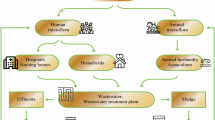
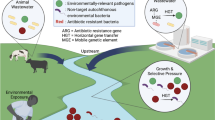
Abstract
Hospital effluents contain a hazardous amalgam of drug residues and infectious agents. Qualitative and quantitative evidence shows that hospital effluents are enriched in antibiotics, multidrug-resistant bacteria, antibiotic resistance genes and genetic vectors which could facilitate the horizontal transfer of these genes. This chapter provides an overview of the current status of antimicrobial resistance (AMR) surveillance in hospital effluents and draws comparisons to other AMR monitoring studies in domestic wastewaters and natural aquatic environments. We discuss approaches and standard tools that have been used to measure levels of AMR contamination and provide insights to the latest developments in the detection and profiling of AMR which have yet to gain traction in present surveillance programs.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Antibiotic residues
- Antibiotic resistance genes
- Antibiotic resistant bacteria
- Hospital effluent
- Resistome
1 Introduction
Antimicrobial resistance (AMR) is a menace in both community and healthcare facility settings. Hospital hygiene limitation and the overuse and misuse of antibiotics are factors contributing to the spread of antimicrobial drug resistance in hospitals [1]. The mode of transmission is complex and can occur between patients through the healthcare environment (surface, air, clothes), contaminated healthcare workers or others [2, 3]. Other sources of transmission are mediated through the use of invasive medical devices during surgical procedures which may result in hospital-acquired infections [4]. The selection pressure is imposed by the constant presence of antibiotics, which in turn accelerates the transfer of antibiotic resistance genes (ARGs) between bacteria by mobile genetic elements (MGEs) [1, 5, 6]. A transmission model of antibiotic-resistant bacteria (ARB) developed by Almagor et al. [6] showed that frequent antibiotic usage heightens the risk of transmission by increasing the vulnerability of susceptible patients and the contagiousness of colonized patients who are treated with antibiotics. The highest likelihood of AMR emergence and dissemination is through human transmission; however, hospital effluents that are loaded with microbes, infectious agents and pharmaceutical waste, originating from human sources, pose a significant public health risk if not sufficiently treated and discharged into receiving environments [7, 8].
On-site hospital wastewater treatment using advanced technologies (membrane bioreactor treatment, ozonation, granulated activated carbon, UV treatment) is capable of reducing ARGs and eliminating antibiotics in hospital effluents [9]; however, in most countries, there are no specific recommended or standardized treatments of hospital wastewaters. Hospital effluents are often routed for release into community wastewater treatment plants and co-treated with domestic wastewaters [10]. In rural areas of India, Nepal and Bangladesh, where wastewater management is inadequate, domestic effluent is directly discharged into receiving water bodies that are used as drinking water sources [11]. Despite the recognized risk of antibiotic resistance emergence and transmission, there is currently a lack of AMR surveillance in hospital wastewater. There is a clear need to survey current methods and practices, which can be applied to assess the spread of AMR from hospital effluents to the environment. This chapter reviews analytical chemistry methods, microbiological techniques and next-generation sequencing platforms which can be used to measure AMR loads in hospital effluents.
2 Antibiotic Residues
2.1 Antibiotic Residues in Hospital Effluents
Hospital effluents are important sources of antibiotics entering into the aquatic environment [12,13,14,15,16,17,18,19]. To date, analytical methods for determination and quantification of target antibiotics in hospital effluents have been well developed and validated, in which high-performance liquid chromatography coupled with tandem mass spectrometry (HPLC-MS/MS) is widely used to identify and quantify antibiotic residues in hospital effluents as well as environmental water samples [20, 21].
2.2 Challenges in Quantification of Antibiotic Residues in Hospital Effluents
To have better understanding of the occurrence, fate and environmental risk of antibiotics in hospital effluents to public health and aquatic ecosystems, the development of robust sensitive analytical methods for simultaneous extraction and analysis of the target antibiotics is critically needed [21]. One of the challenges in the determination of antibiotics in hospital effluents is related to simultaneous extraction of antibiotics from hospital effluent samples, since antibiotics are often present at very low concentrations (ng/L–μg/L) under complex matrices [21]. To date, solid phase extraction (SPE) is widely employed to enrich and purify a wide variety of target antibiotics from environmental samples [21,22,23]. However, apart from enrichment of target antibiotics, solid phase extraction may enrich some interferences that affect HPLC separation and MS/MS detection. In addition, it is challenging to extract simultaneously multiple classes of antibiotics using a single SPE cartridge as the antibiotics belonging to different classes tend to have different physicochemical properties (i.e. log Kow and pKa) and molecular structures. For these reasons, it is difficult to select a suitable single cartridge to extract simultaneously all target antibiotics in environmental samples [21].
The selection of a suitable SPE cartridge plays an important role in enhancing recovery of target antibiotics in environmental samples. Normally, the selection is often based on the physicochemical properties of target antibiotics and SPE adsorbent characteristics [21, 24, 25]. For example, Kasprzyk-Hordern et al. [25] chose a strong cation-exchange mixed-mode polymeric sorbent (Oasis MCX) for the simultaneous extraction of selected antibiotics (including ciprofloxacin, doxycycline, sulfamethoxazole, trimethoprim and erythromycin) and other pharmaceuticals and found that recovery for these analytes ranged from 61.6 to 82.5%. In another study, Babić et al. [24] used the Oasis HLB cartridge to extract seven antibiotics (sulfamethazine, sulfadiazine and sulfaguanidine, trimethoprim, oxytetracycline, enrofloxacin and penicillin G). Theoretically, the use of a specific SPE cartridge for each class of antibiotics may provide a good extraction recovery. However, this approach is time-consuming when analysing a large number of antibiotics with different physicochemical properties, and this approach is quite expensive due to SPE cartridge consumption. In a recent effort, Tran et al. [21] optimized the simultaneous extraction of 20 antibiotics and 2 antimicrobial agents belonging to 10 different classes in environmental water samples via using dual cartridges, Chromabonds HR-X (500 mg, 6 mL) [HR-X] coupled with Chromabonds SB (500 mg, 6 mL) [SB].
In addition to extraction, the detection and quantification of antibiotics in hospital effluents are challenging. To date, the use of HPLC-MS/MS is considered to be the best analytical instrument for the detection and quantification of antibiotics in wastewater matrices as it has high sensitivity and selectivity for target analytes compared to other instruments (i.e. HPLC-UV, HPLC-FID, etc.). However, the matrix effect in wastewater samples may lead to reduced detection sensitivity [13]. For example, in a previous study, Gómez et al. [13] found that significant signal suppression (ca. 85%) was observed for erythromycin when using LC-MS/MS for quantification. Hitherto, matrix effects are often corrected using a matrix-matched standard calibration method [13, 26,27,28,29], but this approach is challenging to apply for routine monitoring of environmental samples because matrices of environmental samples vary from place to place and from time to time. In such circumstances, the selection of a representative blank with a matrix composition similar to the samples is impossible. Therefore, the accuracy of the analytical methods based on matrix-matched standards calibration approach is limited. To tackle the issues regarding the losses of antibiotics during sample preparation (i.e. storage and extraction) and matrix effects during HPLC-MS/MS, the use of isotopically labelled surrogate/internal standards is deemed to be more accurate for quantification of antibiotics in environmental samples in general and hospital effluents in particular [21].
In short, the use of HPLC-MS/MS coupled with isotope dilution is a recommended option to detect and quantify antibiotics in hospital effluents as well as other environmental water samples (i.e. municipal sewage and surface water), because it allows correcting the losses, matrix effects and instrumental fluctuations during analytical processes.
2.3 Occurrence of Antibiotics in Hospital Effluents
The occurrence of multiple classes of antibiotics in hospital effluents has been well documented [13, 14, 17, 30,31,32]. For example, Gómez et al. [13] reported that the concentrations of trimethoprim and erythromycin in hospital effluents in Spain varied from 10 to 30 ng/L. In another study, Duong et al. [14] found that the concentrations of ciprofloxacin and norfloxacin in hospital wastewater in Vietnam ranged from 1.1 to 44 μg/L and from 0.9 to 17 μg/L, respectively. In a recent study, Thai et al. [17] measured the occurrence of beta-lactams, sulfonamides, macrolides, trimethoprim and fluoroquinolone in hospital effluents in Vietnam and found that the concentrations of detected antibiotics ranged substantially from below detection limit (beta-lactams) to over 40 μg/L (fluoroquinolone antibiotics). Similarly, in an earlier study in Singapore, Le et al. [32] found the presence of macrolides, fluoroquinolones, sulfonamides, beta-lactams, lincosamides, tetracycline and trimethoprim in hospital effluents, in which the maximum concentration of macrolide (clarithromycin) and fluoroquinolone (ciprofloxacin) was greater than >70 μg/L while other antibiotic classes such as lincosamides, tetracyclines and beta-lactams were rarely detected, even though beta-lactams are known to be one of the most consumed antibiotic classes. The presence and concentrations of antibiotics in hospital effluents tend to depend on the compound and type and size of hospitals.
