Abstract
Combined methylmalonic aciduria and homocystinuria, cblC type (MMACHC), is the most common inborn error of cellular vitamin B12 metabolism and is caused by mutations in the MMACHC gene. This metabolic disease results in impaired intracellular synthesis of adenosylcobalamin and methylcobalamin, coenzymes for the methylmalonyl-CoA mutase and methionine synthase enzymes, respectively. The inability to produce normal levels of these two coenzymes leads to increased concentrations of methylmalonic acid and homocysteine in plasma and urine, together with normal or decreased concentration of methionine in plasma. Here, we report a novel homozygous deletion mutation (NM_015506.2:c.392_394del) resulting in an in-frame deletion of amino acid Gln131 and late-onset disease in a 23-year-old male. The patient presented with sensory and motoric disabilities, urine and fecal incontinence, and light cognitive impairment. There was an excessive urinary excretion of methylmalonic acid and greatly elevated plasma homocysteine. The clinical symptoms and the laboratory abnormalities responded partly to treatment with hydroxycobalamin, folinic acid, methionine, and betaine. Studies on patient fibroblasts together with spectroscopic activity assays on recombinant MMACHC protein reveal that Gln131 is crucial in order to maintain enzyme activity. Furthermore, structural analyses show that Gln131 is one of only two residues making hydrogen bonds to the tail of cobalamin. Circular dichroism spectroscopy indicates that the 3D structure of the deletion mutant is folded but perturbed compared to the wild-type protein.
Competing interests: None declared
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Introduction
Combined methylmalonic acidemia and homocystinuria, cblC type, is an autosomal recessive inborn error of intracellular cobalamin metabolism that can cause a wide variety of symptoms including developmental, hematologic, neurological, metabolic, ophthalmologic, and dermatologic abnormalities (Martinelli et al. 2011). The clinical picture is divided into two main forms according to the debut age of symptoms: The early onset form starts usually within the first year of life with more serious and general organ manifestations, while the late-onset form is milder and tends to mainly involve the neurological system. The molecular basis for this distinction is poorly understood. The MMACHC disorder, which is the most common inborn error of cobalamin metabolism, is caused by mutations in the MMACHC gene and results in impaired intracellular synthesis of adenosylcobalamin (AdoCbl) and methylcobalamin (MeCbl), coenzymes for the methylmalonyl-CoA mutase and methionine synthase enzymes, respectively. Reduced activity of these two enzymes leads to increased concentrations of methylmalonic acid (MMA) and homocysteine in plasma and urine, together with normal or decreased concentration of methionine in plasma (Thiele and Van Raamsdonk 2006). The gene MMACHC was identified in 2006 and is located on chromosome 1p34.1 (Lerner-Ellis et al. 2006). The gene encodes a protein of 282 amino acids with a predicted molecular weight of 31.7 kDa. Although the exact function of the protein is currently unknown, it appears to play an important role in the synthesis of cobalamin intermediates. For instance, it has been shown that MMACHC can catalyze a reductive decyanation reaction of cyanocobalamin (CNCbl) to yield cob(II)alamin (Kim et al. 2008) and a dealkylation reaction of AdoCbl or MeCbl using glutathione as an electron donor (Hannibal et al. 2009; Kim et al. 2009). Furthermore, MMACHC has been proposed to interact with combined methylmalonic aciduria and homocystinuria cblD type (MMADHC), the downstream protein in the cobalamin pathway (Deme et al. 2012; Plesa et al. 2011). Recently the three-dimensional structure of human MMACHC was determined (Froese et al. 2012; Koutmos et al. 2011). This provides a structural framework to understand the effects of MMACHC mutations and gives new biochemical insight into the catalytic functions of MMACHC.
Here, we have used three-dimensional structure analyses in combination with fibroblast studies and spectroscopic MMACHC activity assay to verify the predicted enzyme impairment of a novel MMACHC deletion mutation from a patient suffering from this inherited cobalamin disorder. This study was performed after receiving written informed consent from the patient involved and in accordance with the guidelines of the institutional ethics committee.
