell Press, Inc. Research in the laboratory of A.C. is supported by grants from the Dr. Miriam and Sheldon Adelson Foundation for Medical research (AMRF), the Israel Science Foundation (ISF), the German-Israeli Foundation for Research and Scientific Development (G.I.F.), the European Union (EU) Network of Excellence (NeOE) Rubicon, an Israel Cancer Research Fund (ICRF) USA Professorship, and a grant from the Foundation for Promotion of Research in the Technion. Y.K.-I. was supported in part by the Zeff Fellowship.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2011 Springer Science+Business Media, LLC
About this paper
Cite this paper
Kravtsova-Ivantsiv, Y., Cohen, S., Ciechanover, A. (2011). Modification by Single Ubiquitin Moieties Rather Than Polyubiquitination Is Sufficient for Proteasomal Processing of the p105 NF-κB Precursor. In: Wallach, D., Kovalenko, A., Feldmann, M. (eds) Advances in TNF Family Research. Advances in Experimental Medicine and Biology, vol 691. Springer, New York, NY. https://doi.org/10.1007/978-1-4419-6612-4_10
Download citation
DOI: https://doi.org/10.1007/978-1-4419-6612-4_10
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4419-6611-7
Online ISBN: 978-1-4419-6612-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)