3 Antibiotic-Resistant Bacteria (ARB)
3.1 ARB in Hospital Effluents
Hospital wastewater contains a mixture of antibiotic residues, disinfectants, metabolized and non-metabolized drugs and bacterial shedding from patients’ excreta [33,34,35]. As a result, hospital wastewater discharged to receiving waters could contribute to AMR dissemination in the natural environment if insufficiently treated [36].
Gram-negative bacteria are of particular concern in hospital settings, with the ability to cause pneumonia, bloodstream, wound, or surgical site infections [4]. In 2017, the World Health Organization (WHO) responded to the burgeoning antimicrobial resistance threat by publishing a priority list of antimicrobial-resistant pathogens which pose problems in human infections, failure to respond to current antibiotic treatment and transmissibility between humans and animals. Within the list, of highest priority are carbapenem-resistant Acinetobacter baumannii and Pseudomonas aeruginosa and carbapenem-resistant, extended spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae [37]. These guidelines provide a context to bacterial targets and patterns of resistance which should be incorporated into surveillance strategies.
3.2 Methods to Detect ARB and Commonly Used Susceptibility Testing Method
To isolate ARB in hospital effluents, wastewater samples are serially diluted and filtered through nitrocellulose membranes to trap biomass or spread-plate on media at dilutions required to capture viable bacterial populations within a countable range. Typically, Luria-Bertani medium [32] or selective media such as MacConkey agar [38] or CHROMagar [39] are used to support growth of viable bacteria. Colonies can then be sub-cultured and taxonomically characterized using Sanger sequencing targeting the 16S rRNA gene, multilocus sequence typing (MLST), MALDI-TOF bacterial identification or whole genome sequencing. Isolates identified, subjected to antibiotic susceptibility testing to determine resistance patterns, permit the determination of the ratio between ARB and total number of viable bacteria, which can be designated as prevalence. Alternatively, antibiotics are supplemented into media to directly select for the ARB growth and expressed as total concentrations of viable ARB.
To determine antibiotic minimal inhibitory concentrations (MIC) of bacterial isolates, manual procedures include the broth dilution tests, the antimicrobial gradient method and the disk diffusion test. Amongst the automated instrument systems, the BD Phoenix Automated Microbiology System (BD Diagnostics), the VITEK 2 System (bioMerieux) and the Sensititre ARIS 2X (Trek Diagnostic Systems) are the most commonly used [40]. Manual procedures such as the disk and gradient diffusion methods allow customization and cost savings. All these techniques provide qualitative assessments using the categories: susceptible (S), intermediate (I) or resistant (R). However, reliable interpretation of MIC values requires constant updating of current clinical breakpoints using either the Clinical and Laboratory Standards Institute (CLSI) or the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines of specific bacterial pathogens [40].
3.3 Occurrence of ARB in Hospital Effluents
The AMR selective pressure is particularly high in hospitals. For example, 20–30% of European inpatients receive an antibiotic treatment during their hospitalization [41], and antibiotics as well as antibiotic-resistant bacteria (ARB) excreted from inpatients receiving treatment contribute to the composition of hospital effluent [42, 43].
A study by Le et al. [32] described various taxa of viable ARB cultured from the effluents of two hospitals in Singapore, which showed resistance to different classes of clinically relevant antibiotics. Concentrations of ARB resistant to amikacin (1.06 × 106 CFU/mL), clindamycin (1.37 × 106 CFU/mL), erythromycin (1.24 × 106 CFU/ml), ciprofloxacin (1.14 × 106 CFU/mL) and tetracycline (1.30 × 106 CFU/mL) were at least one order of magnitude higher than those of meropenem (4.79 × 105 CFU/mL), ceftazidime (8.22 × 105 CFU/mL), vancomycin (9.19 × 105 CFU/mL), chloramphenicol (6.08 × 105 CFU/mL) and co-trimoxazole (2.54 × 105 CFU/mL). Using the same hospital effluent samples, Haller et al. [39] used a selective culture-based screening approach on chromogenic agar to specifically target Gram-negative extended-spectrum beta-lactamase (ESBL)-producing bacteria and bacteria with a decreased susceptibility to carbapenems (carbapenem-resistant bacteria, CRB). Concentrations of ESBL producers ranging from 103 CFU/mL to 106 CFU/mL and mean concentrations of CRB ranging between 103 CFU/mL and 105 CFU/mL were detected in the hospital wastewaters. Amongst the isolated bacterial strains, 35% were resistant to ceftazidime, and 39% were resistant to ceftriaxone (third-generation cephalosporins), while resistance to ertapenem and meropenem were 19 and 26%, respectively [39]. Another study by Korzeniewska and Harnisz [44] described a ceftazidime resistance rate of 81.6% in Enterobacteriaceae strains isolated from hospital wastewater in Poland. Picão et al. [45] measured resistance levels to third-generation cephalosporins ranging between 35 and 79% in hospital sewage in Brazil and levels of meropenem resistance of about 22%, on average.
A wide range of environmental bacteria as well as opportunistic pathogens from the gut microbiota of humans and other animals has been described in hospital wastewater discharge, but this chapter will specifically focus on those published on the WHO priority list.
3.3.1 Extended-Spectrum Beta-Lactamase-Producing Escherichia coli
The concentration of E. coli in hospital and community wastewaters typically lies within the same range (107–108 CFU/100 mL), but concentrations of ESBL-producing E. coli are generally higher in hospital effluents with percentages ranging between 3.8 and 39% [33, 35, 44, 46, 47]. This is attributed to higher incidence and density of carriage amongst inpatients compared to community carriers [48]. In addition, hospital effluents contain large quantities of antibiotics and antiseptic residues that might favour as well as further the development of ESBL-producing E. coli.
3.3.2 Multidrug-Resistant Pseudomonas aeruginosa
P. aeruginosa is increasingly recognized as an emerging opportunistic pathogen of clinical importance. It is a widespread hospital-acquired pathogen responsible for respiratory and urinary tract infections especially in intensive care units, where 15% of healthcare-associated infections are attributed to this pathogen [49]. One of its most worrying characteristics is its low antibiotic susceptibility due to its concerted action of multidrug efflux pumps and low permeability of bacterial cellular envelope, as well as the liability to acquire and express antibiotic resistance genes through plasmids, integrons or other mobile genetic elements. P. aeruginosa is noted for its intrinsic resistance to certain antibiotics and for its ability to acquire genes encoding resistance determinants [50]. Multidrug resistance in P. aeruginosa occurs mainly in clinical settings, which is a result of chromosomal mutations or horizontal gene transfer. As opposed to E. coli, P. aeruginosa is not a commensal human bacterium, and the frequency of carriage amongst inpatients is low [51]. P. aeruginosa is ubiquitous in wastewater; however, the proportion of antibiotic-resistant P. aeruginosa is much higher in hospital than in urban wastewater [49, 52,53,54,55]. Recent studies reported the presence of multidrug-resistant ESBL-producing P. aeruginosa in hospital effluents [39].
3.3.3 Antibiotic Resistant Acinetobacter baumannii
A. baumannii can cause various infections like nosocomial pneumonia, bacteraemia, meningitis, and skin and soft tissue and urinary tract infections. The incidence of serious infections (blood stream infections and ventilator-associated pneumonia) caused by multidrug-resistant A. baumannii ranges between 47 and 93%, with mortality rates between 30 and 70% [56]. The overuse of carbapenems has rapidly resulted in the worldwide dissemination of carbapenem-resistant A. baumannii strains, as reported in studies from Croatia and China [57]. The observation of multidrug-resistant A. baumannii in hospital wastewater has also been previously reported in Brazil, China and Zagreb in Croatia [58,59,60].
3.3.4 Vancomycin-Resistant Enterococci (VRE)
Enterococci are Gram-positive bacteria, which are part of the natural intestinal microbiota of animals and humans, and are released to the environment through sewage or wastewater [61]. Some members of the genus, such as Enterococcus faecalis and Enterococcus faecium, are amongst the major causes of nosocomial infections worldwide [62]. One factor contributing to the pathogenesis of enterococci is their resistance to a broad range of antibiotics. This resistance trend has increased in recent years [63]. Vancomycin is a glycopeptide antibiotic used for serious infections by Gram-positive bacteria when treatment with other antibiotics has failed. The excessive use of this antimicrobial agent has led to the appearance of vancomycin-resistant enterococci (VRE). Concentrations of enterococci in urban and hospital wastewater have been found to be similar, although the proportion of VRE was detected in higher concentrations in hospital than in urban effluents [63,64,65,66]. Varela et al. [66] reported concentrations ranging between 2.50 × 101 and 2.30 × 103 CFU/mL and between 1.60 × 101 and 2.20 × 103 CFU/mL of enterococci resistant to ciprofloxacin and vancomycin, respectively, in hospital effluents. Hospital effluent constitutes a source of enterococci having multiple resistances to antibiotics, presumably from the faeces of patients, because the rules of biosecurity in medical centres would impede other sources of contamination [67].
