Abstract:
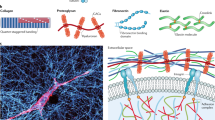
Tissues are structurally and compositionally complex materials that must function in a coordinated fashion at multiple length scales. Many of the structural proteins in soft tissues and in cells form biopolymer networks that provide mechanical benefits and coordinate cell-directed physiological activities. Complicated phenomena operate at multiple scales and are governed to varying degrees by the properties of networks; thus, mechanical models are a necessary tool to unravel the relationships among individual network components and to determine the aggregate properties and functions of cells and tissues. In this work, we review major biopolymers, their function, and the general mechanical behavior of biopolymer gels. We then discuss some network imaging techniques and methods for constructing and modeling networks /in silico/ – including multi-scale methods. Finally, we return to the specific biopolymers, including actin, microtubules, intermediate filaments, spectrin, collagen I and IV, laminin, fibronectin, and fibrin, and discuss what has been learned from the different models. Biopolymer network models, especially when combined with ever-improving experimental methods, have the potential to answer many fundamental questions in mechanobiology
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
19.1 Introduction
Tissues are complex materials composed of cells and extracellular matrix (ECM) proteins that must function in a coordinated fashion at multiple length scales. Biopolymer networks that form the ECM vary in composition and organization in a manner that confers suitable mechanical properties to the tissue and allows tissues to function in their physiological capacity. Mechanical loads and constraints applied to the whole tissue are transmitted down through the matrix and into the cells. The cells, which are stabilized and detect mechanical forces through the cytoskeleton – an intracellular network – then respond through a variety of dynamic activities that can lead to growth, remodeling, and adaptation.
The network architecture provides many beneficial properties to cells and tissues. Networks produce strong and stable structures with a minimum investment in materials. Their open configuration enables various transport processes (e.g. nutrient diffusion) to occur with less hindrance, and permits cell locomotion when appropriate (e.g. white blood cells). Also intrinsic to networks is the ability to communicate signals rapidly and at a distance. In terms of mechanical signaling, such a system may provide a means to coordinate cell behavior within tissues [1, 2].
With many complicated phenomena operating at multiple scales and governed to varying degrees by the properties of networks, mechanical models become a necessary tool for unraveling the relationships between individual network components and the aggregate properties and function of cells and tissues. A sampling of the kinds of questions about cell and tissue function that mechanical models can provide answers to includes:
-
How do tissue material parameters depend on the biopolymers and networks structure, e.g. what components and deformations contribute to the elastic response and where does time-dependent viscous behavior come from?
-
How does the ECM microstructure reorganize to accommodate macroscopic strain? Do fibers rotate, stretch, bend, or buckle?
-
What do those rearrangements mean in terms of mechanical signals that can be sensed by cells?
-
How does a cell sense mechanical force and translate that into a decision to act in a certain way? For example, how do stimuli cause synthesis or degradation?
-
How do mechanical changes in the ECM lead to different diseases, and how might they be prevented?
-
What components and what structural arrangements are necessary to produce a functional engineered-tissue?
The questions listed above are all inherently multiscale, with the tissue scale (∼10–3 m), cell scale (∼10–5 m), ECM fiber scale (∼10–7 m), biomacromolecular scale (∼10–9–10–8 m), and atomic scale (∼10–10 m). Although not all scales necessarily need to be studied to answer every question, any approach to understanding mechanobiology and biomechanics from a structural standpoint must respect their scale-spanning nature.
In this work, which emphasizes network mechanics, we focus on the transitions from biopolymer to cell (as in the cytoskeleton), biopolymer to tissue (as in basement membrane), and fiber to tissue (as in ECM or bioartificial tissues). In each case, the network consists of long, thin units connected in a structured or unstructured manner.
In the next section, we briefly review the major biopolymers and their function, followed by some general mechanical properties of biopolymer gels. We then discuss various methods to analyze images of networks and construct computer models there of. Next, we discuss the current approaches to network modeling in the general case and different methods that have emerged. Finally, we return to the specific biopolymers and discuss what has been learned from the different models.
19.2 Biopolymers of Interest
Cells interact with the physical world, and that interaction depends on networks inside and outside the cell. Inside the cell, a network of actin filaments, microtubules, intermediate filaments, and other proteins come together to form the cytoskeleton. This ensemble of intracellular proteins stabilizes the cell structure and plays a role in many cellular phenomena, including changes in cell shape and cell division. In addition, the cytoskeleton appears to play a prominent role in translating environmental cues, both mechanical and chemical, into a cellular response, which may take the form of biosynthetic activity [3–6] or even programmed cell death (apoptosis) [7, 8].
Outside the cell, a network of proteins, most commonly with collagen as the backbone, forms the extracellular matrix. The ECM composition and organization confers functionality to a tissue and provides a conduit for mechanical signals to alter cellular response. In many tissues, the ECM is in a constant state of turnover. The cells in the host tissue respond to changes in the microenvironment by degrading and synthesizing ECM proteins. Such changes can lead to growth and adaptation, e.g. tissue growth with exercise [9], or can lead to disease, for example glaucomatous damage to the optic nerve head [10] or hypertensive arterial wall thickening [11]. Other-ECM related diseases are congenital and result in impaired tissue function with devastating consequences. In Alport’s syndrome, for example, the genes encoding for a crucial component of the basement membrane malfunction [12]. The basement membrane structure is altered, which greatly impairs the molecular sieve structure of the kidney glomerulus, making it vulnerable to high pressures and more susceptible to proteolytic attack. Consequently, understanding the interplay between molecular interactions and macroscopic tissue mechanics is crucial to understanding many pathologies.
In this section, we introduce and briefly describe some of the monomers that form key intracellular and extracellular networks. The interested reader should consult the references listed in Table 19-1 for a more comprehensive review on each protein.
19.2.1 Intracellular Networks
19.2.1.1 Actin
Actin filaments (F-actin) are composed of a linear chain of G-actin subunits that are constantly and dynamically added to or removed from the ends of F-actin in a manner dependent on the local G-actin concentration. This process enables the cell to reorganize the cytoskeleton, migrate, attach to a substrate, and respond to signaling [13–17]. The G-actin subunits, which are approximately 2–3 nm in diameter [18] form semiflexible F-actin filaments that are approximately 5–7 nm in diameter and can ultimately assemble into hierarchical bundles and networks that span the interior of the cell. For a single actin filament, the stretching stiffness is K s = 4.4×10–8 N [19], the bending stiffness is K b = 7.3×10–26 Nm2, and the persistence length (defined later) is l p = 17 mm [20]. Actin molecules associate readily with divalent cations (Mg2+ and Ca2+ in particular) giving the molecule the capacity to complex with ADP and ATP. The conversion of ATP to ADP via hydrolysis through the ATPase myosin results in a conformational change in the F-actin molecule. This mechanochemical phenomenon driven by myosin has led to the colloquial reference of myosin as a motor protein and gives F-actin the capability to induce mechanical forces within the interior of a cell [21–23]. Furthermore, the protein ARP23 causes branching of actin filaments, contributing to the network structure [24].
19.2.1.2 Microtubules
Microtubules are another major cytoskeletal component critical to cell function. They are created when tubulin, a heterodimer of α-tubulin and β-tubulin, polymerizes to form stiff hollow tubes ∼25 nm in diameter. Microtubules are also controlled by the polymerization/depolymerization of their subunits. Microtubules are involved in a number of cellular processes including vesicle transport and cell division [25]. Their rigidity helps support organelles and maintain cell shape. Microtubules may also oppose the tensile forces generated by F-actin [26, 27].
19.2.1.3 Intermediate Filaments
Intermediate filaments (IFs) comprise a third classification of cytoskeletal components that are more stable structures than F-actin and microtubules [28]. IFs, which are ∼10 nm in diameter, are “intermediate” in size when compared to F-actin and microtubules. Intermediate filaments can be found at the transcellular junctions (e.g. gap junctions, tight junctions, desmosomes and adherens junctions) as well as at anchoring plaques to the extracellular matrix (e.g. focal adhesions and hemidesmosomes). IFs are also linked to F-actin on the interior of a cell creating a pathway for the mechanotransduction of extracellular mechanical phenomena.
19.2.1.4 Spectrin
Spectrin, a cytoskeletal component specific to the red blood cell, is composed of a dimer of either α-spectrin or β-spectrin, both of which are ∼250 kDa. The dimers arrange in an anti-parallel arrangement forming tetramers that associate with short actin filaments (∼15 subunits) creating an inter-triangulated actin-spectrin network conferring mechanical stability and enabling a blood cell to compress and subsequently expand [29, 30]. The spectrin-actin network is intrinsically important to the transport of erythrocytes, allowing the erythrocyte to modulate shape as it passes through narrow capillaries [31].
19.2.2 Extracellular Networks
The ECM functions as a support and anchoring structure for cells and as a means of tissue compartmentalization. The following components represent the major network-forming ECM molecules, and include type I collagen, type IV collagen, laminin, fibrin and fibronectin.
19.2.2.1 Collagen I
Collagen – the most abundant protein in the body – refers to a family of structurally and functionally related proteins that consist of three helically wrapped polypeptide chains [32, 33]. Type I collagen is a fibrillar collagen and accounts for 90% of all collagen (other fibrillar collagens include types II, III, V, IX). The fundamental unit of collagen, tropocollagen, is 280 nm in length and 1.5 nm in diameter [34]. Tropocollagen is composed of three polypeptide chains, or α-chains, that wrap around each other to form a right-handed triple helix. Tropocollagen is secreted into the ECM where it is modified enzymatically, assembled into quarter-staggered subfibrils, and covalently cross-linked [35]. Subfibrils associate laterally into fibrils and fibers, a distinction based mainly on size. Fibril diameters range from 10 nm to several hundred nm. They can organize into higher-order fibril bundles or fibers that can be hundreds of nanometers in diameter [36] and hundreds of micrometers in length [37]. For collagen-I, the properties of the triple helical monomer have been measured to be K s = 5.08×10–10 N, K b = 3.36×10–37 N-m2, and a persistence length l p = 14.5 nm [38]. In aqueous conditions, collagen fibers have been reported to have Young’s Moduli ranging from 32 to 900 MPa [39–42]. The bending stiffness for native collagen fibers ranges from 3×10–15 N-m2 to 6×10–15 N-m2 [43]. Many collagen fibers are heterotypic, meaning they are composed of more than one type of collagen [44]. Due to the complexity of native tissues, reconstituted collagen gels have served as simple but important in vitro tissue models [45–47].
19.2.2.2 Collagen IV
Unlike the fibrillar collagens, type IV collagen assembles into a mesh-like network that serves as the scaffolding for basement membranes. Basement membranes anchor and support endothelial and epithelial cells to connective tissue and provide physical barriers that allow for tissue compartmentalization. Collagen IV is comprised of three polypeptide chains associated as a triple helix and measuring approximately 400 nm in length [12, 48]. Six genetically distinct type IV collagen chains exist and are denoted as α1–α6. The chains assemble specifically forming three heterotrimers [α1(IV)]2,α2(IV), α3(IV), α4(IV),α5(IV), and α5(IV)]2,α6(IV)]. The relative concentration of each heterotrimer is dependent upon the tissue and the functional requirements of the collagen IV network. Type IV collagen interacts cooperatively with a variety of proteins and glycoproteins in forming the membrane [49]. Additionally, collagen IV can be reconstituted in vitro [49].
19.2.2.3 Laminin
Laminin has many functional roles in the ECM that relate primarily to cell attachment, including induction and maintenance of cell polarity, establishment of tissue barriers and compartments, organization of cells into tissues, and prohibition of attachment-induced cell death [50]. Laminin is a cross-shaped heterotrimer of glycoproteins comprised of several combinations of α, β, and γ subunits resulting in 15 distinct heterotrimers. In general, the molecule is comprised of three short arms of ∼37 nm and a long arm of ∼77 nm. All ends of the molecule have a globular domain providing functionality. In vitro, laminin can aggregate into networks in a concentration-dependent and thermally-reversible manner in the presence of divalent ions such as Ca2+ and Mg2+ [48].
19.2.2.4 Fibronectin
Fibronectin (FN) is a cell-secreted, soluble dimer, which polymerizes into an insoluble fibrillar network that facilitates cell attachment to the ECM (collagen types I–III and V, in particular) [51]. The fibronectin subunit, a dimer of polypeptide subunits 60–70 nm in length and 2–3 nm in diameter, associate into dimers that interact directly with integrins on a cell surface [22, 52]. Cytoskeletal tension created across the integrin stretches the FN dimer and exposes FN-FN binding sites to other interstitial FN dimers, resulting in FN fibril and network formation. Because FN directly attaches to the cytoskeleton, and the FN network is assembled by mechanical mediation, it is thought that FN plays an important role in influencing cell shape, organization, and locomotion. Additionally, FN deposition is observed in a wide variety of wound healing processes, usually preceding the deposition of a more permanent collagen matrix [53].
19.2.2.5 Fibrin
Fibrin networks form during blood clotting as part of the wound healing process. Fibrin is formed from the assembly of fibrinogen, a trinodular ∼340 kDa protein present in plasma that is 45 nm in total length [54]. Polymerization is catalyzed by thrombin, which enzymatically cleaves the N-termini of the α and β chains creating the “A” and “B” polymerization sites, respectively. The fibrin monomers arrange in a half-staggered arrangement aligning complimentary bonding sites creating oligomers that arrange in pairs creating dual-stranded protofibrils. Protofibrils associate laterally-forming fibers which ultimately aggregate, constrained by the ionic conditions, into bundles with a paracrystalline structure and distinctive banded pattern [55]. The bundles undergo branching creating a three-dimensional network that is covalently cross-linked by factor VIIIa concomitantly with the release of fibrinopeptide B [56]. In addition to its in vivo function, fibrin has also emerged as an attractive scaffold for tissue engineering [57, 58].
19.2.3 The Mechanical Behavior of Biopolymers
The biopolymers above can be examined in their native state, but frequently are purified and reconstituted in gel form. The in vitro gel is a much simpler system than cells and tissues, while still providing many of the rich mechanical properties observed in the in vivo systems. The bulk properties of gels are frequently measured with a rheometer. A common test performed involves casting a biopolymer gel between two surfaces, often parallel plates, and oscillating one plate back and forth at frequency ω while the other is held fixed. In the small strain limit of a linear viscoelastic material, the stress-strain response is given by
where σ denotes the stress, γ denotes the strain, G ′ denotes the elastic modulus and G ″ denotes the loss modulus. For a perfectly elastic material G ″ = 0, and for a perfectly viscous material G ′ = 0. For typical biopolymers such as actin and collagen, the elastic character of the gel dominates at low frequencies (less than 100 Hz) – G ′ is an order of magnitude greater than G ″ [59, 60] – thus making it possible to measure the elastic properties of the network with these tests. The viscoelastic character of the gels stems from molecular-level rearrangements and fluid-solid interactions (poroelasticity). The viscoelastic behavior is important but will not be discussed here. For more information see [47, 61–65].
The gel’s elastic character depends on the biopolymer concentration c and the cross-links formed. One typically observes power-law scaling of the form \(G'\sim c^x\), where for actin x = 1.4 when no cross-linker is present [66], and x = 2.5 in the presence of very strong and stiff cross-linkers [60]. Another parameter of significance is the ratio of cross-linker to polymer concentration, R. Again, there is typically power-law scaling of the form \(G'\sim R^y\), but a transition point exists, where at small R,y = 0.1, whereas at larger R,y may range from 0.4 [67] to 2.0 [68], depending on the stiffness of the cross-linker. Recently, it has also been observed that when sheared, these materials tend to compress and pull the shear plates together [69].
At small strains, typically less than 10%, it is reasonable to treat biopolymer gels as linear viscoelastic materials. At larger strains, however, they typically stiffen, with a modulus that can increase by 2–3 orders of magnitude [70]. A number of models give alternative explanation for this stiffening. At very large strains, the material breaks, undergoing irreversible deformation. Some materials, including actin, fail earlier at increasing densities [60] while other materials, like collagen, break at the same strain regardless of the density [71]. These mechanical properties are summarized in Figures 19-1, 19-2 and 19-3.