Materials and Methods
Initial Clinical Picture
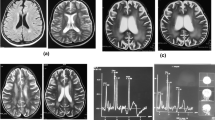
The 23-year-old patient was adopted from an East-Asian country at the age of 8 months. Nothing is known about his parents or relatives. Apart from some possible slightly impaired intellectual disability, he had an unremarkable medical history until January 2009 when he felt tired and worn out and complained of headache. It was interpreted as a flu-like respiratory tract infection. Four weeks later, he was admitted in the local hospital after an acute onset of atactic gait, numbness, and partial paralysis in the lower limbs. Within two days, he developed a fulminant picture of cerebellar edema, verified by CT and MR imaging, and was transferred to another hospital where he was treated with large doses of steroids and transient occipital cranioectomy for pressure relief. A new MR examination demonstrated an extended high intensity lesion in the spinal cord at levels Th9–Th11 and C4–C6. The cerebrospinal fluid contained traces of blood, leukocytes 33 per mm3, and total protein 0.71 g/L. CSF isoelectric focusing yielded negative results. On the basis of the clinical picture and a long series of negative serum analyses, he was suspected to have an acute demyelinating encephalomyelitis (ADEM). Recovery was nearly complete when he, in November 2009, was vaccinated against the swine flu (epidemic influenza A H1N1). Two weeks later, he again experienced the same acute symptoms as in February, except for the cerebellar edema. However, this time the peripheral neurological disabilities progressed until he, in January 2010, ended up in a wheelchair with partial paresis both in upper and lower extremities. He became incontinent to urine and partially lost the control of the defecation. Neurological examination revealed atrophy, partial loss of peripheral sensibility in both upper and lower extremities, and total loss of deep sensibility in lower limbs. There were weak tendon reflexes, however the Achilles tendon reflexes were clonic, and there was a positive bilateral Babinski sign. Neurography revealed incipient sensomotoric polyneuropathy that was not present at the first episode. From the subsequent laboratory investigations, an intracellular defect of cobalamin handling was suspected (see Results and Discussion).
Fibroblast Studies
Methionine and serine formation was measured as described by Fowler et al. (1997). [57Co]Cobalamin incorporation and coenzyme synthesis was performed as previously described (Suormala et al. 2004). Propionate incorporation in intact fibroblasts was studied according to the method of Willard et al. (1976).
Cloning and Site-Directed Mutagenesis of MMACHC
cDNAs encoding full length and a truncated version (residues 1–235) of MMACHC were synthesized with codons optimized for expression in Escherichia coli (E. coli) (GenScript). The cDNAs were subcloned into the EcoRI and HindIII sites of the pETM-11 vector (EMBL collection), which includes an N-terminal His6-tag and a TEV (tobacco etch virus) protease cleavage site in front of the inserted gene.
The QuikChange site-directed mutagenesis kit (Stratagene) was used to introduce the patient’s deletion mutation in the pETM-11-MMACHC construct. The oligonucleotide primers used were designed using the manufacturer’s protocol: 5′-GCTTACTACTACC*GACAAGATGTGG-3′, 5′-CCACATCTTGTC*GGTAGTAGTAAGC-3′ (* indicates the position of the AAC trinucleotide deletion; ΔGln131). Mutant constructs were verified by DNA sequencing.
Expression and Purification of MMACHC
MMACHC constructs were transformed into E. coli BL21(DE3) RIL Codon Plus cells (Stratagene) for overexpression. Cultures were grown in LB medium. Protein expression was induced when the cell density reached an OD600 of ~ 0.80 by addition of isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. Induced cells were grown for 18 h at 18 °C prior to harvesting by centrifugation. Cell pellets were resuspended in 300–500 mM NaCl, 100 mM Hepes pH 7.0, 5 % Glycerol, and 10 mM imidazole; the cells were lysed by sonication. Cellular debris was removed by centrifugation, and the supernatant was applied to Ni-NTA resin equilibrated in lysis buffer and eluted using 300 mM imidazole. The purified protein was dialyzed against 100 mM HEPES pH 7.0, 150 mM NaCl, and 5 % Glycerol; and frozen before further use.