4 Antibiotic Resistance Genes (ARGs)
4.1 ARGs in Hospital Effluents
One of the major aspects of understanding antibiotic resistance in hospital effluents is to detect and quantify ARGs. Molecular techniques such as real-time quantitative PCR (qPCR), including singleplex, multiplex and high throughput, and metagenomics have been employed to identify and quantify ARGs in hospital effluent samples.
4.2 Application of High-Throughput qPCR (HT-qPCR) to Measure ARGs and MGE
Characterizing and quantifying resistomes are a rapid method of assessing AMR pollution. Probes and primers designed to target ARGs that confer resistance to different classes of antibiotics provide quantitative information on evaluating the abundance of genes. Unlike the traditional qPCR approach which is limited to a few targets in one assay, high-throughput qPCR (HT-qPCR) arrays are able to simultaneously quantify hundreds of ARGs and other related MGEs in one run [68]. There are a few commercially available platforms, which includes the Fluidigm Array, the Qiagen Antibiotic Resistance Genes Microbial DNA qPCR Array, OpenArray by Applied Biosystems and the WaferGen Biosystems SmartChip Real-Time PCR. Each system allows customization of primers depending on the ARGs or MGE of interest. The utility of HT-qPCR arrays has been demonstrated in environmental surveys aimed at comparing relative concentrations of ARG contamination across different aquatic sources such as lakes and estuaries [69,70,71], sediments of fish farms [72], drinking water [73] and wastewater treatment plants [74, 75]. Monitoring efforts of ARGs and MGE in hospital wastewaters are predominantly based on data generated from traditional qPCRs targeting the few clinically relevant beta-lactamase genes (e.g. blaKPC, blaNDM, blaCTX-M, blaTEM, blaSHV) [32, 76,77,78,79] with a shifting trend towards using upscaled customized HT-qPCR with increased capacity to detect more ARG targets and markers of MGE [80].
4.3 Prevalent ARGs and MGEs in Hospital Wastewaters Globally and Comparisons with Other Water Sources
High prevalence of sulfonamide (sul) and tetracycline (tet) genes has been detected in various environments and deeply studied by many groups [32, 81, 82]. In addition, investigations on MGEs such as intl1 (class 1 integron-integrase) were included in many studies as integrases have been statistically correlated with anthropogenic sources of ARGs and are potentially involved in ARGs integration in chromosomes or plasmids [32, 81]. However, owing to the rise in importance of beta-lactam resistance and WHO’s recent announcement of the global priority list that consists of beta-lactam-resistant pathogens, there is a shift in trend towards studies focused on the detection and quantification of beta-lactamase genes (e.g. blaSHV, blaCTX-M, blaOXA) [9, 82, 83]. Emerging genes such as blaKPC, blaNDM-1 and mcr-1 are of concern in recent years due to their possible origin from hospitals, their occurrences in plasmids residing in multidrug resistance “superbugs” and the potential for these genes to spread amongst the bacteria community via horizontal gene transfer [84].
A comparison of resistomes in the final effluents of seven European countries (Portugal, Spain, Ireland, Cyprus, Germany, Finland, Norway) using HT-qPCR showed that AMR profiles mirrored patterns of clinical antibiotic resistance prevalence, providing insightful information on country- or region-specific trends of AMR distribution [85]. In a study conducted in China by Li et al. [81], ten tet genes (A, B, C, G, L, M, O, Q, W, X), sul1, sul2 and intl1 were detected in hospital effluents using singleplex qPCR, with intl1 concentrations as high as 1011 gene copies (GC) per mL. Compared to residential area effluents studied in parallel with hospital effluents, the total gene abundances from hospital effluents (1.81 × 1011 GC/mL) were slightly lower as compared to residential effluents (2.79 × 1011 GC/mL). In contrast, Lamba et al. [83] compared 12 hospitals and residential effluents in New Delhi, India, where the gene targets were blaTEM, blaOXA, blaCTX-M and blaNDM-1, and found that all the ARG concentrations had higher relative abundance (normalized to 16S rRNA genes) in hospital effluents than in residential effluents. The differences in ARG abundance could be attributed to antibiotic usage and human demographics where healthy asymptomatic individuals within the community could serve as carriers of ARGs [86].
The effects of discharging untreated hospital effluent into other environments have been evaluated in a few studies. In Tamil Nadu, India, samples were taken from five hospital effluents and five points upstream and downstream of the Cauvery River Basin where genes encoding for beta-lactamase (blaTEM, blaCTX-M, blaSHV, blaNDM-1) and aminoglycosides (aadA) were quantified [79]. Results showed that blaSHV and blaNDM-1 were not present upstream but were detected downstream of the river, indicating that these genes were likely introduced by wastewater. However, it was inconclusive if the genes were derived directly from hospital effluents as the source of wastewater discharge was from a combination of effluents originating from residential areas, industries and hospitals. Rodriguez-Mozaz et al. [82] quantified five ARGs (blaTEM, ermB, qnrS, sul1, tetW) from hospital effluent; influent and effluent of a nearby wastewater treatment facility located in Girona, Spain, that receives the hospital effluent; and water upstream and downstream of the river that receives treated wastewater effluent. All the ARG targets in the hospital effluents were found to be of similar concentrations as compared to wastewater influents, ranging from 3 to 7 log GC/mL, but were significantly higher as compared to the other locations (wastewater effluent, river upstream and downstream) sampled. This implies that the ARG concentrations in domestic wastewater were of similar concentrations as compared to the hospital effluents. On the contrary, a massive study done in the Netherlands by Pallares-Vega et al. [87] concluded that healthcare facilities such as hospitals had minimal impact on the concentrations of ARGs entering wastewater treatment facilities. This could be an effect of dilution by domestic wastewater that have lower ARG concentrations resulting in an overall reduction in the abundance of ARGs. It is however worthwhile to note that within the same study, there was an increase in the relative abundance of broad-host-range IncP-1 type plasmids which are known to carry broad-spectrum ARGs and are transmissible between Gram-negative bacteria. The demographics and antibiotic usage patterns in humans and animal and differences in regulations governing the sale and use of antibiotics differ from one country to the next which could explain varying global trends in AMR.
To understand AMR trends and occurrence patterns, research groups have designed studies to compare ARG concentrations of effluents derived from different ward types across different hospitals locally to facilitate stewardship efforts. For example, a study by Le et al. [32] concluded that effluents from clinical isolation wards had higher concentrations of ARGs compared to general wards. Another study by Li et al. [81] noted significant differences in total resistance gene abundance across seven hospitals, with concentrations ranging from 107 to 1011 GC/mL. A detailed study by Lamba et al. [83] correlated the concentration of blaNDM-1 across 12 different hospitals of different capacities and found that larger hospitals were discharging higher concentrations of blaNDM-1. The authors concluded larger hospitals that receive higher volumes of inpatients likely result in higher AMR output in wastewaters.
In contrast to singleplex qPCR, HT-qPCR is able to profile a wider number of ARG and MGE targets in one run which provides higher throughput to comprehensively assess vectors of AMR in hospital effluents. In contrast to metagenomic profiling, HT-qPCR is more sensitive and requires less starting DNA material (PCR reactions at the nanolitre scale) with the ability to detect concentrations of 10−4 ARGs/16S rRNA gene [72, 88]. There are four main HT-qPCR platforms currently available in the market, with Biomark Dynamic Array (Fluidigm) requiring the lowest reaction volumes (~10 nL) followed by OpenArray (Biosystems ~35 nL reactions), WaferGen SmartChip (WaferGen ~100 nL reactions) and Bio-Rad CFX384 (Biorad ~3,000 nL reactions) [88].