G ′ Properties of actin from. The elastic modulus (G ′) and maximum strain (γ max) for actin networks cross-linked with scurin, as a function of actin concentration (c A) and cross-linker/actin ratio (R). From [204] with permission
Strain stiffening properties of biopolymers. From [70] with permission
Nonaffine-affine transition in mechanical beam networks. G′ scaling with L/λ, where λ = lc(lc/lb)1/3. Note that when the spacing between cross-links is large, the network is much less stiff than the nonaffine network. (Reprinted with permission from Head et al. [143]. Copyright (2003) by the American Physical Society. http://prola.aps.org/abstract/PRE/v68/i6/e061907)
In addition to shear, extension and compression tests have also been conducted on biopolymer networks (mostly collagen) – for a review see [47]. In uniaxial extension, unconstrained network fibers rotate and align with the displacement axis before the network stiffens due to fiber resistance to axial stretch [72–75] thereby producing the characteristic J-shaped stress-strain curve observed in soft tissues [71, 76, 77]. The rapidity of stiffening is dependent on network properties and constraints. For example, a more cross-linked network stiffens faster at a lower extension, as does a network that is constrained transversely from contracting inward when stretched. Soft tissues and collagen gels exhibit a reduction in the peak stress and the amount of hysteresis between the loading/unloading curves that converges to a stable value when the stretch protocol is repeated at the same rate and to the same extent (preconditioning). It has been suggested that this behavior is due to microstructural rearrangements, although the specific cause remains unknown. One possibility is that non-covalent interactions between fibers continue to break as a result of the cyclic stretch until a stable configuration is reached.
The gel response to compression testing is more complicated, and the dissipation mechanisms involve molecular interactions and interstitial flow. Gels are often confined laterally in a chamber and compressed with a porous piston to allow fluid flow out of the gel [65, 78], an experiment inspired by the articular cartilage community [79, 80]. In collagen gels, the network response was found dependent on the time scale of the deformation, with step and ramp tests resulting in fiber collapse near the piston or fiber bending that induced network restructuring throughout the gel [62].
Higher collagen concentration generally translates to better mechanical properties, but again it is unclear whether the underlying cause is more cross-links, larger fibers, or other changes to the network architecture. It is well known that collagen fiber and network architecture is highly dependent on the gelation conditions, including pH, temperature, and ionic concentration [71, 81, 82]. In the absence of cross-linker, it has been found that the storage modulus scales with collagen density c c by \(G'\sim c_c^{\left( {2.45 \pm 0.25} \right)} \) [83, 84]. This is markedly different from the \(G'\sim c_a^{1.4} \) for uncrosslinked actin, which suggests that even in the absence of chemical cross-linkers, the fibers naturally cross-link, a conclusion in agreement with macroscale observations as well [62].
As already noted, tissues are compositionally and architecturally more complex than single-phase biopolymer networks. As a result, other ECM components, including proteoglycans, elastin, laminin, and fibronectin, have been added to collagen gels in order to assess their impact on tissue mechanics [85, 86]. In general, the changes in G ′ and G ″ were concentration dependent with the additives either aggregating collagen fibers together, as was the case with the proteoglycans studied, or thickening the fibers by coating them. Regardless of the macromolecule added, interpreting the results of such experiments is difficult because it is not known how the proteins affect network assembly or how the resulting structure compares to native tissues.
Nevertheless, these studies are relevant to understanding how tissues are built, particularly skin and cartilage, which share similar structural arrangements. Tissues like tendons, on the other hand, are highly organized and cross-linked into a hierarchical structure designed to resist high tensile load and store energy and do not behave like collagen gels. A discussion on the mechanical properties of soft tissues is quite involved and beyond the scope of this review. The interested reader is referred to the works of Fung [76] and Humphrey [87] for more information.
19.3 Network Imaging, Extraction, and Generation
19.3.1 Imaging
19.3.1.1 Fiber-Level Imaging
To produce an accurate representation of a biopolymer network, one must first obtain images of the microstructure. The difficulties involved are many, and each imaging technique has its advantages and disadvantages. The most common means for obtaining microstructural information relies on light level histology techniques [34]. Different colored dyes or stains are applied to thin, fixed sections of tissue to visualize the different matrix components. Histology is relatively inexpensive and easy to do, and it can provide spatial information for multiple species. Histology’s main detractions are that it is labor intensive and only moderate in resolution (submicron). In addition, the information obtained is two-dimensional and prone to artifact, which can arise during fixation, dehydration, sectioning, or staining. More specific staining can be achieved, often through the use of fluorescent antibodies that bind specifically to the target molecule. When the sample is illuminated with a specific bandwidth of light, only the tagged molecules are imaged. Serial sections through a sample can be reconstructed into 3d datasets of the tissue’s microstructure [88, 89], or to obtain fiber orientation [90]. These methods are again labor intensive and often produce artifacts, and the 3D reconstructions are computationally demanding. More importantly, the real-time microstructural response to macroscopic loads is not accessible.
More advanced imaging technologies have emerged, which are also capable of providing 3D data sets. Magnetic resonance imaging (MRI) [91], computed tomography (CT) and micro CT (μCT) [92–94], and optical coherence tomography (OCT) [95–98] have all been applied to tissues, most notably bone. These techniques allow 3-D imaging of living tissues, but are limited in their ability to identify differences in soft tissues and do not provide sufficient resolution to image at the scale of the microstructure [99].
In the case of purified gels, confocal microscopy [75, 100–102], and multiphoton microscopy [82] can be used obtain 3-D images of the networks without destroying their network architecture. Typically, the point spread function of the system is on the order of 500 nm, whereas for collagen, the fiber radius is often less than 100 nm [83]. Thus while fibers of small diameters are visible, the precise radii of the fibers and details of the fibril architecture cannot be resolved.
Electron microscopy (EM) provides a range of techniques useful for visualizing the microstructure in detail because resolution is on the nm scale. EM has been used to directly observe a variety of biomolecules, including type IV collagen [103], laminin [104], and spectrin [105]. Scanning electron microscopy (SEM) is often used because of its superior depth of field, which also has the disadvantage that quantification of fiber dimensions is difficult without resorting to stereoscopic techniques. Transmission electron microscopy (TEM) is much more suited to quantitative measurements because the sample is sectioned into thin slices. TEM can also be combined with other preparatory techniques, such as quick-freeze/deep etch, where a replica coating of the sample is imaged instead of the sample itself [106, 107]. Sample preparation in EM is also difficult and sample artifacts similar to those that occur in light level techniques are also present. Because of the higher resolution, however, artifacts are magnified and present greater difficulties in extracting the true microstructure. Cryo-(SEM) presents a more pristine picture than conventional SEM because water-associated structures are not dehydrated. In this technique, the sample is rapidly frozen so that the water vitrifies [108, 109]. 3-D reconstructions are also possible using electron tomography, which has been used to reconstruct cell structures [110] and single molecules, including collagen and fibrillin [111].
19.3.1.2 Indirect (Population-Level) Imaging
Non-invasive, indirect measurements of the fiber microstructure are possible by probing the sample’s optical properties. Techniques, such as small-angle light scattering (SALS) [112] and polarimetric fiber alignment imaging (PFAI) [72, 113] do not image the fibers directly, but instead make quantitative measurements of the fiber population based on the optical properties of submicroscopic fiber networks. In SALS, the pattern of scattered laser light transmitted through a sample provides a local fiber orientation distribution, which can be used to generate dynamic alignment maps during mechanical testing of tissues [114]. PFAI exploits the birefringent properties of the fiber and the difference in refractive index between fiber and solution to assess principal fiber direction and degree of alignment by measuring the change in amplitude and phase of the elliptically polarized light transmitted through the sample. Consequently, it can only be employed if the biopolymer network is birefringent, as is the case with collagen and fibrin. PFAI has been used extensively to generate 2-D network alignment maps in a variety of static [72, 115, 116] and dynamic [62, 117, 118] bioartificial tissue systems. Indirect techniques do not have the resolution that confocal methods have but they are easier to implement and can survey whole tissue samples under a variety of loading schemes. One limitation, however, is that the samples must be sufficiently transparent (i.e. thin enough). Otherwise sample sectioning or optically clearing the tissue with a hyperosmotic solution may be required [114]. Another issue is that neither method can discriminate between different fiber populations. For example, a fiber orientation distribution obtained from a remodeled fibrin gel cannot distinguish fibrin fibers from newly formed collagen fibers. Regardless of the real-time imaging method used, gaps between scales still exist which can only be addressed with multi-scale computational models and a cohort of imaging techniques.
19.3.2 Network Extraction
Another issue is how to describe the microstructure once an image or representation of the microstructure is obtained. Morphometric and stereologic methods have often been employed to describe tissue microstructure [119, 120]. These descriptors can provide exact quantities, such as volume fraction and number of objects, or distributions, such as fiber length, width, and orientation angle. Several tensor representations have been employed to describe material anisotropy and micro-structural alignment [121, 123]. They can be constructed from image-based measurements, such as mean intercept length [119, 124] or from Fourier transform methods (FTM) [125], and are convenient for capturing the principal direction and strength of fiber alignment.
Other image processing techniques have also been used to extract fiber and network features, as well as to map myocardial fiber orientation [126] and to quantify cytoskeletal reorganization in response to shear [127], stretch [128], and wound healing [129]. Some of these methods involve first thresholding the intensity image into a binary image. Additional processing might include the use of filters for edge detection and gradient calculation [126], or skeletonization and tracking [130] to determine fiber orientation and magnitude, or to reconstruct the network [83, 131, 132]. In addition, Fourier methods [125, 129, 133, 134] and the Hough transform [135, 136] have proven useful for obtaining fiber distributions. The majority of these methods have been developed for 2-D images, and some have been extended to 3-D [83, 131, 132, 137] For more information on image processing techniques see Gonzalez et al. [138].
19.3.3 Model Network Generation
A variety of methods have been implemented to create networks, not all based on measurements of the microstructure. The simplest network model assumes an idealized geometry representative of the material, such as a hexagonal cellular solid unit cell [139]. Another possibility is to use an established algorithm, such as Voronoi tessellation or Delauney triangulation, to subdivide a region into a mesh. Methods of this type are useful but generate more ordered, cellular-solid-like networks, which only share some features with fibrous networks [140, 141]. Consequently, one should consider whether the material to be modeled is more appropriately described as a cellular solid or fiber network when creating the network geometry (Figure 19-4).
Cellular vs. Fibrous Networks. (a) A random cellular network produced via Voronoi tessellation using random points. (b) A random fiber network created from a random growth algorithm. See Huessinger [139] for details on the differences
The generation of random 2-D straight-fiber networks, known as a Mikado model, involves randomly selecting network properties from a distribution function (e.g. uniform, von Mises, etc.). Typically, fiber position and angle are selected randomly, and locations where fibers intersect are made into cross-links [141–144]. Other distributed networks properties can be used to shape the network including fiber length and aspect ratio [145, 146]. In some cases the networks created are periodic, meaning that fibers that overlap the box boundaries are made to wrap back around to the other side [145, 147]. Periodic boundaries are generally used to remove end effects and to more easily impose network properties, such as total fiber length and network volume fraction [145].
These techniques can be extended to generate 3-D networks but an additional angle (ranging from 0 to π) must be included. Automatic 3-D skeletonization algorithms have been developed for extracting the network structure [83, 131, 132] from images and recently, Stein et al. [83] have validated that the architectures extracted by these algorithms have realistic geometric and mechanical properties.
Most of our work has focused on the mechanics of biopolymer gels and tissue equivalents [74, 117, 148–150]. In these studies, 3-D fiber networks were created in a stochastic process that resembles the process of collagen fiber formation in gels. First, a number of seed points are generated inside of a box. A fiber grows bidirectionally from each seed point until intersecting the boundary or another fiber. We have recently set up the method to generate statistically equivalent networks to those obtained from polarized light imaging by adjusting the random direction distribution and checking to match the observed structure [117].
19.3.4 Network Generation via Energy Minimization
Networks can also be generated through energy minimization techniques such as the Metropolis-Hastings (MH) importance sampling algorithm [151, 152] (a Monte Carlo technique). MH is used to generate the structure and interactions of dynamic chemical systems from time-independent and stochastic rules [153–155]. If the rules of the simulation are posed adequately, two differing and commensurate sequences of random numbers should generate statistically equivalent results (i.e. the results will agree to within a small “statistical error”). Consequently, the MH algorithm is a powerful tool to bridge how nanoscale chemical energetics yield macroscopic networks with determinable mechanical properties.
The underlying principle of the MH algorithm is to calculate a thermodynamic minimal average energy \(\left\langle U \right\rangle \), of an ensemble of m molecules, \(\left\{ {n_1 ,..., n_m } \right\} \), at a given temperature, T, using the following equations:
where k is the Boltzmann constant. The difficulty in performing such a calculation is that the normalizing quotient, Q, is generally not known for complex systems such as molecular biofibril networks. To circumvent the lack of knowledge of the normalizing quotient, instead an estimate of \(\left\langle U \right\rangle \) can be made based upon a series of K unique configurations of molecules, \(\Gamma _j \left\{ {n_1 ,...,n_m } \right\} \) for j=1,…,J, such that
As K becomes large, the estimate of \(\left\langle U \right\rangle \) approaches the expected minimum value of the internal energy of an ensemble of molecules. The simulation space must be initially seeded with molecules in a way that precludes infinite energy interactions (e.g. interactions that violate volume exclusion). After initially seeding of the simulation space, an initial energy is calculated based on the thermal properties of the system, the interaction potentials, and distances of the interaction sites. Next a molecule is chosen randomly and displaced a random distance generating a new configuration. The energy of the new configuration is calculated. As long as the quotient of thermodynamic probability of the new configuration is less than a random number ζ, generated on the interval of (0,1), the new configuration and associated energy are accepted. Otherwise, the molecule is returned to the starting position. The acceptance criteria is explicitly demonstrated by
where U(j) denotes the baseline energy prior to reconfiguration and \(U'_j \) is the energy of the new system following the displacement of the randomly chosen molecule.
As the algorithm proceeds, the result is convergence to the minimal energy configuration of a network comprised of the initial fiber set. The algorithm is designed to prohibit convergence upon local energy minima and is sensitive to the level of correlation between random numbers. Consequently, a high-quality pseudo-random-number-generating algorithm is critical to ensure that results are statistically valid (see [153, 154] for a more thorough discussion on this topic). This application of the MH algorithm is thus a means to generate a network from the associated fundamental subunits using an energy minimization approach. An example of a collagen IV network generated using the MH algorithm is demonstrated in Figure 19-5.
Collagen IV Network Generated with MH algorithm. The network is initialized as a collection of α1α1α2 (∼80%) and α3α4α5 (∼20%) monomers randomly selected in the simulation space. System energy is decreased as 7S domains are brought within bonding proximity of other 7S domains (∼3 nm). Over the course of 5×106 Metropolis steps, the system begins to converge upon the global energy minimum of the system. (a) Initial network before energy minimization and (b) network following energy minimization via the Metropolis-Hastings algorithm. Notice the increase in heterogeneity
19.4 General Modeling Approaches for Biopolymer Networks
19.4.1 Definitions
In the following, we define a biopolymer network to be a collection of interconnected fibers. Depending on the biopolymer of interest, a fiber may consist of a true fiber, a fibril, a filament, or a bundle of filaments, which themselves are composed of monomers, depending on the network of interest. Where two fibers interact, we define there to be a node. There are two major types of nodes: entanglements and cross-links. An entanglement is a point at which two or more fibers are in close proximity, such that the possibility of contact alters the deformation properties of the network. A cross-link is defined to be a point where two fibers are chemically linked together. A segment of length \(l_s \) is defined to be a piece of the fiber between two neighboring nodes.