DNA Extraction and Sequencing
Genomic DNA was extracted from EDTA-blood using QIAmp DNA Mini Kit (QIAGEN, Hilden, Germany). Exons 1, 2, 3, and 4 with neighboring intronic and 5′ and 3′ untranslated regions (UTRs) were amplified using AmpliTaq Gold polymerase and Gold buffer (Applied Biosystems; manufactured by Roche, Branchburg, New Jersey, USA), in a MJ Research PTC 200 Thermal Cycler (Watertown, Massachusetts). Sequencing was carried out using Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, USA) and ExoSAP-IT (USB, Cleveland, Ohio, USA) and analyzed on a 3730 DNA Analyzer (Applied Biosystems). The primers used for amplification and sequencing were from Lerner-Ellis et al. (2006). In addition, we used a second set of primers (5′-CGGACAAGGTCATAACTCCC-3′ (sense) and 5′-TCCTTTTCTTGGCAAACCC-3′ (antisense)) spanning exon 3 to eliminate the possibility of allele dropout caused by allele-specific PCR had there been a polymorphism under one of the original PCR primers.
Characterization of Cobalamin Intermediates Using UV-Vis Spectroscopy
Reaction intermediates in the MMACHC catalyzed cleavage of cyanide from cyanocobalamin were followed using UV-Vis spectroscopy. The activity of all MMACHC variants were monitored in the 250–800 nm range using a HP8454 spectrophotometer in the presence of cyanocobalamin and the reductant sodium dithionite. In all experiments, the protein concentration was maintained at about 20 μM in the working buffer consisting of 0.1 M HEPES pH 7.0, 0.5 M NaCl, and 5 % glycerol. Typically, a 0.95 mL protein sample was transferred to a screw-capped septum-sealed cuvette that went through several cycles of evacuation and purified argon gas flushing using a vacuum manifold. UV-Vis spectra were recorded before and after adding 10 μL of a 1 mM anaerobic aqueous cyanocobalamin stock solution to the sealed cuvette using an airtight Hamilton syringe. To initiate the cleavage reaction, 25–50 μL working buffer containing 10 mM sodium dithionite was added to the cuvette. While the products of dithionite oxidation do not absorb light in the 250–800 nm range, dithionite itself absorbs light at 315 nm. Thus we could ensure a reducing environment in the sealed cuvette by adding sodium dithionite until the 315 nm peak had an absorption of 0.8–1. Experiments were carried out with wild-type MMACHC, the deletion mutant ΔGln131, and the truncated variant.
Circular Dichroism Measurements
Circular dichroism spectroscopy was carried out to examine the folds of the wild-type and mutant MMACHC proteins. Samples containing 10–20 μM protein in 10 mM phosphate buffer pH 8.0 and 150 mM NaCl were transferred to 1 mm quarts cuvettes, and spectra were recorded using a Jasco J-810 spectropolarimeter at a scanning speed of 50 nm/min. For each spectrum, buffer background was subtracted and the final spectrum of each sample was the average of five consecutive scans.
Results and Discussion
Ordinary Laboratory Investigations
Measurement (ref. intervals in parentheses) of plasma amino acids revealed a low methionine 6 μmol/L (14–39), prompting a more complete metabolic screening. We found B-Hb 13.2 g/dL (13.4–17.0); normal leukocyte and platelet counts; S-vit B12 559 pmol/L (160–710); S-folate 29 nmol/L (7.1–27); P-homocysteine 158–176 μmol/L (6–16); S-MMA 118 μmol/L (0.07–0.30); P-C3-acylcarnitine 11.2 μmol/L (0.18–0.80); S-free carnitine 7 μmol/L (28–50); S-total carnitine 20 μmol/L (29–59). The urinary organic acid profile (qualitative investigation by GC-MS after methylation) showed substantially increased MMA and somewhat increased methylcitric acid. Further analyses showed the following component/creatinine ratios: U-glycine 1205 μmol/mmol (40–500); U-cystathionine 19 μmol/mmol (<8.1); U-homocystine 31 μmol/mmol (<1.1); U-guanidinoacetate 159 μmol/mmol (8–50).
Studies on Fibroblast Cells from Patient
The formation of methionine and serine in patient fibroblasts and controls grown in normal medium is shown in Table 1. The cells were grown in normal medium and with medium supplemented with different amounts of hydroxocobalamin (OHCbl) for 3 days (Table 1). The patient cells show clearly deficient methionine and serine formation when cells were grown in normal medium, whereas in medium supplemented with varying concentrations of OHCbl, there is a clear increase in the synthesis of methionine and serine. The values after addition of 1000 μg/OHCbl were 2.3 and 0.90 nmol/16 h/mg protein, respectively, i.e., full rescue of activity. Table 2 shows total uptake of [57Co]-CNCbl and cobalamin coenzyme synthesis. In the patient fibroblasts, the total content of radioactive cobalamin is very low and there is virtually no production of either methyl or adenosyl forms of cobalamin coenzymes. Propionate incorporation in intact fibroblasts is given in Table 3. When cells were grown in normal medium for 3 days, there is a clearly deficient propionate fixation. However, there is a substantial increase in cells when the medium is supplemented with a high concentration of OHCbl.