In a study done in Xinxiang City, Central China, Wang et al. [80] fabricated 258 qPCR primers and utilized a HT-qPCR platform to detect 178 unique ARG targets that confer resistance to seven classes of antibiotics and two MGE targets (intl1 and Tn916/Tn1545) to compare concentrations of wastewater from three tertiary public hospitals in the city. A core of 126 ARGs were detected in all three hospital effluents. Concentrations of 12 frequently detected ARGs (tetM, tetO, tetX, ereA, ermA, ermB, sul1, sul2, sul3, qnrA, qnrB, oqxB) were validated by qPCR yielding results of highest concentrations of tetO detected in effluents of two hospitals, with sul1 detected at high abundance in the effluents of the third sampled hospital. Amongst the five MGEs (intI1, intI2, intI3, Tn916/Tn1545, ISCR1), ISCR1 had the highest abundance ranging from 107 to 108 GC/mL. It would be advantageous to use HT-qPCR for routine monitoring of hospital effluents as it is time-efficient, with a low sample volume requirement per reaction, without the reliance on complicated downstream bioinformatics analyses when compared to using metagenomics.
5 Resistomes and Mobile Genetic Elements (MGEs)
5.1 Uncovering Resistomes by Metagenomics
In the field of water research, integrated multi-omics approaches have been used as bio-monitoring tools for water quality assessment to investigate microbial composition, their functional roles and involvement in water contamination [89]. One of the advantages of metagenomics in AMR surveillance is the collective recovery of genomes from microbes in environmental samples which provides genetic insights to microbial composition (bacterial and viral), ARGs and other MGEs (e.g. plasmids, integrons, transposons). The ability to capture entire genomic profiles to track the distribution of ARGs and MGEs in a variety of environments has made it possible to assess and identify AMR hotspots in different aquatic compartments [96], sources and sinks of ARGs in environmental waters [90], ARG removal in the wastewater treatment process [91, 92] and fate and transport of ARGs in environments receiving treated wastewater effluents [93,94,95]. There are a handful of studies which have applied metagenomics as an opportunity to create ARG and microbiome catalogues of hospital wastewaters to identify novel carbapenemases [96] and to classify and resolve differences between municipal wastewaters [97, 98] and waters receiving treated hospital wastewaters [99].
Environmental resistomes are profiled by interrogating metagenomic reads or assembled contigs against one of the publically available ARG databases such as the Comprehensive Antibiotic Resistance Database [100], the Antibiotic Resistance Database [101], Resfams [102], ARG-ANNOT [103], ResFinder [104], MEGARes [105] and ARGs-OAP [106]. The relative abundance of ARGs identified from metagenomic datasets is then calculated by normalizing to the ARG reference sequence length (nucleotide) and to the number of 16S rRNA genes [107] or by coverage normalized to the ARG reference gene and size of metagenomic dataset [108].
5.2 Identifying MGEs
Intercellular mechanism of exchange mediated by MGEs such as plasmids, transposons and integrons play a major role in AMR dissemination as they facilitate the capture, transfer and expression of exogenous ARGs [109, 110]. The mobility of plasmid-borne ARGs and the rates of inter- and intraspecies transfer in hospital effluents are largely unknown. In a laboratory-scale experiment, Chen et al. [111] demonstrated plasmid transferability through mating a ceftazidime-resistant strain of A. baumannii isolated from hospital wastewaters with a ceftazidime susceptible E. coli as a recipient. Whole genome sequencing of plasmids in donor and transconjugants showed highly similar sequences, concluding that plasmid-mediated intraspecies transfer of ARGs had occurred. Interspecies transfer of ESBL-encoding plasmids between microbial community within a hospital sink environments has been inferred [112, 113]. However, there is limited data available on the frequency and environmental cues that trigger ARG transfer in clinical wastewaters.
Class 1 integron gene cassettes, which are frequently carried by human pathogens, often include ARGs acquired by genetic recombination. Hence, insight to their presence in hospital wastewater may contribute to infer about the risks associated with ARG dissemination [96]. There are a range of bioinformatics tools designed for in silico detection of integrons (I-VIP [114], MARA [115], INTEGRALL [116]) and plasmids which are carried by Enterobacteriaceae and Gram-positive bacteria (PlasmidFinder [117]). Inspecting assembled metagenomic datasets (contigs) for co-localization of ARGs and MGE features provides a means of exploring specific mechanisms that mediate the spread of certain ARG types, with this analysis approach proposed as a method to predict ARG mobility incidence in environmental resistomes [118].
5.3 Examples of the Application of Metagenomics to AMR Monitoring in Hospital Wastewaters
There is currently more literature on resistome profiles in hospital wastewaters using either qPCR or HT-qPCR, rather than metagenomics. This could be attributed to better sensitivity (detection limits) offered by qPCR platforms that are target specific and easily interpreted [88]. A metagenomics approach in contrast seizes information of known and unknown DNA sequences in a single run, thus providing a greater depth of sequence information without the restraint of a specific targeted sequence [119]. For example, within the context of AMR monitoring in wastewaters, Le at al. [32] used qPCR to detect the relative abundance of four beta-lactamase ARG targets (blaNDM, blaKPC, blaCTX-M, blaSHV). However, a more in-depth metagenomics analysis of the same samples gave a snapshot of the entire ARG diversity within the hospital wastewater samples and allowed the assembly of entire scaffolds providing information on ARG arrangements and co-occurring MGEs within the same gene neighbourhood [97]. Leveraging on the latest DNA sequencing technologies by combining long-read data from third-generation sequencing platforms (Oxford Nanopore) with Illumina short-read data has yielded better sequencing coverage and assembly of ARG bearing plasmids as described in a study of wastewater treatment plants [120].
AMR metagenomics studies conducted in Singapore [97], the Netherlands [99] and France [98] appear to have a consistent pattern of a core microbiome specific to hospital wastewaters. All three studies reported the predominance of anaerobic human gut bacteria belonging to the order Clostridiales, Bifidobacteriales and Bacteroidales. There were however other dominant bacterial taxa (e.g. Acinetobacter baumannii, Enterobacteriaceae) that contributed to differences in hospital wastewater microbiomes originating from different countries [96,97,98,99], which could explain AMR variations globally [119].
As an extension of the utility of rapid AMR and microbiome profiling, Li et al. [121] demonstrated that by integrating metagenomics datasets from different sources with complementary metadata into machine learning classification models, AMR source contribution could be identified to predict putative sources of ARG contamination. This would be particularly useful in tracking the dissemination of AMR originating from hospital effluents.
5.4 Targeted Metagenomics for Qualitative and Quantitative Resistome Analysis
One of the challenges with the application of metagenomics within complex microbial communities is the detection sensitivity of low abundant bacterial populations that harbour ARG [122]. To overcome the limitation of heterogeneity, Lanza et al. [123] adopted an in-solution targeted capture platform (TCP), a technique used for diagnosis of human-inherited diseases [124] to develop a targeted metagenomic resistome analysis method coined “ResCap”, a TCP based on SeqCapEZ (NimbleGen) technology. The TCP is designed to target ~78,000 nonredundant genes, comprising of ARGs, genes conferring resistance to metals/biocides and relaxase genes as plasmid markers [123]. Briefly, whole-metagenome shotgun libraries are constructed, and DNA captured by the probes are sequenced using Illumina platform and analysed using the ResCap bioinformatics workflow. Comparison of resistomes identified by metagenomic shotgun sequencing versus the ResCap platform showed improved gene abundance detection of 2–83% and increase of gene diversity detection by 300-fold [123]. This underscores the large proportion of ARGs that go undetected by relying on just metagenomics alone. The sensitivity and specificity of the ResCap technology provide qualitative and quantitative means of measuring the levels of ARG contamination which could potentially meet the needs of AMR monitoring and tracking from source to sink.
5.5 Other OMIC Strategies to Study ARG Expression Levels in Hospital Wastewaters
To understand the activity and contribution of ARB to AMR dissemination in linked aquatic environmental sources, a combined OMIC approach of metagenomics and metatranscriptomics was used to detect ARG transcripts in wastewaters from hospital and farm effluents into a receiving river in Cambridge, United Kingdom [125]. The authors reported a significant overexpression of blaGES and blaOXA in hospital effluents over a consistent period of 5 months relative to the two other sampled waters which was considered to be due to the levels of antibiotic usage in hospitals [125].
6 Curbing the Spread of AMR
Antimicrobial stewardship in hospitals is a prescribed intervention strategy by the WHO Global Action Plan to contain the spread of AMR beyond the clinical setting [126]. National or regional surveillance networks that monitor antibiotic usage and resistance using standardized methods will enhance knowledge on the extent of AMR severity, region-specific prevalence trends and associated health outcomes [127]. Antimicrobial peptides, probiotics, phage therapy and phage endolysins have been proposed as alternative replacements of antibiotics. However, safety and efficacy in vivo in humans have yet to be determined [128, 129].
7 Possible Treatment Technologies of Hospital Wastewaters
As hospital effluents are recognized as sources of AMR, there is increased awareness in possibly pretreating effluents before discharging them into municipal sewage. Two published studies have attempted to evaluate the removal efficiency of ARB and/or ARGs and MGE.