We also define various types of polymer networks based on the flexibility of the fibers that make up the network. Flexibility is determined by the persistence length l p of a fiber, which gives the typical length over which a fiber remains straight. For a fiber of length L parameterized by s, l p is given by
where \(\theta \left( s \right) \) is the tangent angle of the fiber with respect to its main axis [156] and \(\left\langle x \right\rangle \) is the expected value of x. It can also be shown that \(l_p = {{K_b } \mathord{\left/ {\vphantom {{K_b } {kT}}} \right. \kern-\nulldelimiterspace} {kT}} \), where K b is the bending stiffness, k is the Boltzmann constant, and T is the temperature [156]. The bending stiffness is a function of the fiber’s Young’s modulus and the moment of inertia of the cross-sectional area. A flexible network, like rubber, is one where \(l_p < < l_s \). Such networks are dominated by the entropic stiffness of the segments [157]. For a semi-flexible polymer network, such as actin, \(l_p \approx l_s \) [63, 64, 158]. These networks are considerably more complex because both mechanical and entropic properties of the fibers play a role in the network dynamics. On the other extreme are mechanical networks, such as collagen-I gels where \(l_p > > l_s \), and the fibers have very long thermal persistence lengths and the entropic effects are negligible [84].
19.4.2 Affine Theory
A variety of approaches exist for modeling biopolymer networks. One common assumption employed is that the network deformation can be described as an affine transformation. An affine transformation preserves the collinearity of points and the ratio between distances. A typical example is that of simple shear, which maps the point (x,y) to (x+γy,y).
Affine theories have been used to describe the properties of flexible gels, such as rubber [157], where the persistence length is much shorter than the distance between nodes. Here, it has been found that the elastic modulus, \(G' \), scales with the cube of the mesh size, \(\xi \). Mesh size is defined as the average of sphere diameter that fits inside the network without touching the fibers. For semiflexible biopolymer networks, the persistence length is on the same order of the mesh size, and the entanglement length l e is used to describe the network. MacKintosh et al. [158] and Morse [63, 64] have developed an affine deformation theory for semiflexible biopolymer networks. By treating the polymer as an entropic worm-like-chain, they derive a force-displacement curve for an individual polymer chain to be:
where dl is the length change of a segment. The force-displacement curve is related to the modulus of the material by assuming there are ξ 2 fibers per area, and that \(dl = \gamma l_e\), where γ is the shear strain. This gives
To relate \(G'\) to the polymer concentration, one must first determine the dependency of K b , ξ, and l e on concentration. MacKintosh et al. [158], assume the fibers do not bundle and thus K b is independent of c. Previous experiments [66] indicate that for non-cross-linked actin, \(\xi \sim c^{ - {1 \mathord{\left/{\vphantom {1 2}} \right.\kern-\nulldelimiterspace} 2}}\). For l e , various relationships have been used. For a densely cross-linked gel, \(l_e \sim \xi\), and \(G'\sim c^{2.5} \). However, the precise relationship between l e and ξ may be more complex. On the other hand, modeling the chain as a fluctuating rod gives \(l_e \sim \xi ^{{4 \mathord{\left/ {\vphantom {4 5}} \right. \kern-\nulldelimiterspace} 5}} \) [158], whereas if one also assumes that the bending stiffness of the polymer depends on the polymer length, as is the case when polymer bending is dominated by shear (discussed below), then \(l_e \sim \xi ^{{4 \mathord{\left/ {\vphantom {4 3}} \right. \kern-\nulldelimiterspace} 3}} \). The affine theory has also been used to explain strain stiffening of biopolymer gels [70] as well as the negative normal stresses observed during shear [69]. Affine-deformation models have also been used to simulate the mechanical response of fibrillar tissues [159], including heart valve [160], cornea [161], skin [162], and articular cartilage [163], accounting for multiple co-existing networks or non-fibrillar tissue components as needed.
While the affine model predicts behavior in line with what has been observed experimentally, it is not clear that the assumption of affine deformation is valid at the length scale of the fibers. The nonaffinity of a deformation can be measured in a number of ways [164] and is typically done by looking at the difference in length, angle, or vector difference between the observed deformation and that predicted for a purely affine deformation. Nonaffine deformations have been observed in practice [72, 102, 114, 117, 165, 166] but disagreement still exists on the applicability of the affine assumption for biopolymer networks [167, 168].
One difficulty with the affine assumption is that the network segments deform independently, and thus the details of network interactions are lost. Such an assumption allows a simpler material description that in some applications may be sufficient for the problem. Within this framework, however, there is no obvious way to account for fiber synthesis or degradation, nor does it allow one to model failure at the cross-links or in individual network fibers. The need for more detailed understanding of networks has led to the development of various non-afifne models, described in the next section.
19.4.3 Nonaffine Models
In modeling non-affine networks, there are three main choices the modeler must make: (1) the constitutive model for the individual fiber segments, (2) the properties of the nodal interactions, and (3) the network organization of the segments. The choice, in part, depends on what type of questions the modeler intends to answer. The individual segments may be treated as linear or nonlinear springs, which only stretch, or as beams or worm-like-chains, which also resist bending and torsion. Additional relationships may be needed to account properly for the bending stiffness if the segment is composed of a bundle of interacting filaments.
Nodes can be treated either as cross-links or entanglements. While macroscopic scaling theories account for both (discussed later), with the exception of Rodney et al. [169], all microstructural models presented here assume that fibers are chemically cross-linked, or sufficiently entangled that on the time scale of interest they are and unable to slip at the nodes. In addition, the analysis is greatly simplified by neglecting steric interactions between fibers, which may contribute to the mechanical response. Nodes may be treated as freely rotating pin joints, welded joints of fixed angle, or linear or torsional springs.
19.4.3.1 Spring Model
We begin by exploring networks of randomly oriented springs, studied by Kellomaki et al. [144]. In this model, each segment acts as a linear spring, and the springs are connected at freely rotating pin joints. Kellomaki et al. [144] showed that under small deformations, such a network is floppy and has zero shear modulus. That is, under small deformations, the network is able to rearrange itself without changing the length of any of the springs. The floppiness of the network can be explained in two ways. One is that for a network cluster to be rigid, it must be composed of triangles that share a common side. Such a structure requires the existence of points where three fibers overlap. In a randomly generated Mikado network, the probability of three fibers intersecting at a single point is almost surely zero, so it is impossible for a stiff cluster of fibers to percolate the network. An alternative framework for analyzing network properties is based on Maxwell counting [170]. Consider a d dimensional space composed of N v vertices connected by N c segments. The condition number is defined to be the average number of segments that connect to a single node and is given by \(z = 2N_c /N_v \). The total number of degrees of freedom in the network (ignoring rigid motions) is given by \(N_f = N_{vd} - N_c \), where d is the spatial dimensionality. For a rigid network \(N_f = 0 \), giving \(N_c = N_{vd} \), and requiring \(z = 2d \), where d is the spatial dimensionalty. For a network of Mikado model structure, even if the free ends are removed (of condition number 1), we are left with vertices that are connected by 2–4 springs. Because the condition number is less than four the network is floppy. An important implication of this model is that the assumptions associated with the affine model discussed above are inconsistent. If a biopolymer gel is modeled as a network of randomly oriented springs, even if the springs are nonlinear, the network cannot resist shear at infinitesimal deformation. In contrast, springs in the affine model stretch immediately. Furthermore, the spring network does not deform affinely because it is able to rearrange itself under small deformations without changing the length of its springs.
Chandran and Barocas [73] have also studied random spring networks with the goal of modeling collagen gels. They studied networks generated by an artificial polymerization algorithm, described above, and these networks also have a condition number that is less than 4. Similar to Kellomaki et al. [144], they find that the network deformations are significantly different from affine and in particular, fibers are likely to reorient rather than stretch, thus leading to smaller stretch ratios than would be seen in the affine case even though the fiber orientation averaged over the entire population remained close to the affine value.
The Mikado and polymerization models are attractive in that the network architecture is reminiscent of biopolymer networks, but their main problem is that networks of zero modulus at small strains are unrealistic. To study rigid spring networks, Wyart et al. [171] explore the strain stiffening properties of networks formed by an alternative algorithm, in which the space is seeded with a number of nodes and then a condition number is imposed by connecting vertices that are close. Buxton and Clarke [172] have also studied beam networks formed in this way. This method allows one to explore the transition from floppy to rigid networks as the condition number increases. However, because this network architecture is not representative of most biopolymer networks, we do not discuss its properties further.
A fourth architecture for modeling biopolymers is the Arruda-Boyce eight-chain network model, used by Palmer and Boyce [173] for modeling actin networks. The model represents the network as a unit cell containing eight segments, each connecting a corner of the box to the center. Incompressibility is imposed on the cell such that even though the network alone is floppy, the network in combination with the incompressibility constraint is stiff. These models have the advantage of being easy to solve, but like the random networks of Wyart et al. above, they are not representative of true biopolymer networks. Nevertheless, by tuning the segment parameters, one can match experimental data for skin [174] and actin networks [173]. In modeling actin, Palmer and Boyce [173] based their force-displacement curves on the theory of MacKintosh [158], described above. This modeling framework allows consideration of prestress, but unfortunately, gives no way to predict it in the network, so the prestress must be fit to each data set individually.
19.4.3.2 Beam Models
In light of the above the result that realistic, spring networks have G ′= 0, it is necessary to account for the bending energy of the segments as well, or, at a minimum, to introduce torsional springs at the nodes [74]. Explicitly accounting for fiber bending is typically accomplished by treating each segment as a worm-like-chain (WLC) that contains both stretching and bending energy. Numerically, segment bending can be implemented either by using a discrete WLC model [175] or a finite element algorithm [145], where the segments are represented as beams having both a stretching stiffness K s , and a bending stiffness K b . Often, the segments are treated as elastic rods using Euler-Bernoulli beam theory, and thus the mechanical stretching stiffness is given by \(K_s = EA \) and \(K_b = EI \), where E is the Young’s modulus of an individual fiber, \(A = \pi R^2 \) is the cross-sectional area, R is the rod radius, and I is the area moment of inertia of the rod. The total energy in a filament is given by,
where the segment is of length L, the transverse displacement is given by u, the curvature is given by \(\nabla ^2 u \), and the axial strain given by ɛ. From the above expressions, one can also define a spring stiffness for a segment. The mechanical stretching stiffness for a cylindrical fiber segment is given by \(k_s = {{K_s } \mathord{\left/ {\vphantom {{K_s } {l_c }}} \right. \kern-\nulldelimiterspace} {l_c }} \), while the bending stiffness is \(k_b = {{K_b } \mathord{\left/ {\vphantom {{K_b } {l_c^3 }}} \right. \kern-\nulldelimiterspace} {l_c^3 }} \), where l c is the mean spacing between nodes.
If the segment consists only of a single isotropic, linear elastic filament of radius r, then \(I = I_{{\textrm{fil}}} = {{\pi r^4 } \mathord{\left/ {\vphantom {{\pi r^4 } 4}} \right. \kern-\nulldelimiterspace} 4} \). However, in many biopolymer networks, including actin and collagen, the segment is in fact a bundle of fibers. If the bundles are tightly coupled together by a stiff and rigid cross-linker, as is the case for actin cross-linked by scurin [177], then a similar formula applies, \(I = {{\pi R^4 } \mathord{\left/ {\vphantom {{\pi R^4 } 4}} \right. \kern-\nulldelimiterspace} 4} \), with R the radius of the bundle. In the case of loose intrasegment coupling, we instead have \(I = N_{{\textrm{fil}}} I_{{\textrm{fil}}} = R^2 /r^2 I_{{\textrm{fil}}} = {{\pi \left( {Rr} \right)^2 } \mathord{\left/ {\vphantom {{\pi \left( {Rr} \right)^2 } 4}} \right. \kern-\nulldelimiterspace} 4} \). In this case, the bundle is much more flexible as \(I\sim R^2 r^2 \) instead of \(R^4 \). There also exists a third, intermediate regime, determined by the nondimensional parameter \(\alpha = {{k_x L^2 } \mathord{\left/ {\vphantom {{k_x L^2 } {\left( {EA\delta } \right)}}} \right. \kern-\nulldelimiterspace} {\left( {EA\delta } \right)}} \) [178, 179], in which k x is the cross-link stiffness at a node, L is the segment length, and δ is the mean spacing between nodes. For small α, the coupling is weak and \(K_b \sim E\left( {Rr} \right)^2 \). For very large α, the coupling is strong and \(K_b \sim ER^4 \). For intermediate values of α, the formula for K b is more complicated, depending upon L, δ, and k x [178]. In the case of actin bundles, three regimes have been observed, depending upon the cross-linker used [179].
The first step in understanding the mechanical properties of these networks is to explore the G ′ behavior as a function of the network density. For the Mikado model, we define two densities: a nondimensional density \(q = {{NL^2 } \mathord{\left/ {\vphantom {{NL^2 } A}} \right. \kern-\nulldelimiterspace} A} \), and a dimensional density \(\rho = NL/A \), where N is the number of fibers of length L (possibly containing multiple segments) in a box of area A, and ρ has units of [1/length]. In two dimensions, it is possible to link l c directly to N, L, and A and at large q, it is given by \(l_c = 2/q\pi \) [180]. The mechanical properties of these networks have been shown to depend critically on q [181], and also on the average number of nodes per segment \({L \mathord{\left/ {\vphantom {L {l_c }}} \right. \kern-\nulldelimiterspace} {l_c }} \) [143]. Here, we choose to describe the network mechanics in terms of \({L \mathord{\left/ {\vphantom {L {l_c }}} \right. \kern-\nulldelimiterspace} {l_c }} \), though the two choices are equivalent [180]. For low \({L \mathord{\left/ {\vphantom {L {l_c }}} \right. \kern-\nulldelimiterspace} {l_c }} \), the system is made up of isolated rods and small, unconnected clusters. In such a system, there is no connected path from one side to the other and \(G' = 0 \). At \({L \mathord{\left/ {\vphantom {L {l_c }}} \right. \kern-\nulldelimiterspace} {l_c }} \)= 5.42 [143], conductivity percolation occurs, meaning that a path exists connecting two opposite sides of the network. In the case that the nodes can resist rotation (e.g. welded joints), the system has also achieved rigidity percolation, and the connected component can resist deformation. In the case that the nodes are treated as freely rotating pin joints, rigidity percolation does not occur until \({L \mathord{\left/ {\vphantom {L {l_c }}} \right. \kern-\nulldelimiterspace} {l_c }} \) = 5.93 [143].
Above rigidity percolation, there are two mechanical regimes, based on whether the deformations are affine or nonaffine. In the case of \(k_b < < k_s \), the fibers are long and thin, and the spacing between cross-links is large. Since the bending stiffness is relatively low, the network responds to deformation by the bending of its fibers, which is inherently non-affine. For freely rotating cross-links, it has been shown both through simulation and through a self consistent analysis of the floppy modes of the system that \(G'\sim k_b \left( {{L \mathord{\left/ {\vphantom {L {l_c }}} \right. \kern-\nulldelimiterspace} {l_c }}} \right)^{3.67} \sim K_b \rho ^{6.67} L^{3.67} \) [141], exhibiting an extreme sensitivity to the network density. The system behaves fundamentally differently when \(k_s > > k_b \). In this regime, deformations are affine and \(G'\sim k_s \sim EA\rho \), and the modulus depends only linearly on density. The critical length at which the transition occurs is given by \(l_{{\textrm{crti}}} = L\left[ {\left( {\rho - \rho _f } \right)L} \right]^{ - 2.84}\). Thus far, this scaling transition has not been fully explored in three-dimensional simulations. However, Huisman et al. [182] have studied artificially generated networks designed to be similar to actin, and Stein et al. [84] have studied collagen networks of realistic architecture. Both have found that at small deformations, the primary mode of energy storage is in bending, and at small strains the deformations are highly non-affine. This result lends further support to the idea that the affine assumption is erroneous for most biopolymer networks.
As discussed in more detail below, cross-linked biopolymer networks typically scale by \(G'\sim c^{2 - 3} \), where c is the polymer concentration. This is quite different than either scaling regime for the Mikado model. Thus the importance of the above results is not in the specific scaling laws derived, but in the observation that there are two distinct mechanical regimes, one dominated by affine stretching and another dominated by nonaffine bending.