Molecular Defect
Sequencing of genomic DNA identified a homozygous deletion of three base pairs in exon 3 of MMACHC; NM_015506.2:c.392_394delAAC (Fig. 1). No other DNA sequence variants were identified in the four exons or their neighboring intronic sequences (60–289 nucleotides), 483 nucleotides of the 5' untranslated region (UTR), or the first 174 nucleotides of the 3' UTR. Since both primer sets yielded identical results for the exon 3 deletion, allelic dropout caused by a sequence variant under a primer was ruled out. The mutation identified deletes the last two nucleotides from the triplet coding for Gln131 and the first nucleotide of the codon for Arg132. However, as the first base in these two codons is identical, the result at the protein level is a deletion of the first and conservation of the second amino acid, producing the in-frame deletion mutant ΔGln131. Gln131 is conserved and lies in a highly conserved region of MMACHC that partly forms the cobalamin-binding pocket.
Characterization of the Reductive Conversion of CNCbl by MMACHC Wild-Type and Deletion Mutant
For the wild-type protein, a decreased intensity of cyanocobalamin bands in the 500–600 nm region of the UV-Vis spectrum is observed when the reaction is initiated by adding the reductant sodium dithionite to the reaction mixture. This observation is expected when cyanide is cleaved from cyanocobalamin. However, the peak typical for the product cob(II)alamin at 414 nm does not appear during the reaction (Fig. 2a), complicating the exact identification of the enzymatic product. In the control experiment, where the MMACHC protein was excluded from the reaction mixture, no change in the cyanocobalamin UV-Vis spectrum was observed, indicating that the enzyme catalyzes a cyanide cleavage reaction in the presence of sodium dithionite (data not shown). The truncated form of MMACHC had catalytic activity similar to the wild-type enzyme (data not shown), whereas the deletion mutant was inactive (Fig. 2b).
Reductive conversion of CNCbl by wild-type or mutant MMACHC. (a) Conversion of CNCbl catalyzed by wt MMACHC in the presence of the reductant sodium dithionite. As CNCbl reacts, absorbance decreases at 550 nm. The inset shows a close up of the spectra around 550 nm. (b) No enzymatic activity can be observed for the deletion mutant ΔGln131
Structural Analysis of the Deletion Mutation ΔGln131 in the MMACHC Protein
We have performed structural analyses of the deletion mutant ΔGln131 based on the recently determined crystal structure of MMACHC in complex with methylcobalamin (Koutmos et al. 2011). The structure showed that MMACHC is comprised of one N-terminal and one C-terminal module connected by a long linker of about nine residues. The N-terminal core module contains a four-stranded antiparallel β-sheet flanked by α-helices and a short antiparallel two-stranded β-sheet, a fold that is characteristic of the NADP oxidase/flavin reductase family. The C-terminal module consists of four α-helices, of which the most C-terminal of the four is unstructured when MMACHC is in complex with methylcobalamin that caps the core N-terminal module. The corrin ring of cobalamin binds in the large cavity that is formed at the interface of the C-terminal cap and the N-terminal core module. The cobalamin tail is positioned in a shallow and narrow cleft on the surface of MMACHC, which is formed between helix F and the two-stranded antiparallel β-sheet consisting of β-strand 1’ and 2’ (Fig. 3). Gln131 is located in the loop connecting the two β-strands, and is one of only two hydrogen bonds between the cobalamin tail and protein in this shallow cleft (Fig. 3). The rest of the interactions are of a hydrophobic nature. It seems clear that Gln131 contributes to tethering the cobalamin tail to the protein and therefore is important for binding and positioning. Thus, a deletion of this residue will most likely disturb the complex formation between cobalamin and MMACHC. The overall folds of the wild-type protein and the deletion mutant were probed by CD-spectroscopy (Fig. 4). Both spectra exhibit features typical for proteins with mixed α-helical and β-sheet secondary structure; however, the spectral minimums are 10 nm apart. This indicates that the deletion mutant's three-dimensional structure is folded but perturbed compared to the wild-type protein.