In Riyadh, Saudi Arabia, Timraz et al. [38] investigated the removal efficiency of wastewater treatment systems placed on site at two different hospitals. Although both treatment plants utilized conventional activated sludge process followed by chlorination, one plant outperformed in terms of log removal values of total viable bacteria and ARGs. The ARGs sul1 and intl1 remained detectable at concentrations of up to 105 GC/mL in hospital effluent from the plant with a more interior removal performance. This observation suggests that operational parameters of wastewater treatment plants play a vital role in removal efficiencies of vectors of AMR.
Paulus et al. [9] compared the removal efficiency between an advanced on-site treatment facility (membrane bioreactor/ozonation/activated carbon/UV treatment), in two Dutch cities which received hospital effluent directly, and a municipal wastewater treatment facility, which received both hospital and residential effluents. Data showed significantly higher removal efficiency for the 13 ARG targets by the advanced treatment as compared to the municipal treatment facility. The study recovered the ARGs blaKPC and vanA only in hospital effluents, which suggests that healthcare facilities are potential sources of these clinically important ARGs.
Other known methods, such as coagulation [130] and the use of biochar [131], have been demonstrated to effectively remove ARB, ARGs and antibiotic residues, although these methods have only been used to treat other types of waste other than hospital effluents. Nevertheless, these methods have the potential to pretreat hospital effluents before discharge into the main sewers.
8 Conclusion
This review covers the detection and quantification of the three main aspects of AMR (antimicrobial residue, antibiotic resistance bacteria and genes) and their occurrences in multiple hospital discharges. There is a need to step up surveillance systems of wastewater discharged from hospitals which are potential drivers for the spread of AMR. Factors which influence differences in AMR occurrence are dependent on the age and size/capacity of the hospital and the severity and types of infections amongst inpatients. The implementation of the latest molecular and OMIC techniques reviewed in this chapter could provide new standardized methods of qualitatively and quantitatively assessing the dissemination of a wider array of ARGs in different aquatic sources. Physical and chemical treatment processes can be put in place to pretreat hospital discharges in order to reduce the spread of AMR into receiving domestic wastewater treatment facilities or natural water bodies.
Abbreviations
- AMR:
-
Antimicrobial resistance
- ARB:
-
Antibiotic-resistant bacteria
- ARGs:
-
Antibiotic resistance genes
- CLSI:
-
Clinical and Laboratory Standards Institute
- CRB:
-
Carbapenem
- ESBL:
-
Extended spectrum beta-lactamase
- EUCAST:
-
European Committee on Antimicrobial Susceptibility Testing
- GC:
-
Gene copy
- HPLC-MS/MS:
-
High-performance liquid chromatography coupled with tandem mass spectrometry
- HT-qPCR:
-
High-throughput quantitative polymerase chain reaction
- MGE:
-
Mobile gene element
- MGEs:
-
Mobile genetic elements
- MLST:
-
Multilocus sequence typing
- qPCR:
-
Quantitative polymerase chain reaction
- SPE:
-
Solid phase extraction
- VRE:
-
Vancomycin-resistant enterococci
- WHO:
-
World Health Organization
References
Weinstein RA (2001) Controlling antimicrobial resistance in hospitals: infection control and use of antibiotics. Emerg Infect Dis 7(2):188–192
Pittet D, Hugonnet S, Harbarth S, Mourouga P, Sauvan V, Touveneau S et al (2000) Effectiveness of a hospital-wide programme to improve compliance with hand hygiene. Lancet 356(9238):1307–1312
Hugonnet S, Harbarth S, Sax H, Duncan RA, Pittet D (2004) Nursing resources: a major determinant of nosocomial infection? Curr Opin Infect Dis 17(4):329–333
Peleg AY, Hooper DC (2010) Hospital-acquired infections due to gram-negative bacteria. N Engl J Med 362(19):1804–1813
Partridge SR, Tsafnat G (2018) Automated annotation of mobile antibiotic resistance in gram-negative bacteria: the Multiple Antibiotic Resistance Annotator (MARA) and database. J Antimicrob Chemother 73(4):883–890
Almagor J, Temkin E, Benenson I, Fallach N, Carmeli Y (2018) The impact of antibiotic use on transmission of resistant bacteria in hospitals: insights from an agent-based model. PLoS One 13(5):1–14
Chereau F, Opatowski L, Tourdjman M, Vong S (2017) Risk assessment for antibiotic resistance in South East Asia. BMJ 358:2–8
Hong P-Y, Julian TR, Pype M-L, Jiang SC, Nelson KL, Graham D et al (2018) Reusing treated wastewater: consideration of the safety aspects associated with antibiotic-resistant bacteria and antibiotic resistance genes. Water 10(3):244
Paulus GK, Hornstra LM, Alygizakis N, Slobodnik J, Thomaidis N, Medema G (2019) The impact of on-site hospital wastewater treatment on the downstream communal wastewater system in terms of antibiotics and antibiotic resistance genes. Int J Hyg Environ Health 222(4):635–644. https://doi.org/10.1016/j.ijheh.2019.01.004
Hocquet D, Muller A, Bertrand X (2016) What happens in hospitals does not stay in hospitals: antibiotic-resistant bacteria in hospital wastewater systems. J Hosp Infect 93(4):395–402. https://doi.org/10.1016/j.jhin.2016.01.010
WHO, UNICEF (2014) A snapshot of drinking water and sanitation in WHO South-East Asia Region, pp 1–24
Kummerer K (2001) Drugs in the environment: emission of drugs, diagnostic aids and disinfectants into wastewater by hospitals in relation to other sources – a review. Chemosphere 45:957–969
Gómez MJ, Petrović M, Fernández-Alba AR, Barceló D (2006) Determination of pharmaceuticals of various therapeutic classes by solid-phase extraction and liquid chromatography-tandem mass spectrometry analysis in hospital effluent wastewaters. J Chromatogr A 1114(2):224–233
Duong HA, Pham NH, Nguyen HT, Hoang TT, Pham HV, Pham VC et al (2008) Occurrence, fate and antibiotic resistance of fluoroquinolone antibacterials in hospital wastewaters in Hanoi, Vietnam. Chemosphere 72(6):968–973
Kosma CI, Lambropoulou DA, Albanis TA (2010) Occurrence and removal of PPCPs in municipal and hospital wastewaters in Greece. J Hazard Mater 179(1–3):804–817. https://doi.org/10.1016/j.jhazmat.2010.03.075
Al Aukidy M, Verlicchi P, Voulvoulis N (2014) A framework for the assessment of the environmental risk posed by pharmaceuticals originating from hospital effluents. Sci Total Environ 493:54–64. https://doi.org/10.1016/j.scitotenv.2014.05.128
Thai PK, Ky LX, Binh VN, Nhung PH, Nhan PT, Hieu NQ et al (2018) Occurrence of antibiotic residues and antibiotic-resistant bacteria in effluents of pharmaceutical manufacturers and other sources around Hanoi, Vietnam. Sci Total Environ 645:393–400. https://doi.org/10.1016/j.scitotenv.2018.07.126
Zhang X, Zhang T, Fang HHP (2009) Antibiotic resistance genes in water environment. Appl Microbiol Biotechnol 82:397–414
Tran NH, Reinhard M, Gin KYH (2018) Occurrence and fate of emerging contaminants in municipal wastewater treatment plants from different geographical regions – a review. Water Res 133:182–207. https://doi.org/10.1016/j.watres.2017.12.029
Tran NH, Hu J, Ong SL (2013) Simultaneous determination of PPCPs, EDCs, and artificial sweeteners in environmental water samples using a single-step SPE coupled with HPLC-MS/MS and isotope dilution. Talanta 113:82–92. https://doi.org/10.1016/j.talanta.2013.03.072
Tran NH, Chen H, Do TV, Reinhard M, Ngo HH, He Y et al (2016) Simultaneous analysis of multiple classes of antimicrobials in environmental water samples using SPE coupled with UHPLC-ESI-MS/MS and isotope dilution. Talanta 159:163–173. https://doi.org/10.1016/j.talanta.2016.06.006
García-Galán MJ, Díaz-Cruz MS, Barceló D (2010) Determination of 19 sulfonamides in environmental water samples by automated on-line solid-phase extraction-liquid chromatography-tandem mass spectrometry (SPE-LC-MS/MS). Talanta 81(1–2):355–366