19.4.3.3 Entropic Beam Model
An additional level of detail that can be added to the model is the entropic component of the stretching stiffness of an individual filament. In this framework, the stretching stiffness k s of a segment is modeled as two springs connected in series: an elastic element \(k_{{\textrm{el}}} = {{EA} \mathord{\left/ {\vphantom {{EA} {l_c }}} \right. \kern-\nulldelimiterspace} {l_c }} \), and an entropic element given by \(k_{{\textrm{en}}} = {{K_b^2 } \mathord{\left/ {\vphantom {{K_b^2 } {kTl_c^4 }}} \right. \kern-\nulldelimiterspace} {kTl_c^4 }} \). The total stretching stiffness is given by \(k_s^{ - 1} = k_{{\textrm{el}}}^{ - 1} + k_{{\textrm{en}}}^{ - 1} \) and is dominated by the more compliant of the two elements. In the case that the entropic stiffness is weakest, we have \(k_s \sim l_c^{ - 4} \), which is markedly different from \(k_s \sim l_c^{ - 1} \) for purely mechanical networks. Fibrous networks exhibit G ′ scaling that is very sensitive to the cross-link behavior. For rigid cross-links, \(G'\sim Ak_s + Bk_b \), where rather than acting in series, the stretching elements now act in parallel, with the stronger of the two dominating the elastic response. For flexible cross-links, however, in the case of inextensible fibers (\(k_{{\textrm{el}}} \to \infty \)), \(k_b^{0.5} k_s^{0.5} \). For these systems too, it is found that there is a critical average segment length l crit, such that for networks with \(l_c < l_{{\textrm{crit}}} \), the deformations are affine and when \(l_c > l_{{\textrm{crit}}} \), the deformations are nonaffine. Thus including the entropic properties of the network gives qualitatively different scaling laws for G ′, but the nonaffine/affine transition is still present.
19.4.4 Finite Strain
19.4.4.1 Strain Stiffening
The above section focused on the small-strain behavior of networks. Soft tissues, in particular, routinely deform beyond the small strain limit, ranging anywhere from 2% to over 40% strain depending on the tissue [76]. The typical load-deformation response of cells and soft tissues in uniaxial tension is non-linear, starting with a long, extensible toe region, followed by a linear and then exponential increase in the force. The stiffening observed at high strains is universal, but the cause of strain stiffening is unclear; the proposed mechanisms underlying it are dependent on the type of model used, and the mechanisms may be different for different biopolymers.
Storm et al. [70] have used the affine theory developed by MacKintosh [158] to show that the strain stiffening of a large number of polymers could qualitatively be explained by an affine deformation of a network of strain stiffening filaments. Similar behavior can also be produced by network reorganizations in which fibers are free to rotate with the deformation, both in spring [74, 149] and beam models [84, 146, 147, 182, 183]. It is likely that the precise nature of strain stiffening depends upon the specific properties of the biopolymers, as well as the manner in which they are organized. Tissues are denser and more cross-linked than gels, and their strain stiffening response may derive from both molecular entropic effects and ECM geometry. In some instances, the scale of a problem involving tissues may warrant the use of affine theories [166], although experiments show that at least some tissue fiber deformations are not affine [114, 166, 184].
19.4.5 Bridging Scales – Multiscale Behavior of Networks
19.4.5.1 Representative Volume Element
A common approach used in relating the macroscopic behavior of a material to its microstructure is to find a region in which the microstructure is structurally typical to the entire sample [185]. Such a region is referred to as a representative volume element (RVE). RVEs possess a characteristic length scale that is at least an order of magnitude larger (and preferably larger) than the length scale of heterogeneity in the microstructure. As a result, the whole material can be subdivided into a repeating array of RVEs, joined at their boundaries. Because the RVE is periodic and it is similar mechanically to the whole material, analysis can be conducted on the RVE alone.
Once an RVE has been selected, the analysis that follows assumes that the microstructure deforms continuously with the macroscopic strain field (the affine assumption). Such is the approach taken with cellular solid models [139, 186], which have been used to study a variety of materials, including metals and plastics [139], bone [187–189], and connective tissue in the optic nerve head [190]. Cellular networks can be setup with idealized, regular geometries that permit analytical solutions, or they can be created with irregular structures and probed with the finite element method [141, 191]. Either way, the bulk properties of the material can be related to the microstructure.
The analysis, however, is not limited to the behavior of one archetypal RVE. The RVE mechanical response can vary spatially, as in homogenization theory, where RVEs in the material develop different levels of strain to accommodate the inhomogeneous macroscopic displacement field [192–194]. The strategies to link scales in soft tissues are more challenging because large deformations are possible; hence, techniques based on a small strain assumption, such as many forms of homogenization, will fail. More importantly, linking strategies that rely on periodicity cannot incorporate macroscopic heterogeneity.
Our group has developed a multi-scale computational model that relies on the method of volume averaging [195] to link the macroscopic level to the microscopic level [74, 117, 149]. Because a material average volume is formulated (i.e. a volume that deforms with the material) large deformations are easily addressed. Furthermore, macroscopic heterogeneity, manifested as regional differences in the local ECM microstructure, can be accommodated naturally by employing different RVE network structures, provided that the regional differences are larger than the scale of the RVE. To clarify, the RVE domain should be bigger than the scale of microscopic gradients but smaller than that of macroscopic gradients [196]. As a result, the model provides a means to study the dependency of macroscopic tissue mechanics on the underlying ECM microstructure, which for our purposes is typically represented as a network of collagen fibers contained within an RVE [74, 117, 149, 150]. Consequently, the remainder of this discussion applies to collagen fiber networks, but other networks (e.g. electrospun fibers [195]) can also be examined with the method provided their attributes are accounted for in the fiber constitutive equation and volume averaging equation detailed below.
19.4.5.2 Volume Averaging
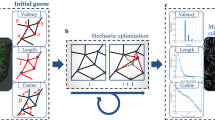
In the model, the macroscopic domain is represented with a Galerkin finite element (FE) model (Figure 19-6). However, in place of a macroscopic constitutive equation, the stress needed for the FE solution is obtained by solving the force balance on the fiber network contained within an RVE. The RVEs are centered at the FE integration points, and their boundaries are displaced based on the macroscopic deformation field. Boundary displacements produce forces in the fibers that are transmitted via fiber crosslinks, with the result that the fibers in the network reorganize to achieve force equilibrium. The network fiber stress is averaged over the RVE to obtain the macroscopic average Cauchy stress tensor, which is then used in the macroscopic stress balance to determine the new macroscopic deformation field, and the process iterates until convergence is achieved.
Multiscale modeling with volume averaging. The multiscale model relies on volume averaging theory to link scales. The macroscopic problem is represented using the Galerkin finite element method. RVEs containing fiber networks are centered at the integration points in the element, and the RVE boundaries are deformed with the macroscopic deformation field. Fiber forces in the network are volume averaged and the resulting macroscopic stress tensor is used in the macroscopic stress balance to solve for the new macroscopic displacement field. This process iterates going back and forth between scales until convergence is achieved
The method utilizes three basic equations: (1) a constitutive equation to relate fiber stress to fiber strain (2) an equation that relates the average macroscopic stress to the volume average of the local fiber stresses (3) and an equation for the force balance at the macroscopic level. A fourth expression to incorporate rotational stiffness at the nodes can also be used [74]. A number of constitutive equations have been proposed for collagen fibers [160, 193, 198] that represent the fiber as strong in tension and weak in compression. In previous work [73, 149], we have found that the fiber constitutive equation used only has a minor influence on the macroscopic behavior. For convenience, we employ an exponential constitutive equation [160] to relate the fiber force, F, as
where E f and B are constitutive constants, and A f is the fiber cross-sectional area. The Green’s strain of the fiber, ɛ f , is given in terms of the fiber stretch ratio, λ f , as \(\varepsilon _f = 0.5\left( {\lambda _f^2 - 1} \right) \). Equation (19-10), at the low-strain limit, reduces to a linear model with elastic modulus E f .
In volume averaging [195, 199], the macroscopic Cauchy stress tensor, S ij , is determined by averaging the microscopic stress field, s ij , over the RVE volume, V,
Here we use index notation with uppercase and lowercase letters to refer to macroscopic and microscopic variables, respectively. The microscopic stress can be rewritten as \(s_{ij} = s_{kj} \delta _{ik} \), where δ ik is the Kronecker delta. Because the gradient of the direction vector, x, is equivalent to \(\delta _{ik} \) (\(\nabla x = x_{i,j} = \delta _{ij} \)), Eq. (19-11) can be rewritten as
The second term on the RHS vanishes because microscopic equilibrium requires that \(s_{kj,k} = 0 \). Applying the divergence theorem allows the macroscopic stress to be calculated as integral of the RVE surface tractions, t j , over the RVE surface,
The tractions occur at the locations where network segments intersect the RVE boundary (cross-links). For thin segments, x varies little over the segment-boundary intersection. Thus, \(\smash{\int\limits_{} {n_k s_{kj} x_i dA \approx x_i \int\limits_{} {n_k s_{kj} dA} } = x_i F_j} \), and the components of S ij are given in terms of the crosslink positions, x, and forces, F, as
The final equation needed is the macroscopic stress balance. Since the averaging volume is material and changes with the macroscopic displacement, additional terms must be incorporated (see Chandran [74]). The advantages of material description are (1) it is consistent with how the microstructure deforms, (2) it satisfies the mass balance implicitly, and (3) the macroscopic gradients are naturally applied as the boundary conditions of the RVE [74].
The macroscopic stress balance is given as
where u is RVE boundary displacement and n is the unit normal vector. The right hand side of Eq. (19-8) arises from the correlation between inhomogeneous displacement of the RVE boundary and local inhomogeneities in the stress field. In the case of a fixed RVE, the RHS would be zero.
19.5 Applications to Biopolymers
Now that the general methods used to model biopolymer networks have been discussed, we examine the application of network models to specific problems.
19.5.1 Actin
Actin is a popular choice for microstructural analysis due to its critical role in a number of cellular events and biological processes, including cell motility [179]. Of particular interest is the wide spectrum of actin cross-linkers, whose effect on network formation and mechanics has important implications for normal cell function. When no cross-linker is present, the networks are extremely compliant (\(G' < 0.5 \) Pa) and elasticity scales with concentration as \(G'\sim c_a^{7/5} \) [200]. The addition of a cross-linker can bring the modulus to 100 Pa or larger, clearly demonstrating their importance in network formation. Cross-linkers, however, can serve two functions. First, they can group individual filaments into a larger bundle, which can strongly influence the bending stiffness [179, 201]. Second, they can connect filaments and bundles together to form a network. Cross-linkers vary in length with shorter molecules, such as scurin and fascin, forming relatively tight bundles whereas longer molecules, such as filamin and α-actinin form looser bundles. Heavy meromyosin cross-links while forming no bundles at all. The effect that various cross-linkers have on the actin networks is summarized in Table 19-2. Remarkably, scurin [60, 68], fascin [202], and HMM [203] all have similar effects in terms of the scaling of G ′ with respect to the actin concentration and cross-linker ratio, with \(G'\sim c_a^{\left( {2.35 \pm 0.15} \right)} R^{\left( {1.6 \pm 0.4} \right)} \). An additional parameter that is tracked is the critical strain \(\gamma _{{\textrm{crit}}} \sim R^{\left( { - 0.7 \pm 0.3} \right)} \), which indicates the onset of strain stiffening, and again is relatively similar for the three different cross-linkers.
Many models exist that have been used to explain some of these data, including the affine stretching model of worm-like chains [68, 158], the nonaffine 8-chain model [173], and the nonaffine bending model [167, 202]. All three models have also been able to explain the strain stiffening behavior of biopolymers [70, 147, 173]. In vivo, turnover of the actin network may contribute to its apparent viscosity. That is, a stressed fiber may disassemble and be replaced by new, unstressed fibers. The significance of this phenomenon varies with cell type, phenotype, and activity. While non-affinity has been directly observed at short length scales in scurin-cross-linked actin networks [102], the community does not yet agree upon whether such nonaffinity is sufficient to invalidate the affine theory [168]. The 8-chain model of Palmer and Boyce [173] requires one to refit the network prestress at each actin density. Thus their model makes the prediction that lower density networks have higher degrees of prestress, but such a prediction has yet to be validated.
Finally, it has been observed that the maximum strain that a gel can withstand decreases with increasing density [204]. This is hypothesized to be due to a shortening of the space between cross-links, which according to entropic stiffening hypothesis, means that the fibers reach their maximum state of strain sooner [158].
19.5.2 Microtubules, IFs, and the Cytoskeleton
Microtubules and IFs have been cast into gels and subjected to rheology tests to determine their individual mechanical characteristics [205, 206]. However, the networks were formed in vitro from purified monomer and may differ substantially from those formed inside a cell. It is important to understand the individual properties of these proteins, but how they integrate with actin to form the cytoskeleton is the ultimate goal, and much remains to be learned.
Wang and Stamenovic explore the contribution of IFs to cellular mechanics by measuring cell stiffness to applied stress in adherent wild-type and vimentin-deficient fibroblasts through magnetic twisting cytometry [207]. At high applied stress (>> 10 dynes/cm2), the stiffness of the vimentin-deficient fibroblasts is much smaller than the wild-type fibroblasts, while at a stress of 10 dynes/cm2, the stiffness is comparable. A six-strut tensegrity model (discussed below) was able to replicate the stiffening that resulted from cytoskeletal fiber realignment.
Microtubules and IFs are integrated into the cytoskeleton, and therefore can affect the properties of the whole cell. One perspective on the structure-function relationship between the cell, its cytoskeleton, and the extracellular matrix is the hypothesis that the cell is a tensegrity (tensional integrity) structure [26, 208, 209]. In this model, the stability of the cytoskeleton is derived from a balance between a continuous filament network (actin and intermediate filaments) under tension and isolated compression-resistant elements (microtubules and thick actin bundles). Without internal tension, or “prestress”, which can be generated through the cell’s contractile machinery, the cytoskeleton would collapse. External forces, which are transmitted from the ECM to the cytoskeleton through focal adhesions, cause the cytoskeleton to reorganize and stabilize until equilibrium is achieved. Support for this view appears to be based largely on its intuitive appeal and model predictions that match cell stiffening behavior with increasing tension and surface attachments [210, 211]. The non-linear behavior observed derives from geometric changes in the network, a behavior also observable in random fiber networks without compression elements. Although the concept of tensegrity is attractive, and experimental evidence shows that microtubules can buckle under cellular loading [212], the exact nature of the complex interactions that define cytoskeletal mechanics remains poorly understood.
19.5.3 Spectrin
Spectrin has been studied as triangulated networks of Hookean springs of non-zero force-free length and finite maximum length [212]. Triangulated networks are generated from infinitely thin hard rods with six-fold vertices. Under compression, the six-fold symmetric structures undergo a phase transition to two-fold network symmetry as studied analytically at zero temperature or through Monte Carlo simulation with a non-zero temperature [212]. In subsequent studies, a six-fold symmetric network of polymer chains, representing the actin-spectrin cytoskeleton of the erythrocyte, is generated and the geometrical and elastic properties are determined [214] and found to be in agreement with the shear modulus, of 6.6×10–3 dynes/cm at 25°C, for the erythrocyte cytoskeleton as determined from micromechanical techniques [215].
With respect to modeling the macroscopic erythrocyte structure, an intertriangulated network of chains becomes unwieldy. Consequently, Boey, et al., represented the spectrin chains using a worm-like chain potential providing a tractable and more physically realistic representation of an intertriangulated spectrin network than the original Hookean spring representation [214]. An ensemble-averaging technique was applied to non-axisymmetrical deformed shapes, analogous to an erythrocyte undergoing micropipette aspiration, demonstrating how the triangulated mesh of the spectrin-actin cytoskeleton imposes the macroscopic geometry of the erythrocyte [216]. Discher and colleagues’ simulation results from three structural models of the spectrin network attached to a bilayer suggest that the network exists in a prestressed condition of compression balanced through tension created by the lipid bilayer [216]. Lee et al. later confirmed that the prestressed erythryocyte membrane is capable of sustaining large anisotropic strains using fluorescently-patterned photobleaching of a rhodamine phalloidin-labeled spectrin-actin cytoskeleton [214, 217].