Cartoon representation of the structure of MMACHC in complex with MeCbl. MMACHC is comprised of one N-terminal (light blue) and one C-terminal module (light green) connected by a long linker of about nine residues (purple). The corrin ring of cobalamin (yellow) binds in a large cavity that is formed at the interface of the C-terminal cap and the N-terminal core module. The cobalamin tail is positioned in a shallow and narrow cleft on the surface of MMACHC, which is formed between helix F and the two-stranded antiparallel β-sheet consisting of β-strand 1′ and 2′. Deletion of amino acid Gln131 abolishes a hydrogen bond between the cobalamin tail and the MMACHC protein (PDB id: 3SC0; (Koutmos et al. 2011)
Treatment, Laboratory Response, and Further Clinical Course
Once the diagnosis was established, the patient was substituted daily with 1 mg OHCbl i.m. and perorally 3 × 400 mg methionine, 4 mg calcium folinic acid, 2 × 6 g betaine (Cystadane®), and 3 × 3.75 mg carnitine (Biocarn®). There was an immediate decrease in the disease markers, S-MMA: 32.0, 8.1, and 6.0 μmol/L; P-total homocysteine: 93, 43, and 44 μmol/L; P-methionine 29, n.a., and 25 μmol/L; C3-carnitine: 3.5, n.a., and 1.7 μmol/L 1 month, 1 year, and 2 years respectively, after start of treatment, (n.a. = not analyzed). He gradually regained control over the bladder and anal sphincters, and peripheral sensibility was somewhat improved. At present he normally prefers to use a wheelchair, but he is able to walk up to 100 m without support. The slow and gradual improvement in symptoms parallels the change in the disease markers and is quite consistent with what is observed in other patients with a late-onset cblC disorder. An analogous mode of onset and a similar clinical picture have been described in by (Wang et al. 2012).
Conclusion
We have identified a novel deletion mutation of Gln131 in a patient with late-onset methylmalonic acidemia and homocystinuria cblC type defect. Spectroscopic studies of recombinant mutant protein in addition to patient fibroblasts studies confirm functional deficiency of intracellular cobalamin metabolism. Furthermore, analysis of the recently determined crystal structure of MMACHC in complex with MeCbl shows that amino acid Gln131 is in close proximity to MeCbl and interacts with the vitamin through a hydrogen bond which seems essential for the activity of the protein. The mutation reported here, which deletes amino acid Gln131, also sheds light on the importance of a stable association of the coenzyme precursor in the binding pocket of MMACHC for conserved functional activity. Late-onset cblC disorder has a rare occurrence, and the mutation found in this particular patient has not been described elsewhere. It is therefore difficult to predict the onset mode solely from the genetic defect. Our experiments indicate that the mutated protein possibly retains some of its structural integrity and that some of its function can be restored by appropriate coenzyme access. Patients with late-onset cblC disorder can be remarkably normal until some triggering factors take place. These can be dietary changes, rapid growth, metabolic derangement, and fever (Martinelli et al. 2011; Tsai et al. 2007), but how they intervene is largely unknown. To which extent the initial virus infection and subsequent swine flu vaccination contributed to elicit the clinical symptoms in this particular patient with a late-onset manifestation of the disorder remains to be elucidated.