Tran NH, Chen H, Reinhard M, Mao F, Gin KYH (2016) Occurrence and removal of multiple classes of antibiotics and antimicrobial agents in biological wastewater treatment processes. Water Res 104:461–472. https://doi.org/10.1016/j.watres.2016.08.040
Babić S, Ašperger D, Mutavdžić D, Horvat AJM, Kaštelan-Macan M (2006) Solid phase extraction and HPLC determination of veterinary pharmaceuticals in wastewater. Talanta 70(4):732–738
Kasprzyk-Hordern B, Dinsdale RM, Guwy AJ (2007) Multi-residue method for the determination of basic/neutral pharmaceuticals and illicit drugs in surface water by solid-phase extraction and ultra performance liquid chromatography-positive electrospray ionisation tandem mass spectrometry. J Chromatogr A 1161(1–2):132–145
Löffler D, Ternes TA (2003) Determination of acidic pharmaceuticals, antibiotics and ivermectin in river sediment using liquid chromatography-tandem mass spectrometry. J Chromatogr A 1021(1–2):133–144
Cha JM, Yang S, Carlson KH (2006) Trace determination of β-lactam antibiotics in surface water and urban wastewater using liquid chromatography combined with electrospray tandem mass spectrometry. J Chromatogr A 1115(1–2):46–57
Tong L, Li P, Wang Y, Zhu K (2009) Analysis of veterinary antibiotic residues in swine wastewater and environmental water samples using optimized SPE-LC/MS/MS. Chemosphere 74(8):1090–1097. https://doi.org/10.1016/j.chemosphere.2008.10.051
Chen M, Yi Q, Hong J, Zhang L, Lin K, Yuan D (2015) Simultaneous determination of 32 antibiotics and 12 pesticides in sediment using ultrasonic-assisted extraction and high performance liquid chromatography-tandem mass spectrometry. Anal Methods 7(5):1896–1905
Martins AF, Vasconcelos TG, Henriques DM, Frank CS, König A, Kümmerer K (2008) Concentration of ciprofloxacin in Brazilian hospital effluent and preliminary risk assessment: a case study. Clean Soil Air Water 36(3):264–269
Verlicchi P, Galletti A, Petrovic M, BarcelÓ D (2010) Hospital effluents as a source of emerging pollutants: an overview of micropollutants and sustainable treatment options. J Hydrol 389(3–4):416–428. https://doi.org/10.1016/j.jhydrol.2010.06.005
Le T-H, Ng C, Chen H, Yi XZ, Koh TH, Barkham TMS et al (2016) Occurrences and characterization of antibiotic-resistant bacteria and genetic determinants of hospital wastewater in a tropical country. Antimicrob Agents Chemother 60(12):7449–7456
Chagas TPG, Seki LM, Cury JC, Oliveira JAL, Dávila AMR, Silva DM et al (2011) Multiresistance, beta-lactamase-encoding genes and bacterial diversity in hospital wastewater in Rio de Janeiro, Brazil. J Appl Microbiol 111(3):572–581
Chang H-H, Cohen T, Grad YH, Hanage WP, O’Brien TF, Lipsitch M (2015) Origin and proliferation of multiple-drug resistance in bacterial pathogens. Microbiol Mol Biol Rev 79(1):101–116
Galvin S, Boyle F, Hickey P, Vellinga A, Morris D, Cormican M (2010) Enumeration and characterization of antimicrobial-resistant Escherichia coli bacteria in effluent from municipal, hospital, and secondary treatment facility sources. Appl Environ Microbiol 76(14):4772–4779
Proia L, Von Schiller D, Sànchez-Melsió A, Sabater S, Borrego CM, Rodríguez-Mozaz S et al (2016) Occurrence and persistence of antibiotic resistance genes in river biofilms after wastewater inputs in small rivers. Environ Pollut 210:121–128
Harbarth S, Kahlmeter G, Kluytmans J, Mendelson M, Hospital GS, Town C et al (2017) Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/. Accessed Feb 26 2020
Timraz K, Xiong Y, Al QH, Hong P (2017) Removal of bacterial cells, antibiotic resistance genes and integrase genes by on-site hospital wastewater treatment plants: surveillance of treated hospital effluent quality. Environ Sci Water Res Technol 3:293
Haller L, Chen H, Ng C, Le TH, Koh TH, Barkham T et al (2018) Occurrence and characteristics of extended-spectrum β-lactamase- and carbapenemase- producing bacteria from hospital effluents in Singapore. Sci Total Environ 615:1119–1125. https://doi.org/10.1016/j.scitotenv.2017.09.217
Jorgensen JH, Ferraro MJ (2009) Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis 49(11):1749–1755. http://www.ncbi.nlm.nih.gov/pubmed/19857164
Ansari F, Erntell M, Goossens H, Davey P (2009) The European Surveillance of Antimicrobial Consumption (ESAC) Point-Prevalence Survey of antibacterial use in 20 European hospitals in 2006. Clin Infect Dis 49(10):1496–1504
Yang CM, Lin MF, Liao PC, Yeh HW, Chang BV, Tang TK et al (2009) Comparison of antimicrobial resistance patterns between clinical and sewage isolates in a regional hospital in Taiwan. Lett Appl Microbiol 48(5):560–565
Prado T, Pereira WC, Silva DM, Seki LM, Carvalho APDA, Asensi MD (2008) Detection of extended-spectrum β-lactamase-producing Klebsiella pneumoniae in effluents and sludge of a hospital sewage treatment plant. Lett Appl Microbiol 46(1):136–141
Korzeniewska E, Harnisz M (2013) Extended-spectrum beta-lactamase (ESBL)-positive Enterobacteriaceae in municipal sewage and their emission to the environment. J Environ Manage 128:904–911. https://doi.org/10.1016/j.jenvman.2013.06.051
Picão RC, Cardoso JP, Campana EH, Nicoletti AG, Petrolini FVB, Assis DM et al (2013) The route of antimicrobial resistance from the hospital effluent to the environment: focus on the occurrence of KPC-producing Aeromonas spp. and Enterobacteriaceae in sewage. Diagn Microbiol Infect Dis 76(1):80–85. https://doi.org/10.1016/j.diagmicrobio.2013.02.001
Bréchet C, Plantin J, Sauget M, Thouverez M, Talon D, Cholley P et al (2014) Wastewater treatment plants release large amounts of extended-spectrum β-lactamase-producing Escherichia coli into the environment. Clin Infect Dis 58(12):1658–1665
Kwak YK, Colque P, Byfors S, Giske CG, Möllby R, Kühn I (2015) Surveillance of antimicrobial resistance among Escherichia coli in wastewater in Stockholm during 1 year: does it reflect the resistance trends in the society? Int J Antimicrob Agents 45(1):25–32. https://doi.org/10.1016/j.ijantimicag.2014.09.016
Ruppé E, Lixandru B, Cojocaru R, Büke Ç, Paramythiotou E, Angebault C et al (2013) Relative fecal abundance of extended-spectrum-β-lactamase-producing Escherichia coli strains and their occurrence in urinary tract infections in women. Antimicrob Agents Chemother 57(9):4512–4517
Slekovec C, Plantin J, Cholley P, Thouverez M, Talon D, Bertrand X et al (2012) Tracking down antibiotic-resistant Pseudomonas aeruginosa isolates in a wastewater network. PLoS One 7(12):e49300
Strateva T, Yordanov D (2009) Pseudomonas aeruginosa – a phenomenon of bacterial resistance. J Med Microbiol 58(9):1133–1148
Bertrand X, Thouverez M, Talon D, Boillot A, Capellier G, Floriot C et al (2001) Endemicity, molecular diversity and colonisation routes of Pseudomonas aeruginosa in intensive care units. Intensive Care Med 27(8):1263–1268
Tuméo E, Gbaguidi-Haore H, Patry I, Bertrand X, Thouverez M, Talon D (2008) Are antibiotic-resistant Pseudomonas aeruginosa isolated from hospitalised patients recovered in the hospital effluents? Int J Hyg Environ Health 211(1–2):200–204
Schwartz T, Volkmann H, Kirchen S, Kohnen W, Schön-Hölz K, Jansen B et al (2006) Real-time PCR detection of Pseudomonas aeruginosa in clinical and municipal wastewater and genotyping of the ciprofloxacin-resistant isolates. FEMS Microbiol Ecol 57(1):158–167
Spindler A, Otton LM, Fuentefria DB, Corção G (2012) Beta-lactams resistance and presence of class 1 integron in Pseudomonas spp. isolated from untreated hospital effluents in Brazil. Antonie van Leeuwenhoek 102(1):73–81
Fuentefria DB, Ferreira AE, Corção G (2011) Antibiotic-resistant Pseudomonas aeruginosa from hospital wastewater and superficial water: are they genetically related? J Environ Manage 92(1):250–255. https://doi.org/10.1016/j.jenvman.2010.09.001
Clark NM, Zhanel GG, Lynch JP (2016) Emergence of antimicrobial resistance among Acinetobacter species: a global threat. Curr Opin Crit Care 22(5):491–499