Additionally, the equilibrium shape of the human erythrocyte has been investigated using spectrin-level energetics [218]. Li and colleagues populated spherical and biconcave structures with spectrin networks capable of 2,3,…,9 element junctions. After the initial shape was populated, cytosol was removed allowing the shape to deflate with fixed spectrin connectivities. Coarse-grained molecular dynamics was employed to find the equilibrium shape of the deflated RBC employing a worm-like chain free energy model for the spectrin tetramer links. As a consequence of the hypothesis that spectrin networks are constantly undergoing remodeling at some sufficiently small characteristic time scale, Li et al., employed a liquefied network structure evolution algorithm to relax the in-plane shear elastic energy of the macroscopic network shape which permits the evolution to discocyte and stomacyte shapes based on the approach of Discher and colleagues [213, 214, 216, 217].
19.5.4 Collagen I
Similarly to actin, collagen I has been modeled extensively due to its abundance and central structural role in many tissues. For this review, we focus on network models, where the fibers are modeled as tension resisting springs [73] or tension and moment resisting beams [84]. As with the networks applied to actin, these models show that significant matrix restructuring occurs in a highly non-affine manner. In addition, uniaxial deformations applied to random networks result in nonlinear stiffening, which arises from fiber rotations that gradually lead to fiber stretch. In the small-strain limit, most of the energy in the network is stored in fiber and cross-link bending (when bending is accounted for). At large strains the primary energy storage mode is fiber stretching, and thus the large strain modulus scales linearly with density [84].
Our multiscale model was originally developed for collagen gels, which appear smooth and continuous on the macroscopic scale but are in fact composed of discrete fibers. The model was applied to collagen gels [74, 149] and extended to complex geometries [219] by means of a sophisticated computational environment. A representative example can be seen in Figure 19-7, which shows the stretching of a sample with inhomogeneous structure. The model was also applied to electrospun synthetic polymer networks [197], demonstrating its generality.
Heterogeneous test sample. A model system was constructed with a highly aligned central “wound” region and a more isotropic surrounding region. The sample was stretched uniaxially to 30% strain. The highly aligned central region deformed less than the surrounding isotropic region. Images are 2-D projections of a 3-D result
As noted earlier, an important challenge in such modeling is acquiring structural information, and as the sophistication of the model increases, so too does its need for structural specification. Sander et al. [117] have recently shown that the multiscale scheme can be specified based on polarized-light imaging, with each finite element in the model set at the average orientation measured for it in a sample. The sample was subsequently stretched in an off-axis hold test, which was concurrently simulated. The force and strain results were compared between the model and experiment, and agreement was good with fitting parameters based only on the constitutive equation for the fibers (an example of this kind of simulation is presented in Figure 19-8).
Image based multiscale modeling. One quadrant of a cell-seeded, compacted collagen gel was tested mechanically while microstructural orientation and strength of alignment was measured using polarized light. A unique 3-D, interconnected fiber network was generated for each finite element to match the average experimental values in the corresponding location in the experiment. (a) The model predictions (red) are overlaid on the experiment in white. For an off-axis hold test, in which the horizontal axis remained stationary while the vertical axis was displaced to a stretch ratio of 1.3, the model reasonably captured the local kinematics with some obvious differences. (b) The networks underwent significant restructuring in a location dependent manner. The network near the top right corner started off with moderate alignment in the horizontal direction. Fibers were free to rotate towards the vertical and stretch to accommodate the strain created by movement of the vertical grip. (c) This network, which was already aligned in the vertical direction increased in alignment with the stretch
Because many tissues contain multiple components, a rule-of-mixtures model was applied to arterial wall mechanics [150], with a network phase representing the collagen fibrils in the vessel wall and an incompressible continuous phase representing the non-collagenous tissue (Figure 19-9 shows typical simulation results and comparison to Experiment). The model agreed well with experimental data on decellularized porcine carotid artery [220] and contained an important result: it was predicted that the non-collagenous material is in compression even when the vessel is inflated. That is, the role of the non-collagenous material in the model is to prevent vessel wall and/or lumen collapse, not solely to provide elastic recoil.
Simulated inflation of a decellularized artery [150]. (a) The simulation involves four steps: an initial open artery is constructed, it is closed to produce a prestressed artery, the artery is stretched longitudinally, and finally it is inflated. (b) Pressure-diameter curve for the simulation (line) and experiment (dots, [220]) are in good agreement with only two fitting parameters, the fiber stiffness and the matrix stiffness
In some tissues, it has been observed that collagen fibrils/fibers/bundles are undulated and possess “crimp”, and it has been hypothesized that their nonlinear stress-strain response is in part the result of collagen fibers straightening out [221]. When undulations were incorporated into a network model, the effect was to delay the stiffening response [147].
Affine theories for collagen have also fit well to some experimental data when fiber-level properties are not required to match fiber level measurements [160, 222, 223].
19.5.5 Type IV Collagen
In spite of the importance of basement membrane, structural modeling has lagged behind that of collagen I or actin. A notable exception is the recent work of Burd [224]. Burd examined two phenomenological models (a linear elastic model and a Fung-type hyperelastic model), concluding neither model correlates satisfactorily with the mechanical properties of the collagen IV network present in the human lens capsule. Instead, a regular hexagonal lattice of pin-jointed bars was created and subsequently perturbed randomly to create an irregular polygonal structure, as used by Cavalcante et al. [225] for collagen-elastin networks. Representing the lens capsule as an irregular lattice of collagen IV embedded in a hyperelastic sheet correlated well with published data.
19.5.6 Fibronectin, Laminin, and the ECM
DiMilla and colleagues studied cell migration through a mathematical model elucidating the dependence of migration velocity on cellular mechanics and surface receptors binding with complementary ligands present in the extracellular substratum such as fibronectin and laminin [226]. They conclude that cytoskeletal force generation, cellular polarity, and adhesion dynamics are required for persistent cell motility. The model predicts how cell speed varies with a variety of phenomena including cytoplasmic rheology, intracellular contractile force, receptor/ligand densities and kinetics [226]. Ingber and Wang explored how the mechanical tension and the extracellular matrix through adhesion to the ligand fibronectin influence cytoskeletal mechanics [227]. Magnetic beads were coated with fibronectin of varying density, and a mechanical tension was applied to adherent endothelial cells through a magnetic twisting device. Ingber and Wang find that the cytoskeletal stiffness increases proportionally with the applied stress controlled by the magnetic twisting device and fibronectin density. A model is presented coupling cell mechanics to the applied stress [227]. Bischofs et al. developed a tension-elasticity model to correlate cell morphology as a consequence of adhesion to fibronectin-patterned substrates [227]. Bischofs derived a modified Laplace law from analytical results and computer simulation to describe filamentous network mechanics and contractility [227]. Such results demonstrate the coupling of extracellular adhesion influences cytoskeletal organization and cell shape.
19.6 Summary
Two major challenges, in our opinion, remain for the community. The first is describing the segments and nodes in the network. Much work is now being done at the molecular scale [229], but we do not yet have network-level models that can incorporate molecular-level information. A similar rise in single-molecule and single-fiber experimental methods [19, 20, 41, 230] has not yet resulted in better fiber models in networks, which are still largely restricted to simple descriptions (springs, beams, or worm-like chains, none of which truly captures the full range of behaviors seen at the molecular/fiber level). The challenge is even greater when nodes are considered. Pin joints, rigid crosses, and moment springs are all idealizations that do not capture the complex molecular interactions that characterize the junction between two fibers, and they are assuredly different for different biopolymers. It is expected that future scientists, armed with improved computer hardware and software, will incorporate our new knowledge of molecular detail as it arises.
The other major challenge, as suggested by our choice of title, is understanding how a network of many components can be interpreted in terms the one continuous material it appears to be on the macroscopic scale. We have seen in this paper how numerous researchers have explored the relationship between fibers and how the network properties vary with, for example, fiber or cross-link density, but a vast majority of the work has focused on these phenomena from a physical or material standpoint, working on the assumption of macroscopic homogeneity and often emphasizing a single component. While these studies represent a critical first step in any analysis, we need methods to incorporate multiple components and, as often occurs in biological systems, heterogeneity at all length scales. Only when these phenomena are understood will we truly be able to understand the behavior of the one tissue in terms of its many components.
19.7 Nomenclature
- A :
-
Cross-sectional area of a fiber, [m2]
- B :
-
Constitutive constant relating to fiber force
- c :
-
Concentration of polymer, [mol/L]
- d :
-
Spatial dimension
- dl :
-
Differential change in fiber length, [m]
- E :
-
Young’s modulus of an individual fiber, [Pa]
- U(j):
-
Exact energy of an ensemble of molecules in configuration j, [J]
- E f :
-
Fiber constitutive constant, [N]
- U ʹ j :
-
Exact energy of an ensemble of molecules in configuration j with a slight perturbation to one randomly chosen molecule, [J]
- 〈U〉:
-
Expected average energy of an ensemble of molecules, [J]
- F :
-
Force along a fiber, [N]
- G ʹ(ω):
-
Elastic modulus, [Pa]
- G ʹʹ(ω):
-
Loss modulus, [Pa]
- H :
-
Total energy of a filament, [J]
- I :
-
Area moment of inertia of a rod, [m4]
- I fil :
-
Area moment of inertia of a filament, [m4]
- J :
-
The last configuration for an ensemble of molecules undergoing an energy minimization scheme
- K b :
-
Bending stiffness of a material, [N•m2]
- K s :
-
Stretching stiffness of a material, [N]
- j :
-
A series of configurations for an ensemble of molecules
- k :
-
Boltzmann constant, 1.3806503×10–23 [J/K]
- k b :
-
Bending stiffness of an individual fiber segment of length l s , [N/m]
- k el :
-
Elastic component of stretching stiffness of an individual fiber of length l s , [N/m]
- k en :
-
Entropic component of stretching stiffness of an individual fiber of length l s , [N/m]
- k s :
-
Stretching stiffness of an individual fiber in a network, [N]
- k x :
-
Cross-link stiffness at a node, [N]
- L :
-
Length of a segment in a fiber network, [m]
- l c :
-
Mean displacement between nodes in a network, [m]
- l crit :
-
Critical length between cross-links through which a transition between affine and non-affine deformations, [m]
- l e :
-
Entanglement length for a fiber, [m]
- l p :
-
Persistence length for a fiber, [m]
- l s :
-
Length of a fiber segment [m]
- m :
-
General representation to denote the last molecule in an ensemble of molecules
- N:
-
Number of fibers in a network
- N c :
-
Number of connecting segments for N vertices in a d dimensional space
- N f :
-
Number of network degrees of freedom in a network
- N fil :
-
Number of filaments in a network
- N ν :
-
Number of nodes in a network
- n i :
-
Denotes the ith molecule of an ensemble of m molecules
- n k :
-
Normal vector
- Q :
-
Normalization quotient for calculating the average energy of an ensemble of molecules
- q :
-
A non-dimensional network density
- R :
-
Ratio of cross-linking component to actin concentration in a solution, or the radius of an individual fiber, [m]
- r :
-
Radius of an elastic filament, [m]
- S ij :
-
Cauchy stress tensor, [Pa]
- s :
-
Axial parameterization of fiber length, [m]
- s ij :
-
Microscopic stress field, [Pa]
- T :
-
Absolute temperature, [K]
- t :
-
Time, [sec]
- t j :
-
Surface traction, [Pa]
- u :
-
Transverse displacement, [m]
- u k,i :
-
Boundary displacement, [m]
- ∇2 u :
-
Curvature, [m–1]
- V :
-
Volume, [m3]
- x :
-
Power law exponent relating biopolymer concentration to elastic modulus
- y :
-
Power law exponent relating cross-linker-polymer ratio and elastic modulus
- z :
-
Condition number of a network (average number of fibers that connect to a node)
- α :
-
Dimensionless parameter
- δ :
-
Mean spacing between nodes in a fiber network, [m]
- δ ik :
-
Kronecker delta
- ɛ :
-
Axial strain
- ɛ f :
-
Green’s strain of a fiber
- γ(ωt):
-
Shear strain
- \(\dot\gamma\left({\omega t} \right)\) :
-
Shear strain rate, [s–1]
- \(\dot{\gamma} \omega t\) \(\Gamma_ k \left\{ {n_1 ,...,n_m } \right\}\) :
-
An individual configuration of m molecules
- σ(ω,t):
-
Stress, [Pa]
- λ f :
-
Stretch ratio of a fiber
- θ :
-
Angular parameterization of the tangent angle of a fiber with respect to the longitudinal fiber axis, [radians]
- ρ :
-
Total fiber length per unit area of a 2-D Mikado network, [m–1]
- ρ c :
-
Critical fiber length per unit area of a 2-D Mikado network at which percolation occurs, [m–1]
- ξ :
-
A random number on the interval (0,1), or the mesh size of a network
- ω :
-
Frequency, [radians/s]
REFERENCES
V. Vogel and G. Baneyx, “The tissue engineering puzzle: a molecular perspective,” Annu. Rev. Biomed. Eng., vol. 5, pp. 441–463, 2003.
B. I. Shraiman, “Mechanical feedback as a possible regulator of tissue growth,” Proc. Natl. Acad. Sci. USA, vol. 102, pp. 3318–3323, Mar 1, 2005.
Z. H. Syedain, J. S. Weinberg and R. T. Tranquillo, “Cyclic distension of fibrin-based tissue constructs: evidence of adaptation during growth of engineered connective tissue,” Proc. Natl. Acad. Sci. USA, vol. 105, pp. 6537–6542, May 6, 2008.
F. Grinnell, “Fibroblast biology in three-dimensional collagen matrices,” Trends Cell Biol., vol. 13, pp. 264–269, May, 2003.
F. Grinnell and C. H. Ho, “Transforming growth factor beta stimulates fibroblast-collagen matrix contraction by different mechanisms in mechanically loaded and unloaded matrices,” Exp. Cell Res., vol. 273, pp. 248–255, Feb 15, 2002.
M. Eastwood, V. C. Mudera, D. A. McGrouther and R. A. Brown, “Effect of precise mechanical loading on fibroblast populated collagen lattices: morphological changes,” Cell Motil. Cytoskeleton, vol. 40, pp. 13–21, 1998.
S. Nakagawa, P. Pawelek and F. Grinnell, “Long-term culture of fibroblasts in contracted collagen gels: effects on cell growth and biosynthetic activity,” J. Invest. Dermatol., vol. 93, pp. 792–798, Dec, 1989.
F. Grinnell, M. Zhu, M. A. Carlson and J. M. Abrams, “Release of mechanical tension triggers apoptosis of human fibroblasts in a model of regressing granulation tissue,” Exp. Cell Res., vol. 248, pp. 608–619, May 1, 1999.
M. Kjaer, “Role of extracellular matrix in adaptation of tendon and skeletal muscle to mechanical loading,” Physiol. Rev., vol. 84, pp. 649–698, Apr, 2004.
C. F. Burgoyne, J. C. Downs, A. J. Bellezza, J. K. Suh and R. T. Hart, “The optic nerve head as a biomechanical structure: a new paradigm for understanding the role of IOP-related stress and strain in the pathophysiology of glaucomatous optic nerve head damage,” Prog. Retin. Eye Res., vol. 24, pp. 39–73, 2005.
R. L. Gleason and J. D. Humphrey, “A mixture model of arterial growth and remodeling in hypertension: altered muscle tone and tissue turnover,” J. Vasc. Res., vol. 41, pp. 352–363, Jul–Aug, 2004.
J. Khoshnoodi, V. Pedchenko and B. G. Hudson, “Mammalian collagen IV,” Microsc. Res. Tech., vol. 71, pp. 357–370, May, 2008.
C. Frieden, “Polymerization of actin: mechanism of the Mg2+-induced process at pH 8 and 20 degrees C,” Proc. Natl. Acad. Sci. USA, vol. 80, pp. 6513–6517, Nov, 1983.
C. Frieden, “Actin and tubulin polymerization: the use of kinetic methods to determine mechanism,” Annu. Rev. Biophys. Biophys. Chem., vol. 14, pp. 189–210, 1985.
C. Neidl and J. Engel, “Exchange of ADP, ATP and 1: N6-ethenoadenosine 5'-triphosphate at G-actin. Equilibrium and kinetics,” Eur. J. Biochem., vol. 101, pp. 163–169, Nov 1, 1979.