References
Deme JC, Miousse IR, Plesa M, Kim JC, Hancock MA, Mah W, Rosenblatt DS, Coulton JW (2012) Structural features of recombinant MMADHC isoforms and their interactions with MMACHC, proteins of mammalian vitamin B(12) metabolism. Mol. Genet, Metab
Fowler B, Whitehouse C, Wenzel F, Wraith JE (1997) Methionine and serine formation in control and mutant human cultured fibroblasts: evidence for methyl trapping and characterization of remethylation defects. Pediatr Res 41:145–151
Froese DS, Krojer T, Wu X, Shrestha R, Kiyani W, von DF, Gravel RA, Oppermann U, Yue WW (2012) Structure of MMACHC reveals an arginine-rich pocket and a domain-swapped dimer for its B(12) processing function. Biochemistry 51:5083–5090
Hannibal L, Kim J, Brasch NE, Wang S, Rosenblatt DS, Banerjee R, Jacobsen DW (2009) Processing of alkylcobalamins in mammalian cells: a role for the MMACHC (cblC) gene product. Mol Genet Metab 97:260–266
Kim J, Gherasim C, Banerjee R (2008) Decyanation of vitamin B12 by a trafficking chaperone. Proc Natl Acad Sci U S A 105:14551–14554
Kim J, Hannibal L, Gherasim C, Jacobsen DW, Banerjee R (2009) A human vitamin B12 trafficking protein uses glutathione transferase activity for processing alkylcobalamins. J Biol Chem 284:33418–33424
Koutmos M, Gherasim C, Smith JL, Banerjee R (2011) Structural basis of multifunctionality in a vitamin B12-processing enzyme. J Biol Chem 286:29780–29787
Lerner-Ellis JP, Tirone JC, Pawelek PD, Dore C, Atkinson JL, Watkins D, Morel CF, Fujiwara TM, Moras E, Hosack AR, Dunbar GV, Antonicka H, Forgetta V, Dobson CM, Leclerc D, Gravel RA, Shoubridge EA, Coulton JW, Lepage P, Rommens JM, Morgan K, Rosenblatt DS (2006) Identification of the gene responsible for methylmalonic aciduria and homocystinuria, cblC type. Nat Genet 38:93–100
Martinelli D, Deodato F, onisi-Vici C (2011) Cobalamin C defect: natural history, pathophysiology, and treatment. J Inherit Metab Dis 34:127–135
Plesa M, Kim J, Paquette SG, Gagnon H, Ng-Thow-Hing C, Gibbs BF, Hancock MA, Rosenblatt DS, Coulton JW (2011) Interaction between MMACHC and MMADHC, two human proteins participating in intracellular vitamin B(1)(2) metabolism. Mol Genet Metab 102:139–148
Suormala T, Baumgartner MR, Coelho D, Zavadakova P, Kozich V, Koch HG, Berghauser M, Wraith JE, Burlina A, Sewell A, Herwig J, Fowler B (2004) The cblD defect causes either isolated or combined deficiency of methylcobalamin and adenosylcobalamin synthesis. J Biol Chem 279:42742–42749
Thiele J, Van Raamsdonk JM (2006) Gene discovery in methylmalonic aciduria and homocystinuria. Clin Genet 69:402–403
Tsai AC, Morel CF, Scharer G, Yang M, Lerner-Ellis JP, Rosenblatt DS, Thomas JA (2007) Late-onset combined homocystinuria and methylmalonic aciduria (cblC) and neuropsychiatric disturbance. Am J Med Genet A 143A:2430–2434
Wang X, Sun W, Yang Y, Jia J, Li C (2012) A clinical and gene analysis of late-onset combined methylmalonic aciduria and homocystinuria, cblC type, in China. J Neurol Sci 318:155–159
Willard HF, Ambani LM, Hart AC, Mahoney MJ, Rosenberg LE (1976) Rapid prenatal and postnatal detection of inborn errors of propionate, methylmalonate, and cobalamin metabolism: a sensitive assay using cultured cells. Hum Genet 34:277–283
Acknowledgments
This work was supported by grants from University of Oslo and the Norwegian Research Council of Norway and the South-East Health Authority of Norway. B.F. was supported by a grant from Swiss National Science Foundation (320000_122568 and 31003A_138521). P.H.B. also received grants from “Legatet til Henrik Homans Minde” and “Dr. Fürst medisinske laboratoriums fond til klinisk kjemisk og klinisk fysiologisk forskning”.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Additional information
Communicated by: Matthias Baumgartner
Take-Home Message
Take-Home Message
A single amino acid deletion in MMACHC causes late-onset combined methylmalonic acidemia and homocystinuria, cblC type, and neurological damage.
Rights and permissions
Copyright information
© 2013 SSIEM and Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Backe, P.H. et al. (2013). Novel Deletion Mutation Identified in a Patient with Late-Onset Combined Methylmalonic Acidemia and Homocystinuria, cblC Type. In: Zschocke, J., Gibson, K., Brown, G., Morava, E., Peters, V. (eds) JIMD Reports - Volume 11. JIMD Reports, vol 11. Springer, Berlin, Heidelberg. https://doi.org/10.1007/8904_2013_225
Download citation
DOI: https://doi.org/10.1007/8904_2013_225
Received:
Revised:
Accepted:
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-37327-5
Online ISBN: 978-3-642-37328-2
eBook Packages: MedicineMedicine (R0)