Kovacic A, Music MS, Dekic S, Tonkic M, Novak A, Rubic Z et al (2017) Transmission and survival of carbapenem-resistant Acinetobacter baumannii outside hospital setting. Int Microbiol 20(4):165–169
Ferreira AE, Marchetti DP, De Oliveira LM, Gusatti CS, Fuentefria DB, Corção G (2011) Presence of OXA-23-producing isolates of Acinetobacter baumannii in wastewater from hospitals in Southern Brazil. Microb Drug Resist 17(2):221–227. http://www.liebertpub.com/doi/10.1089/mdr.2010.0013
Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S et al (2006) Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents 27(4):351–353
Zhang C, Qiu S, Wang Y, Qi L, Hao R, Liu X et al (2013) Higher isolation of NDM-1 producing Acinetobacter baumannii from the sewage of the hospitals in Beijing. PLoS One 8(6):6–11
Farrell DJ, Morrissey I, de Rubeis D, Robbins M, Felmingham D (2003) A UK multicentre study of the antimicrobial susceptibility of bacterial pathogens causing urinary tract infection. J Infect 46(2):94–100
Harwood VJ, Delahoya NC, Ulrich RM, Kramer MF, Whitlock JE, Garey JR et al (2004) Molecular confirmation of Enterococcus faecalis and E. faecium from clinical, faecal and environmental sources. Lett Appl Microbiol 38(6):476–482
Leclercq R, Oberlé K, Galopin S, Cattoir V, Budzinski H, Petit F (2013) Changes in enterococcal populations and related antibiotic resistance along a medical center-wastewater treatment plant-river Continuum. Appl Environ Microbiol 79(7):2428–2434
Caplin JL, Hanlon GW, Taylor HD (2008) Presence of vancomycin and ampicillin-resistant Enterococcus faecium of epidemic clonal complex-17 in wastewaters from the south coast of England. Environ Microbiol 10(4):885–892
Narciso-Da-Rocha C, Varela AR, Schwartz T, Nunes OC, Manaia CM (2014) BlaTEM and vanA as indicator genes of antibiotic resistance contamination in a hospital-urban wastewater treatment plant system. J Glob Antimicrob Resist 2(4):309–315
Varela AR, Ferro G, Vredenburg J, Yanik M, Vieira L, Rizzo L et al (2013) Vancomycin resistant enterococci: from the hospital effluent to the urban wastewater treatment plant. Sci Total Environ 450–451:155–161
Nuñez L, Tornello C, Puentes N, Espigares E, Moreno E, Espigares M et al (2016) Hospital effluent constitutes a source of vancomycin-resistant enterococci. Ars Pharm 57(3):121–126
Karkman A, Do TT, Walsh F, Virta MPJ (2018) Antibiotic-resistance genes in waste water. Trends Microbiol 26(3):220–228. https://doi.org/10.1016/j.tim.2017.09.005
Liu L, Su JQ, Guo Y, Wilkinson DM, Liu Z, Zhu YG et al (2018) Large-scale biogeographical patterns of bacterial antibiotic resistome in the waterbodies of China. Environ Int 117:292–299. https://doi.org/10.1016/j.envint.2018.05.023
Zheng J, Zhou Z, Wei Y, Chen T, Feng W, Chen H (2018) High-throughput profiling of seasonal variations of antibiotic resistance gene transport in a peri-urban river. Environ Int 114:87–94. https://doi.org/10.1016/j.envint.2018.02.039
Zhu YG, Zhao Y, Li B, Huang CL, Zhang SY, Yu S et al (2017) Continental-scale pollution of estuaries with antibiotic resistance genes. Nat Microbiol 2:16270. https://doi.org/10.1038/nmicrobiol.2016.270
Muziasari WI, Pitkänen LK, Sørum H, Stedtfeld RD, Tiedje JM, Virta M (2017) The resistome of farmed fish feces contributes to the enrichment of antibiotic resistance genes in sediments below Baltic sea fish farms. Front Microbiol 7:1–10
Xu L, Ouyang W, Qian Y, Su C, Su J, Chen H (2016) High-throughput profiling of antibiotic resistance genes in drinking water treatment plants and distribution systems. Environ Pollut 213:119–126. https://doi.org/10.1016/j.envpol.2016.02.013
An XL, Su JQ, Li B, Ouyang WY, Zhao Y, Chen QL et al (2018) Tracking antibiotic resistome during wastewater treatment using high throughput quantitative PCR. Environ Int 117:146–153. https://doi.org/10.1016/j.envint.2018.05.011
Karkman A, Johnson TA, Lyra C, Stedtfeld RD, Tamminen M, Tiedje JM et al (2016) High-throughput quantification of antibiotic resistance genes from an urban wastewater treatment plant. FEMS Microbiol Ecol 92(3):1–7
Proia L, Anzil A, Borrego C, Farrè M, Llorca M, Sanchis J et al (2018) Occurrence and persistence of carbapenemases genes in hospital and wastewater treatment plants and propagation in the receiving river. J Hazard Mater 358:33–43. https://doi.org/10.1016/j.jhazmat.2018.06.058
Proia L, Adriana A, Jessica S, Carles B, Marinella F, Marta L et al (2018) Antibiotic resistance in urban and hospital wastewaters and their impact on a receiving freshwater ecosystem. Chemosphere 206:70–82
Szekeres E, Baricz A, Chiriac CM, Farkas A, Opris O, Soran ML et al (2017) Abundance of antibiotics, antibiotic resistance genes and bacterial community composition in wastewater effluents from different Romanian hospitals. Environ Pollut 225:304–315
Devarajan N, Laffite A, Mulaji CK, Otamonga J, Mpiana PT, Mubedi JI et al (2016) Occurrence of antibiotic resistance genes and bacterial markers in a tropical river receiving hospital and urban wastewaters. PLoS One 11(2):1–14
Wang Q, Wang P, Yang Q (2018) Occurrence and diversity of antibiotic resistance in untreated hospital wastewater. Sci Total Environ 621:990–999. https://doi.org/10.1016/j.scitotenv.2017.10.128
Li J, Cheng W, Xu L, Strong PJ, Chen H (2015) Antibiotic-resistant genes and antibiotic-resistant bacteria in the effluent of urban residential areas, hospitals, and a municipal wastewater treatment plant system. Environ Sci Pollut Res 22:4587–4596
Rodriguez-Mozaz S, Chamorro S, Marti E, Huerta B, Gros M, Sànchez-Melsió A et al (2015) Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res 69:234–242
Lamba M, Graham DW, Ahammad SZ (2017) Hospital wastewater releases of carbapenem-resistance pathogens and genes in urban India. Environ Sci Technol 51:13906–13912
Sun D, Jeannot K, Xiao Y, Knapp CW (2019) Editorial: horizontal gene transfer mediated bacterial antibiotic resistance. Front Microbiol 10:1933
Pärnänen KMM, Narciso-Da-Rocha C, Kneis D, Berendonk TU, Cacace D, Do TT et al (2019) Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci Adv 5(3):eaau9124
Lu X, Hu Y, Zhou H, Wang X, Du Y, Li Z et al (2017) MCR-1.6, a new MCR variant carried by an IncP plasmid in a colistin-resistant Salmonella enterica Serovar Typhimurium isolate from a healthy individual. Antimicrob Agents Chemother 61(5):1–5
Pallares-Vega R, Blaak H, van der Plaats R, de Roda Husman AM, Leal LH, van Loosdrecht MCM et al (2019) Determinants of presence and removal of antibiotic resistance genes during WWTP treatment: a cross-sectional study. Water Res 161:319–328. https://linkinghub.elsevier.com/retrieve/pii/S0043135419304981
Waseem H, Jameel S, Ali J, Ur Rehman HS, Tauseef I, Farooq U et al (2019) Contributions and challenges of high throughput qPCR for determining antimicrobial resistance in the environment: a critical review. Molecules 24(1):E163
Tan BF, Ng C, Nshimyimana JP, Loh LL, Gin KYH, Thompson JR (2015) Next-generation sequencing (NGS) for assessment of microbial water quality: current progress, challenges, and future opportunities. Front Microbiol 6:1027
Watts JEM, Schreier HJ, Lanska L, Hale MS (2017) The rising tide of antimicrobial resistance in aquaculture: sources, sinks and solutions. Mar Drugs 15(6):1–16
Gupta SK, Shin H, Han D, Hur HG, Unno T (2018) Metagenomic analysis reveals the prevalence and persistence of antibiotic- and heavy metal-resistance genes in wastewater treatment plant. J Microbiol 56(6):408–415
Guo J, Li J, Chen H, Bond PL, Yuan Z (2017) Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res 123:468–478. https://doi.org/10.1016/j.watres.2017.07.002
Bondarczuk K, Piotrowska-Seget Z (2019) Microbial diversity and antibiotic resistance in a final effluent-receiving lake. Sci Total Environ 650:2951–2961