A. Wegner, “Spontaneous fragmentation of actin filaments in physiological conditions,” Nature, vol. 296, pp. 266–267, Mar 18, 1982.
A. Wegner and P. Savko, “Fragmentation of actin filaments,” Biochemistry, vol. 21, pp. 1909–1913, Apr 13, 1982.
A. Patkowski, W. Eimer, J. Seils, G. Schneider, B. M. Jackusch and T. Dorfmueller, “The molecular dimensions of G-actin in solution as studied by dynamic light scattering,” Biopolymers, vol. 30, pp. 1281–1287, 1990.
H. Kojima, A. Ishijima and T. Yanagida, “Direct measurement of stiffness of single actin filaments with and without tropomyosin by in vitro nanomanipulation.” Proc. Natl. Acad. Sci. USA, vol. 91, pp. 12962, 1994.
A. Ott, M. Magnasco, A. Simon and A. Libchaber, “Measurement of the persistence length of polymerized actin using fluorescence microscopy,” Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics, vol. 48, pp. R1642–R1645, Sep, 1993.
W. Kabsch and J. Vandekerckhove, “Structure and function of actin,” Annu. Rev. Biophys. Biomol. Struct., vol. 21, pp. 49–76, 1992.
H. Lodish, A. Berk, S. L. Zipursky, P. Matsudaira, D. Baltimore and J. E. Darnell, Molecular Cell Biology, 4th ed. New York: W.H. Freeman & Company, 1999.
D. Boal, Mechanics of the Cell. Cambridge: Cambridge University Press, 2002, 406pp.
P. Cossart, “Actin-based motility of pathogens: the Arp2/3 complex is a central player,” Cell. Microbiol., vol. 2, pp. 195–205, Jun, 2000.
M. K. Gardner and D. J. Odde, “Modeling of chromosome motility during mitosis,” Curr. Opin. Cell Biol., vol. 18, pp. 639–647, Dec, 2006.
D. E. Ingber, “Tensegrity: the architectural basis of cellular mechanotransduction,” Annu. Rev. Physiol., vol. 59, pp. 575–599, 1997.
D. Stamenovic and N. Wang, “Invited review: engineering approaches to cytoskeletal mechanics,” J. Appl. Physiol., vol. 89, pp. 2085–2090, Nov, 2000.
H. Herrmann, H. Bar, L. Kreplak, S. V. Strelkov and U. Aebi, “Intermediate filaments: from cell architecture to nanomechanics,” Nat. Rev. Mol. Cell Biol., vol. 8, pp. 562–573, Jul, 2007.
D. E. Discher, R. Winardi, P. O. Schischmanoff, M. Parra, J. G. Conboy and N. Mohandas, “Mechanochemistry of protein 4.1’s spectrin-actin-binding domain: ternary complex interactions, membrane binding, network integration, structural strengthening,” J. Cell Biol., vol. 130, pp. 897–907, Aug, 1995.
C. Picart, P. Dalhaimer and D. E. Discher, “Actin protofilament orientation in deformation of the erythrocyte membrane skeleton,” Biophys. J., vol. 79, pp. 2987–3000, Dec,2000.
D. E. Discher and N. Mohandas, “Kinematics of red cell aspiration by fluorescence-imaged microdeformation,” Biophys. J., vol. 71, pp. 1680–1694, Oct, 1996.
K. E. Kadler, “Extracelluar matrix 1: fibril-forming collagens” Protein Prof., vol. 2, pp. 491–619, 1995.
P. Fratzl, Collagen: Structure and Mechanics. New York: Springer, 2008, 506 pp.
L. C. Junqueira, J. Carneiro and R. O. Kelley, Basic Histology, 9th ed. New York: McGraw-Hill, 1998.
F. H. Silver, J. W. Freeman and G. P. Seehra, “Collagen self-assembly and the development of tendon mechanical properties,” J. Biomech., vol. 36, pp. 1529–1553, Oct, 2003.
A. O. Brightman, B. P. Rajwa, J. E. Sturgis, M. E. McCallister, J. P. Robinson and S. L. Voytik-Harbin, “Time-lapse confocal reflection microscopy of collagen fibrillogenesis and extracellular matrix assembly in vitro,” Biopolymers, vol. 54, pp. 222–234, Sep, 2000.
K. E. Kadler, D. F. Holmes, J. A. Trotter and J. A. Chapman, “Collagen fibril formation,” Biochem. J., vol. 316 (Pt 1), pp. 1–11, May 15, 1996.
Y. L. Sun, Z. P. Luo, A. Fertala and K. N. An, “Direct quantification of the flexibility of type I collagen monomer,” Biochem. Biophys. Res. Commun., vol. 295, pp. 382–386, Jul 12, 2002.
J. S. Graham, A. N. Vomund, C. L. Phillips and M. Grandbois, “Structural changes in human type I collagen fibrils investigated by force spectroscopy,” Exp. Cell Res., vol. 299, pp. 335–342, Oct 1, 2004.
H. Miyazaki and K. Hayashi, “Tensile tests of collagen fibers obtained from the rabbit patellar tendon,” Biomed. Microdev., vol. 2, pp. 151–157, 1999.
J. A. van der Rijt, K. O. van der Werf, M. L. Bennink, P. J. Dijkstra and J. Feijen, “Micromechanical testing of individual collagen fibrils,” Macromol. Biosci., vol. 6, pp. 697–702, Sep 15, 2006.
Z. L. Shen, M. R. Dodge, H. Kahn, R. Ballarini and S. J. Eppell, “Stress-strain experiments on individual collagen fibrils,” Biophys. J., vol. 95, pp. 3956–3963, 2008.
L. Yang, K. O. van der Werf, C. F. Fitie, M. L. Bennink, P. J. Dijkstra and J. Feijen, “Mechanical properties of native and cross-linked type I collagen fibrils,” Biophys. J., vol. 94, pp. 2204–2211, Mar 15, 2008.
D. J. S. Hulmes, “Collagen diversity, synthesis and assembly,” in Collagen: Structure and Mechanics, P. Fratzl, Ed. New York: Springer, 2008, pp. 15–47.
G. Orsini, A. Ruggeri, Jr, A. Mazzoni, V. Papa, M. Piccirilli, M. Falconi, R. Di Lenarda and L. Breschi, “Immunohistochemical identification of type I and type III collagen and chondroitin sulphate in human pre-dentine: a correlative FEI-SEM/TEM study,” Int. Endod. J., vol. 40, pp. 669–678, Sep, 2007.
C. Guidry and F. Grinnell, “Studies on the mechanism of hydrated collagen gel reorganization by human skin fibroblasts,” J. Cell. Sci., vol. 79, pp. 67–81, Nov, 1985.
E. A. Sander and V. H. Barocas, “Biomimetic collagen tissues: collagenous tissue engineering and other applications,” in Collagen: Structure and Mechanics, P. Fratzl, Ed. New York: Springer, 2008, pp. 475–504.
P. D. Yurchenco and J. C. Schittny, “Molecular architecture of basement membranes,” FASEB J., vol. 4, pp. 1577–1590, Apr 1, 1990.
P. D. Yurchenco, “Assembly of basement membranes,” Ann. NY Acad. Sci., vol. 580, pp. 195–213, 1990.
J. H. Miner and P. D. Yurchenco, “Laminin functions in tissue morphogenesis,” Annu. Rev. Cell Dev. Biol., vol. 20, pp. 255–284, 2004.
R. B. Colvin, “Fibronectin in wound healing,” in Fibronectin, D. F. Mosher, Ed. Newyork: Academic press, pp. 213–254, 1989.
I. Wierzbicka-Patynowski and J. E. Schwarzbauer, “The ins and outs of fibronectin matrix assembly,” J. Cell. Sci., vol. 116, pp. 3269–3276, Aug 15, 2003.
M. K. Magnusson and D. F. Mosher, “Fibronectin: structure, assembly, and cardiovascular implications,” Arterioscler. Thromb. Vasc. Biol., vol. 18, pp. 1363–1370, Sep, 1998.
W. E. Fowler, R. R. Hantgan, J. Hermans and H. P. Erickson, “Structure of the fibrin protofibril,” Proc. Natl. Acad. Sci. USA, vol. 78, pp. 4872–4876, Aug, 1981.
J. W. Weisel and C. Nagaswami, “Computer modeling of fibrin polymerization kinetics correlated with electron microscope and turbidity observations: clot structure and assembly are kinetically controlled,” Biophys. J., vol. 63, pp. 111–128, Jul, 1992.
M. W. Mosesson, “Fibrinogen and fibrin polymerization: appraisal of the binding events that accompany fibrin generation and fibrin clot assembly,” Blood Coagul. Fibrin., vol. 8, pp. 257–267, Jul, 1997.
M. R. Neidert, E. S. Lee, T. R. Oegema and R. T. Tranquillo, “Enhanced fibrin remodeling in vitro with TGF-beta1, insulin and plasmin for improved tissue-equivalents,” Biomaterials, vol. 23, pp. 3717–3731, Sep, 2002.
E. D. Grassl, T. R. Oegema and R. T. Tranquillo, “Fibrin as an alternative biopolymer to type-I collagen for the fabrication of a media equivalent,” J. Biomed. Mater. Res., vol. 60, pp. 607–612, Jun 15, 2002.
V. H. Barocas, A. G. Moon and R. T. Tranquillo, “The fibroblast-populated collagen microsphere assay of cell traction force – Part 2: Measurement of the cell traction parameter,” J. Biomech. Eng., vol. 117, pp. 161–170, May, 1995.
M. L. Gardel, J. H. Shin, F. C. MacKintosh, L. Mahadevan, P. A. Matsudaira and D. A. Weitz, “Scaling of F-actin network rheology to probe single filament elasticity and dynamics,” Phys. Rev. Lett., vol. 93, pp. 188102, Oct 29, 2004.
O. Lieleg, M. M. Claessens, C. Heussinger, E. Frey and A. R. Bausch, “Mechanics of bundled semiflexible polymer networks,” Phys. Rev. Lett., vol. 99, pp. 088102, Aug 24, 2007.
P. L. Chandran and V. H. Barocas, “Microstructural mechanics of collagen gels in confined compression: poroelasticity, viscoelasticity, and collapse,” J. Biomech. Eng., vol. 126, pp. 152–166, Apr, 2004.
D. C. Morse, “Viscoelasticity of concentrated isotropic solutions of semiflexible polymers. 1. Model and stress tensor,” Macromolecules, vol. 31, pp. 7030–7043, 1998.
D. C. Morse, “Viscoelasticity of concentrated isotropic solutions of semiflexible polymers. 2. Linear response,” Macromolecules, vol. 31, pp. 7044–7067, 1998.
D. M. Knapp, V. H. Barocas, A. G. Moon, K. Yoo, L. R. Petzold and R. T. Tranquillo, “Rheology of reconstituted type I collagen gel in confined compression,” J. Rheol., vol. 41, pp. 971–993, 1997.
C. F. Schmidt, M. Baermann, G. Isenberg and E. Sackmann, “Chain dynamics, mesh size, and diffusive transport in networks of polymerized actin: a quasielastic light scattering and microfluorescence study,” Macromolecules, vol. 22, pp. 3638–3649, 1989.
B. Wagner, R. Tharmann, I. Haase, M. Fischer and A. R. Bausch, “Cytoskeletal polymer networks: the molecular structure of cross-linkers determines macroscopic properties,” Proc. Natl. Acad. Sci. USA, vol. 103, pp. 13974–13978, Sep 19, 2006.
J. H. Shin, M. L. Gardel, L. Mahadevan, P. Matsudaira and D. A. Weitz, “Relating microstructure to rheology of a bundled and cross-linked F-actin network in vitro,” Proc. Natl. Acad. Sci. USA, vol. 101, pp. 9636–9641, Jun 29, 2004.
P. A. Janmey, M. E. McCormick, S. Rammensee, J. L. Leight, P. C. Georges and F. C. MacKintosh, “Negative normal stress in semiflexible biopolymer gels,” Nat. Mater., vol. 6, pp. 48–51, Jan, 2007.
C. Storm, J. J. Pastore, F. C. MacKintosh, T. C. Lubensky and P. A. Janmey, “Nonlinear elasticity in biological gels,” Nature, vol. 435, pp. 191–194, May 12, 2005.
B. A. Roeder, K. Kokini, J. E. Sturgis, J. P. Robinson and S. L. Voytik-Harbin, “Tensile mechanical properties of three-dimensional type I collagen extracellular matrices with varied microstructure,” J. Biomech. Eng., vol. 124, pp. 214–222, Apr, 2002.
T. T. Tower, M. R. Neidert and R. T. Tranquillo, “Fiber alignment imaging during mechanical testing of soft tissues,” Ann. Biomed. Eng., vol. 30, pp. 1221–1233, Nov–Dec, 2002.
P. L. Chandran and V. H. Barocas, “Affine versus non-affine fibril kinematics in collagen networks: theoretical studies of network behavior,” J. Biomech. Eng., vol. 128, pp. 259–270, Apr, 2006.
P. L. Chandran and V. H. Barocas, “Deterministic material-based averaging theory model of collagen gel micromechanics,” J. Biomech. Eng., vol. 129, pp. 137–147, Apr, 2007.
B. A. Roeder, K. Kokini, J. P. Robinson and S. L. Voytik-Harbin, “Local, three-dimensional strain measurements within largely deformed extracellular matrix constructs,” J. Biomech. Eng., vol. 126, pp. 699–708, Dec, 2004.
Y. C. Fung, Biomechanics: Mechanical Properties of Living Tissues, 2nd ed. New York: Springer-Verlag, 1993, 568 pp.
L. Krishnan, J. A. Weiss, M. D. Wessman and J. B. Hoying, “Design and application of a test system for viscoelastic characterization of collagen gels,” Tissue Eng., vol. 10, pp. 241–252, Jan–Feb, 2004.
T. S. Girton, V. H. Barocas and R. T. Tranquillo, “Confined compression of a tissue-equivalent: collagen fibril and cell alignment in response to anisotropic strain,” J. Biomech. Eng., vol. 124, pp. 568–575, Oct, 2002.
V. C. Mow, S. C. Kuei, W. M. Lai and C. G. Armstrong, “Biphasic creep and stress relaxation of articular cartilage in compression: Theory and experiments,” J. Biomech. Eng., vol. 102, pp. 73–84, 1980.
J. K. Suh, Z. Li and S. L. Woo, “Dynamic behavior of a biphasic cartilage model under cyclic compressive loading,” J. Biomech., vol. 28, pp. 357–364, 1995.
G. C. Wood and M. K. Keech, “The formation of fibrils from collagen solutions. 1. The effect of experimental conditions: kinetic and electron-microscope studies,” Biochem. J., vol. 75, pp. 588–598, Jun, 1960.
C. B. Raub, V. Suresh, T. Krasieva, J. Lyubovitsky, J. D. Mih, A. J. Putnam, B. J. Tromberg and S. C. George, “Noninvasive assessment of collagen gel microstructure and mechanics using multiphoton microscopy,” Biophys. J., vol. 92, pp. 2212–2222, Mar 15, 2007.
A. M. Stein, D. A. Vader, L. M. Jawerth, D. A. Weitz and L. M. Sander, “An algorithm for extracting the network geometry of 3d collagen gels,” J. Microscopy, vol. 232, pp. 463–475, 2008.
A. M. Stein, D. A. Vader, D. A. Weitz and L. M. Sander, “The micromechanics of collagen I gels,” Arxiv, vol. 0807.2805, 2008.
S. Hsu, A. M. Jamieson, and J. Blackwell, “Viscoelastic studies of extracellular matrix interactions in a model native collagen gel system,” Biorheology, vol. 31, pp. 21–36, 1994.
R. M. Kuntz and W. M. Saltzman, “Neutrophil motility in extracellular matrix gels: mesh size and adhesion affect speed of migration,” Biophys. J., vol 72, pp. 1472–1480, 1997.
J. D. Humphrey, Cardiovascular Solid Mechanics. New York: Springer, 2002, 776pp.
C. F. Burgoyne, J. C. Downs, A. J. Bellezza and R. T. Hart, “Three-dimensional reconstruction of normal and early glaucoma monkey optic nerve head connective tissues,” Invest. Ophthalmol. Vis. Sci., vol. 45, pp. 4388–4399, Dec, 2004.
P. Bajcsy, S. C. Lee, A. Lin and R. Folberg, “Three-dimensional volume reconstruction of extracellular matrix proteins in uveal melanoma from fluorescent confocal laser scanning microscope images,” J. Microsc., vol. 221, pp. 30–45, Jan, 2006.
P. J. Elbischger, H. Bischof, P. Regitnig and G. A. Holzapfel, “Automatic analysis of collagen fiber orientation in the outermost layer of human arteries,” Pattern Anal. Appl., vol. 7, pp. 269–284, 2004.
B. van Rietbergen, S. Majumdar, W. Pistoia, D. C. Newitt, M. Kothari, A. Laib and P. Ruegsegger, “Assessment of cancellous bone mechanical properties from micro-FE models based on micro-CT, pQCT and MR images,” Technol. Health Care, vol. 6, pp. 413–420, Dec, 1998.
B. Van Rietbergen, R. Muller, D. Ulrich, P. Ruegsegger and R. Huiskes, “Tissue stresses and strain in trabeculae of a canine proximal femur can be quantified from computer reconstructions,” J. Biomech., vol. 32, pp. 443–451, Apr, 1999.
H. H. Bayraktar and T. M. Keaveny, “Mechanisms of uniformity of yield strains for trabecular bone,” J. Biomech., vol. 37, pp. 1671–1678, Nov, 2004.
R. Muller, A. Nazarian, P. Schneider, M. Stauber, P. Thurner, G. H. van Lenthe and R. Voide, “Functional micro-imaging at the interface of bone mechanics and biology,” in Mechanics of Biological Tissue G. A. Holzapfel and R. W. Odgen, Eds. Germany: Springer-Verlag, 2006, pp. 473–487.
C. Hitzenberger, E. Gotzinger, M. Sticker, M. Pircher and A. Fercher, “Measurement and imaging of birefringence and optic axis orientation by phase resolved polarization sensitive optical coherence tomography,” Opt. Exp., vol. 9, pp. 780–790, 2001.
M. Brezinski and J. Fujimoto, “Optical coherence tomography: high-resolution imaging innontransparent tissue,” IEEE J. Select. Topics Quant. Electron., vol. 5, pp. 1185–1192, 1999.
J. Rogowska, N. A. Patel, J. G. Fujimoto and M. E. Brezinski, “Optical coherence tomographic elastography technique for measuring deformation and strain of atherosclerotic tissues,” Heart, vol. 90, pp. 556–562, May, 2004.
M. K. Al-Qaisi and T. Akkin, “Polarization-sensitive optical coherence tomography based on polarization-maintaining fibers and frequency multiplexing,” Opt. Express, vol. 16, pp. 13032–13041, 2008.
W. Sun, A. Darling, B. Starly and J. Nam, “Computer-aided tissue engineering: overview, scope and challenges,” Biotechnol. Appl. Biochem., vol. 39, pp. 29–47, Feb, 2004.
P. Friedl and E. Brocker, “Biological confocal reflection microscopy: reconstruction of three-dimensional extracellular matrix, cell migration, and matrix reorganization,” in Image Analysis: Methods and Applications D. Hader, Ed.. Boca Raton: CRC Press, 2001, pp. 9–22.
S. L. Voytik-Harbin, B. A. Roeder, J. E. Sturgis, K. Kokini and J. P. Robinson, “Simultaneous mechanical loading and confocal reflection microscopy for three-dimensional microbiomechanical analysis of biomaterials and tissue constructs,” Microsc. Microanal., vol. 9, pp. 74–85, Feb. 2003.
J. Liu, G. H. Koenderink, K. E. Kasza, F. C. Mackintosh and D. A. Weitz, “Visualizing the strain field in semiflexible polymer networks: strain fluctuations and nonlinear rheology of F-actin gels,” Phys. Rev. Lett., vol. 98, pp. 198304, May 11, 2007.
P. D. Yurchenco and H. Furthmayr, “Self-assembly of basement membrane collagen,” Biochemistry, vol. 23, pp. 1839–1850, Apr 10, 1984.
A. S. Charonis, E. C. Tsilibary, P. D. Yurchenco and H. Furthmayr, “Binding of laminin to type IV collagen: a morphological study,” J. Cell Biol., vol. 100, pp. 1848–1853, Jun, 1985.
D. M. Shotton, B. E. Burke and D. Branton, “The molecular structure of human erythrocyte spectrin. Biophysical and electron microscopic studies,” J. Mol. Biol., vol. 131, pp. 303–329, Jun 25, 1979.
E. Katayama, “Quick-freeze deep-etch electron microscopy of the actin-heavy meromyosin complex during the in vitro motility assay,” J. Mol. Biol., vol. 278, pp. 349–367, May 1, 1998.
D. Overby, J. Ruberti, H. Gong, T. F. Freddo and M. Johnson, “Specific hydraulic conductivity of corneal stroma as seen by quick-freeze/deep-etch,” J. Biomech. Eng., vol. 123, pp. 154–161, Apr, 2001.
S. Erlandsen, C. Frethem and Y. Chen, “Field emission scanning electron microscopy (FESEM) Entering the 21st century: nanometer resolution and molecular topography of cell structure,” J. Histotechnol., vol. 23, pp. 249–259, 2000.
Y. Chen, L. Zardi and D. M. P. Peters, “high-resolution cryo-scanning electron microscopy study of the macromolecular structure of fibronectin fibrils,” Scanning, vol. 19, pp. 349–355, 1997.
B. F. McEwen and M. Marko, “The emergence of electron tomography as an important tool for investigating cellular ultrastructure,” J. Histochem. Cytochem., vol. 49, pp. 553–564, May, 2001.
C. Baldock, C. J. Gilpin, A. J. Koster, U. Ziese, K. E. Kadler, C. M. Kielty and D. F. Holmes, “Three-dimensional reconstructions of extracellular matrix polymers using automated electron tomography,” J. Struct. Biol., vol. 138, pp. 130–136, Apr–May, 2002.
P. L. Kronick and M. S. Sacks, “Quantification of vertical-fiber defect in cattle hide by small-angle light scattering,” Connect. Tissue Res., vol. 27, pp. 1–13, 1991.
T. T. Tower and R. T. Tranquillo, “Alignment maps of tissues: I. Microscopic elliptical polarimetry,” Biophys. J., vol. 81, pp. 2954–2963, Nov, 2001.
K. L. Billiar and M. S. Sacks, “A method to quantify the fiber kinematics of planar tissues under biaxial stretch,” J. Biomech., vol. 30, pp. 753–756, Jul, 1997.
M. R. Neidert and R. T. Tranquillo, “Tissue-engineered valves with commissural alignment,” Tissue Eng., vol. 12, pp. 891–903, Apr, 2006.
B. C. Isenberg, C. Williams and R. T. Tranquillo, “Small-diameter artificial arteries engineered in vitro,” Circ. Res., vol. 98, pp. 25–35, Jan 6, 2006.
E. A. Sander, T. Stylianopoulos, R. T. Tranquillo and V. H. Barocas, “Image-based biomechanics of collagen-based tissue equivalents: multiscale models compared to fiber alignment predicted by polarimetric imaging,” IEEE Eng. Med. Biol. Mag., vol. 28, pp. 10–18, 2009.
C. Jhun, M.C. Evans, V. H. Barocas and R. T. Tranquillo, “Planar biaxial mechanical behavior of bioartificial tissues possessing prescribed fiber alignment,” J. Biomech. Eng., vol. 131:081006, 2009.
E. E. Underwood, Quantitative Stereology. Reading, MA: Addison-Wesley, 1970, 274pp.
C. Howard and M. Reed, Unbiased Stereology: Three-Dimensional Measurement in Microscopy. New York: Springer, 1998.
S. C. Cowin, “Wolff’s law of trabecular architecture at remodeling equilibrium,” J. Biomech. Eng., vol. 108, pp. 83–88, Feb, 1986.
A. Odgaard, “Three-dimensional methods for quantification of cancellous bone architecture,” Bone, vol. 20, pp. 315–328, 1997.
V. H. Barocas and R. T. Tranquillo, “An anisotropic biphasic theory of tissue-equivalent mechanics: the interplay among cell traction, fibrillar network deformation, fibril alignment, and cell contact guidance,” J. Biomech. Eng., vol. 119, pp. 137–145, May, 1997.
T. P. Harrigan and R. W. Mann, “Characterization of microstructural anisotropy in orthotropic materials using a second rank tensor,” J. Mat. Sci., vol. 19, pp. 761–767, 1984.
E. A. Sander and V. H. Barocas, “Comparison of 2D fiber network orientation measurement methods,” J. Biomed. Mater. Res. A., vol. 88, pp. 322–331, 2008.
W. J. Karlon, J. W. Covell, A. D. McCulloch, J. J. Hunter and J. H. Omens, “Automated measurement of myofiber disarray in transgenic mice with ventricular expression of ras,” Anat. Rec., vol. 252, pp. 612–625, Dec, 1998.
W. J. Karlon, P. P. Hsu, S. Li, S. Chien, A. D. McCulloch and J. H. Omens, “Measurement of orientation and distribution of cellular alignment and cytoskeletal organization,” Ann. Biomed. Eng., vol. 27, pp. 712–720, Nov–Dec, 1999.
M. Yoshigi, E. B. Clark and H. J. Yost, “Quantification of stretch-induced cytoskeletal remodeling in vascular endothelial cells by image processing,” Cytometry A., vol. 55, pp. 109–118, Oct, 2003.
W. M. Petroll, H. D. Cavanagh, P. Barry, P. Andrews and J. V. Jester, “Quantitative analysis of stress fiber orientation during corneal wound contraction,” J. Cell. Sci., vol. 104 (Pt 2), pp. 353–363, Feb, 1993.
B. Pourdeyhimi and R. Dent, “Measuring fiber orientation in nonwovens: Part II: Direct tracking,” Text. Res. J., vol. 66, pp. 747–753, 1996.
J. Wu, B. Rajwa, D. L. Filmer, C. M. Hoffmann, B. Yuan, C. Chiang, J. Sturgis and J. P. Robinson, “Automated quantification and reconstruction of collagen matrix from 3D confocal datasets,” J. Microsc., vol. 210, pp. 158–165, May, 2003.
J. Wu, B. Rajwa, D. L. Filmer, C. M. Hoffmann, B. Yuan, C. S. Chiang, J. Sturgis and J. P. Robinson, “Analysis of orientations of collagen fibers by novel fiber-tracking software,” Microsc. Microanal., vol. 9, pp. 574–580, Dec, 2003.
Y. Xia and K. Elder, “Quantification of the graphical details of collagen fibrils in transmission electron micrographs,” J. Microsc., vol. 204, pp. 3–16, Oct, 2001.
J. P. Marquez, “Fourier analysis and automated measurement of cell and fiber angular orientation distributions.” Int. J. Solids Struct., vol. 43, pp. 6413–6423, 2006.
E. Ghassemieh, M. Acar and H. Versteeg, “Microstructural analysis of non-woven fabric using scanning electron microscopy and image processing. Part 2: Application to hydroentangled fabrics.” Proc. Instn. Mech. Engrs. L., vol. 216, pp. 211–218, 2002.
B. Xu and L. Yu, “Determining fiber orientation distribution in nonwovens with hough transform techniques,” Text. Res. J., vol. 67, pp. 563–571, 1997.
A. Takahashi, R. Kita, T. Shinozaki, K. Kubota and M. Kaibara, “Real space observation of three-dimensional network structure of hydrated fibrin gel,” Coll. Polym. Sci., vol. 281, pp. 832–838, 2003.
R. C. Gonzalez, R. E. Woods and S. L. Eddins, Digital Image Processing using MATLAB. Upper Saddle River, NJ: Pearson Prentice Hall, 2004, 609pp.
L. J. Gibson and M. F. Ashby, Cellular Solids: Structures and Properties, 2nd ed. Oxford: Pergamon Press, 1997.
C. Heussinger and E. Frey, “Stiff polymers, foams, and fiber networks,” Phys. Rev. Lett., vol. 96, pp. 017802, Jan 13, 2006.
C. Heussinger and E. Frey, “Role of architecture in the elastic response of semiflexible polymer and fiber networks,” Phys. Rev. E. Stat. Nonlin Soft Matter Phys., vol. 75, pp. 011917, Jan, 2007.
B. Pourdeyhimi, R. Ramanathan and R. Dent, “Measuring fiber orientation in nonwovens: Part I: Simulation,” Text. Res. J., vol. 66, pp. 713–722, 1996.
D. A. Head, A. J. Levine and F. C. MacKintosh, “Distinct regimes of elastic response and deformation modes of cross-linked cytoskeletal and semiflexible polymer networks,” Phys. Rev. E. Stat. Nonlin Soft Matter Phys., vol. 68, pp. 061907, Dec, 2003.
M. Kellomaki, J. Astrom and J. Timonen, “Rigidity and dynamics of random spring networks,” Phys. Rev. Lett., vol. 77, pp. 2730–2733, Sep 23, 1996.
A. M. Sastry, C. Cheng and C. W. Wang, “Mechanics of stochastic fibrous networks,” J. Thermoplast. Compos. Mater., vol. 11, pp. 288–296, 1998.
C. W. Wang, L. Berhan and A. M. Sastry, “Structure, mechanics and failure of stochastic fibrous networks: Part I: Microscale considerations,” J. Eng. Mater. Technol., vol. 122, pp. 450–468, 2000.
P. R. Onck, T. Koeman, T. van Dillen and E. van der Giessen, “Alternative explanation of stiffening in cross-linked semiflexible networks,” Phys. Rev. Lett., vol. 95, pp. 178102, Oct 21, 2005.
B. Agoram and V. H. Barocas, “Coupled macroscopic and microscopic scale modeling of fibrillar tissues and tissue equivalents,” J. Biomech. Eng., vol. 123, pp. 362–369, Aug, 2001.
T. Stylianopoulos and V. H. Barocas, “Volume-averaging theory for the study of the mechanics of collagen networks,” Comput. Methods Appl. Mech. Eng., 2007.
T. Stylianopoulos and V. H. Barocas, “Multiscale, structure-based modeling for the elastic mechanical behavior of arterial walls,” J. Biomech. Eng., vol. 129, pp. 611–618, Aug, 2007.
N. Metropolis, A. Rosenbluth, M. Rosenbluth, A. Teller and E. Teller, “Perspective on ‘‘Equation of state calculations by fast computing machines’,” J. Chem. Phys., vol. 21, pp. 1087–1092, 1953.
W. Hastings, “Monte Carlo sampling methods using Markov chains and their applications,” Biometrika, vol. 57, pp. 97–109, 1970.
M. Allen and D. Tildesley, Computer Simulation of Liquids. USA: Oxford University Press, 1989.
D. P. Landau and K. Binder, A Guide to Monte Carlo Simulations in Statistical Physics. New York: Cambridge University Press, 2005.
F. J. Vesely, Computational Physics: An Introduction. New York: Kluwer Academic/Plenum Publishers, 2001.
J. Howard, Mechanics of Motor Proteins and the Cytoskeleton. Sunderland, MA: Sinauer Associates, 2001, 367pp.
M. Doi and S. F. Edwards, The Theory of Polymer Dynamics. New York: Oxford University Press, 1986.
F. C. MacKintosh, J. Kas and P. A. Janmey, “Elasticity of semiflexible biopolymer networks,” Phys. Rev. Lett., vol. 75, pp. 4425–4428, Dec 11, 1995.
Y. Lanir, “Constitutive equations for fibrous connective tissues,” J. Biomech., vol. 16, pp. 1–12, 1983.
K. L. Billiar and M. S. Sacks, “Biaxial mechanical properties of the native and glutaraldehyde-treated aortic valve cusp: Part II – A structural constitutive model,” J. Biomech. Eng., vol. 122, pp. 327–335, Aug, 2000.
P. M. Pinsky, D. van der Heide and D. Chernyak, “Computational modeling of mechanical anisotropy in the cornea and sclera,” J. Cataract Refract. Surg., vol. 31, pp. 136–145, Jan, 2005.