Chu BTT, Petrovich ML, Chaudhary A, Wright D, Murphy B, Wells G et al (2018) Metagenomics reveals the impact of wastewater treatment plants on the dispersal of microorganisms and genes in aquatic sediments. Appl Environ Microbiol 84(5):1–15. https://www.scopus.com/inward/record.uri?eid=2-s2.0-85042018572&doi=10.1128%2FAEM.02168-17&partnerID=40&md5=bbeaa6cf5e779312033180c3c5f713eb%0Ahttp://ovidsp.ovid.com/ovidweb.cgi?T=JS&PAGE=reference&D=emexb&NEWS=N&AN=625138131
Rizzo L, Manaia C, Merlin C, Schwartz T, Dagot C, Ploy MC et al (2013) Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ 447:345–360. https://doi.org/10.1016/j.scitotenv.2013.01.032
Marathe NP, Berglund F, Razavi M, Pal C, Dröge J, Samant S et al (2019) Sewage effluent from an Indian hospital harbors novel carbapenemases and integron-borne antibiotic resistance genes. Microbiome 7(1):1–11
Ng C, Tay M, Tan B, Le TH, Haller L, Chen H et al (2017) Characterization of metagenomes in urban aquatic compartments reveals high prevalence of clinically relevant antibiotic resistance genes in wastewaters. Front Microbiol 9:1–12
Buelow E, Rico A, Gaschet M, Lourenço J, Kennedy SP, Wiest L et al (2019) Classification of hospital and urban wastewater resistome and microbiota over time and their relationship to the eco-exposome:1–33. https://doi.org/10.1101/697433
Buelow E, Bayjanov JR, Majoor E, Willems RJL, Bonten MJM, Schmitt H et al (2018) Limited influence of hospital wastewater on the microbiome and resistome of wastewater in a community sewerage system. FEMS Microbiol Ecol 94(7):1–9
Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK et al (2017) CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45(D1):D566–D573
Liu B, Pop M (2009) ARDB – antibiotic resistance genes database. Nucleic Acids Res 37(Suppl 1):443–447
Gibson MK, Forsberg KJ, Dantas G (2015) Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J 9(1):207–216. https://doi.org/10.1038/ismej.2014.106
Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L et al (2014) ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58(1):212–220
Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O et al (2012) Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67(11):2640–2644
Lakin SM, Dean C, Noyes NR, Dettenwanger A, Ross AS, Doster E et al (2017) MEGARes: an antimicrobial resistance database for high throughput sequencing. Nucleic Acids Res 45(D1):D574–D580
Yin X, Jiang XT, Chai B, Li L, Yang Y, Cole JR et al (2018) ARGs-OAP v2.0 with an expanded SARG database and Hidden Markov Models for enhancement characterization and quantification of antibiotic resistance genes in environmental metagenomes. Bioinformatics 34(13):2263–2270
Li B, Yang Y, Ma L, Ju F, Guo F, Tiedje JM et al (2015) Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J 9(11):2490–2502. https://doi.org/10.1038/ismej.2015.59
Ma L, Xia Y, Li B, Yang Y, Li LG, Tiedje JM et al (2016) Metagenomic assembly reveals hosts of antibiotic resistance genes and the shared resistome in pig, chicken and human feces. Environ Sci Technol 50(1):420–427
Partridge SR, Kwong SM, Firth N, Jensen SO (2018) Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31(4):1–61
Gillings M, Boucher Y, Labbate M, Holmes A, Krishnan S, Holley M et al (2008) The evolution of class 1 integrons and the rise of antibiotic resistance. J Bacteriol 190(14):5095–5100
Chen H, Gu X, Ng C, Haller L, Charles FR, Gin KY (2019) Draft genome sequences of a ceftazidime-resistant Acinetobacter baumannii donor and a conjugal Escherichia coli recipient with acquired resistance. Microbiol Resour Announc 8(13):9–10
Boyd D, Taylor G, Fuller J, Bryce E, Embree J, Gravel D et al (2015) Complete sequence of four multidrug-resistant MOB Q1 plasmids harboring bla GES-5 isolated from Escherichia coli and Serratia marcescens persisting in a hospital in Canada. Microb Drug Resist 21(3):253–260
Weingarten RA, Johnson RC, Conlan S, Ramsburg AM, Dekker JP, Lau AF et al (2018) Genomic analysis of hospital plumbing reveals diverse reservoir of bacterial plasmids conferring carbapenem resistance. MBio 9(1):1–16
Zhang AN, Li LG, Ma L, Gillings MR, Tiedje JM, Zhang T (2018) Conserved phylogenetic distribution and limited antibiotic resistance of class 1 integrons revealed by assessing the bacterial genome and plasmid collection. Microbiome 6(1):1–14
Partridge SR, Tsafnat G, Coiera E, Iredell JR (2009) Gene cassettes and cassette arrays in mobile resistance integrons: review article. FEMS Microbiol Rev 33(4):757–784
Moura A, Soares M, Pereira C, Leitão N, Henriques I, Correia A (2009) INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25(8):1096–1098
Carattoli A, Zankari E, Garciá-Fernández A, Larsen MV, Lund O, Villa L et al (2014) In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58(7):3895–3903
Ju F, Beck K, Yin X, Maccagnan A, McArdell CS, Singer HP et al (2019) Wastewater treatment plant resistomes are shaped by bacterial composition, genetic exchange, and upregulated expression in the effluent microbiomes. ISME J 13(2):346–360. https://doi.org/10.1038/s41396-018-0277-8
Hendriksen RS, Munk P, Njage P, van Bunnik B, McNally L, Lukjancenko O et al (2019) Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat Commun 10(1):1124
Che Y, Xia Y, Liu L, Li AD, Yang Y, Zhang T (2019) Mobile antibiotic resistome in wastewater treatment plants revealed by Nanopore metagenomic sequencing. Microbiome 7(1):1–13
Li L-G, Yin X, Zhang T (2018) Tracking antibiotic resistance gene pollution from different sources using machine-learning classification. Microbiome 6(1):93. https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-018-0480-x
Lynch MDJ, Neufeld JD (2015) Ecology and exploration of the rare biosphere. Nat Rev Microbiol 13(4):217–229. https://doi.org/10.1038/nrmicro3400
Lanza VF, Baquero F, Martínez JL, Ramos-Ruíz R, González-Zorn B, Andremont A et al (2018) In-depth resistome analysis by targeted metagenomics. Microbiome 6(1):1–14
Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C et al (2009) Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461(7261):272–276. https://doi.org/10.1038/nature08250
Rowe WPM, Baker-Austin C, Verner-Jeffreys DW, Ryan JJ, Micallef C, Maskell DJ et al (2017) Overexpression of antibiotic resistance genes in hospital effluents over time. J Antimicrob Chemother 72(6):1617–1623
van Dijck C, Vlieghe E, Cox JA (2018) Antibiotic stewardship interventions in hospitals in low-and middle-income countries: a systematic review. Bull World Health Organ 96(4):266–280
Vong S, Anciaux A, Hulth A, Stelling J, Thamlikitkul V, Gupta S et al (2017) Using information technology to improve surveillance of antimicrobial resistance in South East Asia. BMJ 358:j3781
Dickey SW, Cheung GYC, Otto M (2017) Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat Rev Drug Discov 16(7):457–471. https://doi.org/10.1038/nrd.2017.23
Gilmer DB, Schmitz JE, Thandar M, Euler CW, Fischetti VA (2017) The phage lysin PlySs2 decolonizes Streptococcus suis from murine intranasal mucosa. PLoS One 12(1):1–13
Li N, Sheng GP, Lu YZ, Zeng RJ, Yu HQ (2017) Removal of antibiotic resistance genes from wastewater treatment plant effluent by coagulation. Water Res 111:204–212
Ye M, Sun M, Feng Y, Wan J, Xie S, Tian D et al (2016) Effect of biochar amendment on the control of soil sulfonamides, antibiotic-resistant bacteria, and gene enrichment in lettuce tissues. J Hazard Mater 309:219–227. https://doi.org/10.1016/j.jhazmat.2015.10.074
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Ng, C., Chen, H., Tran, N.H., Haller, L., Gin, K.YH. (2020). Antibiotic Resistance in Municipal Wastewater: A Special Focus on Hospital Effluents. In: Manaia, C., Donner, E., Vaz-Moreira, I., Hong, P. (eds) Antibiotic Resistance in the Environment . The Handbook of Environmental Chemistry, vol 91. Springer, Cham. https://doi.org/10.1007/698_2020_471
Download citation
DOI: https://doi.org/10.1007/698_2020_471
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-55064-6
Online ISBN: 978-3-030-55065-3
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)