Y. Lanir, “The rheological behavior of the skin: experimental results and a structural model,” Biorheology, vol. 16, pp. 191–202, 1979.
T. M. Quinn and V. Morel, “Microstructural modeling of collagen network mechanics and interactions with the proteoglycan gel in articular cartilage,” Biomech. Model. Mechanobiol., vol. 6, pp. 73–82, Jan, 2007.
H. Hatami-Marbini and R. Picu, “Scaling of nonaffine deformation in random semiflexible fiber networks,” Phys. Rev. E, vol. 77, pp. 62103, 2008.
A. M. Pizzo, K. Kokini, L. C. Vaughn, B. Z. Waisner and S. L. Voytik-Harbin, “Extracellular matrix (ECM) microstructural composition regulates local cell-ECM biomechanics and fundamental fibroblast behavior: a multidimensional perspective,” J. Appl. Physiol., vol. 98, pp. 1909–1921, May, 2005.
D. G. Hepworth, A. Steven-fountain, D. M. Bruce and J. F. Vincent, “Affine versus non-affine deformation in soft biological tissues, measured by the reorientation and stretching of collagen fibres through the thickness of compressed porcine skin,” J. Biomech., vol. 34, pp. 341–346, Mar, 2001.
C. Heussinger, B. Schaefer and E. Frey, “Nonaffine rubber elasticity for stiff polymer networks,” Phys. Rev. E. Stat. Nonlin Soft Matter Phys., vol. 76, pp. 031906, Sep, 2007.
Q. Wen, A. Basu, J. P. Winer, A. Yodh and P. A. Janmey, “Local and global deformations in a strain-stiffening fibrin gel,” New J. Phys., vol. 9, pp. 428, 2007.
D. Rodney, M. Fivel and R. Dendievel, “Discrete modeling of the mechanics of entangled materials,” Phys. Rev. Lett., vol. 95, pp. 108004, Sep 2, 2005.
J. C. Maxwell, “On the calculation of the equilibrium and stiffness of frames,” Philosophical Magazine, vol. 27, pp. 598–604, 1864.
M. Wyart, H. Liang, A. Kabla and L. Mahadevan, “Elasticity of soft particles and colloids near Random Close Packing,” Arxiv, vol. 0806.4653, 2008.
G. A. Buxton and N. Clarke, ““Bending to stretching” transition in disordered networks,” Phys. Rev. Lett., vol. 98, pp. 238103, Jun 8, 2007.
J. S. Palmer and M. C. Boyce, “Constitutive modeling of the stress–strain behavior of F-actin filament networks,” Acta Biomaterialia, vol. 4, pp. 597–612, 2008.
J. E. Bischoff, E. M. Arruda and K. Grosh, “Finite element modeling of human skin using an isotropic, nonlinear elastic constitutive model,” J. Biomech., vol. 33, pp. 645–652, Jun, 2000.
I. Klapper and H. Qian, “Remarks on discrete and continuous large-scale models of DNA dynamics,” Biophys. J., vol. 74, pp. 2504–2514, 1998.
K. J. Bathe, “Finite Element Procedures,” New Jersery: Prentice-Hall, 1996.
J. H. Shin, L. Mahadevan, P. T. So and P. Matsudaira, “Bending stiffness of a crystalline actin bundle,” J. Mol. Biol., vol. 337, pp. 255–261, Mar 19, 2004.
C. Heussinger, M. Bathe and E. Frey, “Statistical mechanics of semiflexible bundles of wormlike polymer chains,” Phys. Rev. Lett., vol. 99, pp. 048101, Jul 27, 2007.
M. Bathe, C. Heussinger, M. M. Claessens, A. R. Bausch and E. Frey, “Cytoskeletal bundle mechanics,” Biophys. J., vol. 94, pp. 2955–2964, Apr 15, 2008.
D. A. Head, F. C. MacKintosh and A. J. Levine, “Nonuniversality of elastic exponents in random bond-bending networks,” Phys. Rev. E. Stat. Nonlin Soft Matter Phys., vol. 68, pp. 025101, Aug, 2003.
J. Wilhelm and E. Frey, “Elasticity of stiff polymer networks,” Phys. Rev. Lett., vol. 91, pp. 108103, Sep 5, 2003.
E. M. Huisman, T. van Dillen, P. R. Onck and E. Van der Giessen, “Three-dimensional cross-linked F-actin networks: relation between network architecture and mechanical behavior,” Phys. Rev. Lett., vol. 99, pp. 208103, Nov 16, 2007.
C. Wang and A. Sastry, “Structure, mechanics and failure of stochastic fibrous networks: Part II—Network simulations and application,” J. Eng. Mater. Technol., vol. 122, pp. 460, 2000.
K. K. Brewer, H. Sakai, A. M. Alencar, A. Majumdar, S. P. Arold, K. R. Lutchen, E. P. Ingenito and B. Suki, “Lung and alveolar wall elastic and hysteretic behavior in rats: effects of in vivo elastase treatment,” J. Appl. Physiol., vol. 95, pp. 1926–1936, Nov, 2003.
R. Hill, “Elastic properties of reinforced solids: some theoretical principles,” J. Mech. Phys. Solids, vol. 11, pp. 357–372, 1963.
L. J. Gibson, “Biomechanics of cellular solids,” J. Biomech., vol. 38, pp. 377–399, Mar, 2005.
L. J. Gibson, “The mechanical behaviour of cancellous bone,” J. Biomech., vol. 18, pp. 317–328, 1985.
S. Vajjhala, A. M. Kraynik and L. J. Gibson, “A cellular solid model for modulus reduction due to resorption of trabeculae in bone,” J. Biomech. Eng., vol. 122, pp. 511–515, 2000.
E. A. Sander, D. A. Shimko, K. C. Dee and E. A. Nauman, “Examination of apparent and cellular level properties of human vertebral cancellous bone using combined cellular solid models,” Biomech. Model. Mechanobiol., vol. 2, pp. 97–107, Nov, 2003.
E. A. Sander, J. C. Downs, R. T. Hart, C. F. Burgoyne and E. A. Nauman, “A cellular solid model of the lamina cribrosa: mechanical dependence on morphology,” J. Biomech. Eng., vol. 128, pp. 879–889, Dec, 2006.
M. J. Silva and L. J. Gibson, “The effects of non-periodic microstructure and defects on the compressive strength of two-dimensional cellular solids,” Int. J. Mech. Sci., vol. 39, pp. 549–563, 1997.
D. A. Puleo and A. Nanci, “Understanding and controlling the bone-implant interface,” Biomaterials, vol. 20, pp. 2311–2321, 1999.
M. H. Schwartz, P. H. Leo and J. L. Lewis, “A microstructural model for the elastic response of articular cartilage,” J. Biomech., vol. 27, pp. 865–873, Jul, 1994.
R. G. Breuls, B. G. Sengers, C. W. Oomens, C. V. Bouten and F. P. Baaijens, “Predicting local cell deformations in engineered tissue constructs: a multilevel finite element approach,” J. Biomech. Eng., vol. 124, pp. 198–207, Apr, 2002.
S. Nemat-Nasser and M. Hori, Micromechanics: Overall Properties of Heterogeneous Materials, 2nd ed. Amsterdam: North Holland, 1999.
M. Oda and K. Iwashita, Mechanics of Granular Materials: An Introduction. Rotterdam: Aa Balkema, 1999.
T. Stylianopoulos, C. A. Bashur, A. S. Goldstein, S. A. Guelcher and V. H. Barocas, “Computational predictions of the tensile properties of electrospun fiber meshes: effect of fiber diameter and fiber orientation.” J. Mech. Behav. Biomed. Mat., vol. 1, pp. 326–333, 2008.
J. Soulhat, M. Buschmann and A. Shirazi-Adl, “A fibril-network-reinforced biphasic model of cartilage in unconfined compression,” J. Biomech. Eng., vol. 121, pp. 340, 1999.
D. A. Drew, “Averaged field equations for two-phase media,” Stud. Appl. Math., vol. L, pp. 133–166, 1971.
B. Hinner, M. Tempel, E. Sackmann, K. Kroy and E. Frey, “Entanglement, elasticity, and viscous relaxation of actin solutions,” Phys. Rev. Lett., vol. 81, pp. 2614–2617, 1998.
M. Claessens, R. Tharmann, K. Kroy and A. Bausch, “Microstructure and viscoelasticity of confined semiflexible polymer networks,” Nat. Phys., vol. 2, pp. 186, 2006.
O. Lieleg and A. Bausch, “Cross-linker unbinding and self-similarity in bundled cytoskeletal networks,” Phys. Rev. Lett., vol. 99, pp. 158105, 2007.
R. Tharmann, M. M. Claessens and A. R. Bausch, “Viscoelasticity of isotropically cross-linked actin networks,” Phys. Rev. Lett., vol. 98, pp. 088103, Feb 23, 2007.
M. L. Gardel, J. H. Shin, F. C. MacKintosh, L. Mahadevan, P. Matsudaira and D. A. Weitz, “Elastic behavior of cross-linked and bundled actin networks,” Science, vol. 304, pp. 1301–1305, May 28, 2004.
P. A. Janmey, U. Euteneuer, P. Traub and M. Schliwa, “Viscoelastic properties of vimentin compared with other filamentous biopolymer networks,” J. Cell Biol., vol. 113, pp. 155–160, 1991.
J. Leterrier, J. Kas, J. Hartwig, R. Vegners and P. Janmey, “Mechanical effects of neurofilament cross-bridges. Modulation by phosphorylation, lipids, and interactions with F-actin,” J. Biol. Chem., vol. 271, pp. 15687, 1996.
N. Wang and D. Stamenovic, “Contribution of intermediate filaments to cell stiffness, stiffening, and growth,” Am. J. Physiol. – Cell Physiol., vol. 279, pp. 188–194, 2000.
N. Wang, J. P. Butler and D. E. Ingber, “Mechanotransduction across the cell surface and through the cytoskeleton,” Science, vol. 260, pp. 1124–1127, May 21, 1993.
D. E. Ingber, “Opposing views on tensegrity as a structural framework for understanding cell mechanics,” J. Appl. Physiol., vol. 89, pp. 1663–1670, Oct, 2000.
M. F. Coughlin and D. Stamenovic, “A tensegrity model of the cytoskeleton in spread and round cells,” J. Biomech. Eng., vol. 120, pp. 770–777, 1998.
D. Stamenovic and D. Ingber, “Models of cytoskeletal mechanics of adherent cells,” Biomech. Model. Mechanobiol., vol. 1, pp. 95–108, 2002.
C. P. Brangwynne, F. C. MacKintosh, S. Kumar, N. A. Geisse, J. Talbot, L. Mahadevan, K. K. Parker, D. E. Ingber and D. A. Weitz, “Microtubules can bear enhanced compressive loads in living cells because of lateral reinforcement,” J. Cell Biol., vol. 173, pp. 733, 2006.
D. E. Discher, D. H. Boal and S. Boey, “Phase transitions and anisotropic responses of planar triangular nets under large deformation,” Phys. Rev. E, vol. 55, pp. 4762–4772, 1997.
S. K. Boey, D. H. Boal and D. E. Discher, “Simulations of the erythrocyte cytoskeleton at large deformation. I. Microscopic models,” Biophys. J., vol. 75, pp. 1573–1583, 1998.
R. Waugh and E. A. Evans, “Thermoelasticity of red blood cell membrane,” Biophys. J., vol. 26, pp. 115–131, Apr, 1979.
D. E. Discher, D. H. Boal and S. K. Boey, “Simulations of the erythrocyte cytoskeleton at large deformation. II. Micropipette aspiration,” Biophys. J., vol. 75, pp. 1584–1597, 1998.
J. C. M. Lee, D. T. Wong and D. E. Discher, “Direct measures of large, anisotropic strains in deformation of the erythrocyte cytoskeleton,” Biophys. J., vol. 77, pp. 853–864, 1999.
J. Li, M. Dao, C. Lim and S. Suresh, “Spectrin-level modeling of the cytoskeleton and optical tweezers stretching of the erythrocyte,” Biophys. J., vol. 88, pp. 3707–3719, 2005.
X. Luo, T. Stylianopoulos, V. H. Barocas and M. S. Shephard, “Multiscale computation for bioartificial soft tissues with complex geometries,” Eng. Comput., vol. 25, pp. 87–95, 2008.
N. Stergiopulos, S. Vulliemoz, A. Rachev, J. J. Meister and S. E. Greenwald, “Assessing the homogeneity of the elastic properties and composition of the pig aortic media,” J. Vasc. Res., vol. 38, pp. 237–246, May–Jun, 2001.
M. Frisen, M. Magi, I. Sonnerup and A. Viidik, “Rheological analysis of soft collagenous tissue. Part I: theoretical considerations,” J. Biomech., vol. 2, pp. 13–20, Mar, 1969.
P. S. Robinson, “Development of a functional tissue-engineered heart valve replacement,” PhD dissertation, University of Minnesota, 2007.
S. Thomopoulos, G. M. Fomovsky, P. L. Chandran and J. W. Holmes, “Collagen fiber alignment does not explain mechanical anisotropy in fibroblast populated collagen gels,” J. Biomech. Eng., vol. 129, pp. 642, 2007.
H. J. Burd, “A structural constitutive model for the human lens capsule,” Biomech. Model. Mechanobiol, Jul 13, 2008.
F. S. A. Cavalcante, S. Ito, K. Brewer, H. Sakai, A. M. Alencar, M. P. Almeida, J. S. Andrade, A. Majumdar, E. P. Ingenito and B. Suki, “Mechanical interactions between collagen and proteoglycans: implications for the stability of lung tissue,” J. Appl. Physiol., vol. 98, pp. 672–679, 2005.
P. A. DiMilla, K. Barbee and D. A. Lauffenburger, “Mathematical model for the effects of adhesion and mechanics on cell migration speed.” Biophys. J., vol. 60, pp. 15, 1991.
N. Wang and D. E. Ingber, “Control of cytoskeletal mechanics by extracellular matrix, cell shape, and mechanical tension,” Biophys. J., vol. 66, pp. 2181–2189, 1994.
I. B. Bischofs, F. Klein, D. Lehnert, M. Bastmeyer and U. S. Schwarz, “Filamentous network mechanics and active contractility determine cell and tissue shape,” Biophys. J., vol. 95, pp. 3488–3496, 2008.
M. J. Buehler, “Hierarchical nanomechanics of collagen fibrils: Atomistic and molecular modeling,” in Collagen: Structure and Mechanics, Anonymous. New York: Springer, 2008, pp. 175–248.
S. J. Eppell, B. N. Smith, H. Kahn and R. Ballarini, “Nano measurements with micro-devices: mechanical properties of hydrated collagen fibrils,” J. R. Soc. Interface, vol. 3, pp. 117–121, Feb 22, 2006.
A. Desai and T. J. Mitchison, “Microtubule polymerization dynamics,” Annu. Rev. Cell Dev. Biol., vol. 13, pp. 83–117, 1997.
V. Bennett and D. Gilligan, “The spectrin-based membrane skeleton and micron-scale organization of the plasma membrane,” Annu. Rev. Cell Biol., vol. 9, pp. 27–66, 1993.
J. P. Collet, D. Park, C. Lesty, J. Soria, C. Soria, G. Montalescot, and J. W. Weisel, “Influence of fibrin network conformation and fibrin fiber diameter on fibrinolysis speed: dynamic and structural approaches by confocal microscopy,” Aterioscler. Thromb. Vasc. Biol., vol. 20, pp. 1354–1361, 2000.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2010 Springer Science+Business Media B.V.
About this chapter
Cite this chapter
Sander, E., Stein, A., Swickrath, M., Barocas, V. (2010). Out of Many, One: Modeling Schemes for Biopolymer and Biofibril Networks. In: Dumitrica, T. (eds) Trends in Computational Nanomechanics. Challenges and Advances in Computational Chemistry and Physics, vol 9. Springer, Dordrecht. https://doi.org/10.1007/978-1-4020-9785-0_19
Download citation
DOI: https://doi.org/10.1007/978-1-4020-9785-0_19
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-1-4020-9784-3
Online ISBN: 978-1-4020-9785-0
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)