Abstract
As emerging contaminants, both antibiotic-resistant bacteria and antibiotic resistance genes have captured global attention, imposing adverse effects on the surrounding environment. Effluents released into the surface water are the primary sources of emerging contaminants that contribute to soil and groundwater pollution. Wastewater treatment plants are the hotspots for the spread of antibiotic resistance through horizontal gene transfer. Given the prevalence of genetic elements and resistance genes detected in waste water samples, a concerning scenario emerges, exacerbating the challenges associated with antibiotic treatment in the healthcare system. In this chapter, we focus on the methods employed for the detection of antibiotic resistance genes in wastewater and the latest research in this area. By delving into public health implications, we analyze the potential routes of exposure to antibiotic genes, the increasing threat of antibiotic resistance in healthcare settings, and the broader environmental and societal consequences.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
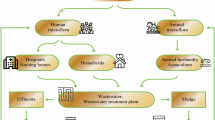
Worldwide for decades, antibiotics have been predominantly used for the efficient management of infectious diseases. The spread of antibiotic resistance stands as a paramount global concern, presenting a formidable and complex challenge to public health. Antibiotic-resistant diseases significantly heighten the risk to public health. They are anticipated to become even more prevalent in the future, putting individuals in jeopardy, even during routine infections and minor surgical procedures. Common infections caused by AMR microbes result in high mortality (Laxminarayan et al., 2013). Recently, the Global Antimicrobial Surveillance System (GLASS) of the World Health Organization (WHO) reported that many regions of the world are grappling with increased levels of resistance in numerous serious and common bacterial infections (WHO, 2019). According to reports from the US Center for Disease Control and Prevention (CDC), 35000 deaths occur every year due to antimicrobial resistance (AMR) in the United States (CDC report, 2019). Antibiotic resistant bacteria (ARB) led to approximately 671,689 infections that caused overall 33,000 deaths in European Economic activity countries in 2015 (Cassini et al., 2019). The situation is worsening in developing countries where the demand for antibiotics is driven by the high incidence of infectious diseases (Lirermore, 1995). Despite extensive measures taken by certain nations to curtail antibiotic usage, there have been noticeable increases in antibiotic resistance rates. This underscores the intricate nature of the public and environmental concerns associated with this issue (O’neill, 2014). Antibiotic resistance is developing at a faster rate than the production of new antibiotics suggesting a challenging situation arising day by day for identifying new antibiotics (Ling et al., 2015). As most antibiotics are not completely metabolized within humans and animals, about 30–90% of these drugs are expelled via urine and feces into water bodies and soil. Industrial and municipal wastewater entering wastewater treatment plants (WWTPs), along with sewage sludge, can contain a diverse range of contaminants, including heavy metals, antibiotics, various chemicals, organic matter, nutrients, bacteria, viruses, and more. These contaminants, originating from domestic, industrial, and commercial sources, contribute to the potential reservoir of antibiotic resistance genes (ARGs), which can further exacerbate the issue of antibiotic resistance. The specific composition and prevalence of ARGs in wastewater is influenced by local factors, such as industrial practices, healthcare protocols, and regional regulations. Effective monitoring and management of ARGs in wastewater are crucial components of addressing the broader challenge of antibiotic resistance in the environment and its implications for public health. The main drivers of antibiotic-resistant bacteria (ARB) and multidrug-resistant (MDR) bacteria are the improper use and dispersal of antibiotics. These bacteria carry an evolved set of antibiotic-resistant genes (ARGs). Both ARGs and ARB are present in air (aerosols) and water wastewater treatment plants (WWTPs) and may pose a risk of dispersion to nearby workers. Moreover, these WWTPs are not designed to eliminate ARB or ARGs, but contain antibiotics that contribute to the development of antibiotic resistance. The contribution of residual antibiotics toward the development of antibiotic-resistant bacteria and their impact on the ecological system are the two serious concerns. WWTPs are considered as the hotspot for ARGs and play a major role in horizontal gene transfer further leading to the dispersal of ARGs in the broader sense. In WWTPs, the evolution of resistance and dissemination of ARGs is not evidenced clearly. In sludge of various WWTPs, many of the antibiotics like sulfamethoxazole, norfloxacin, tetracycline, ofloxacin, and ciprofloxacin are present in higher amounts. Antibiotics from the sewage of households and hospitals create selective pressure on both ARB and ARGs preceding their dispersal into the surroundings (Martínez, 2009).
The effluents (antibiotics, ARB, ARGs) present in WWTPs get released into surface water then enter the environment. Antibiotic resistance is developed by a different mechanisms in bacteria that makes antibiotic ineffective. The genes responsible are situated either on a bacterial chromosome or extrachromosomal region and they can be transmitted via horizontal or vertical gene transfer. Horizontal gene transfer through conjugation is more common in high-density bacteria which may also form biofilms.The spread and proliferation of ARGs are complex and can vary widely in different contexts and environments. Efforts to mitigate the spread of ARGs are a crucial part of combating antibiotic resistance and preserving the effectiveness of antibiotics, as, ARB remian the prime concern in circulating antibiotic resistance in the microbial system (Larson, 2007). Due to the improper use of antibiotics, excretion via humans and animals, the compounds, their metabolites, and transformed products get deposited in hospital and industrial wastewater. The observed concentration of tetracycline has been as high as 1300 ng/L and 1420 ng/L in WWTPs and effluents respectively (Minh et al., 2009). One of the sulfonamides i.e., sulfamethoxazole was identified with a concentration of 5597 ng/L (Peng et al., 2008) in effluent and 6000 ng/L (Batt et al., 2006) in a wastewater plant. Both the resistant genes of tetracycline (tet A, B, C, G, L, M) and sulfonamide (sull) have been investigated in WWTPs (Szczepanowski et al., 2009). The overall findings confirmed the effluents responsible for anthropogenic sources in an environment that further outspread the resistance to non-resistant bacteria (LaPara et al., 2011; Pruden et al., 2012).
A study reported the detection of NSAID (Non-steroidal anti-inflammatory drugs) in municipal WWTPs with the concentration of 0.42–367 mg/kg whereas in livestock WWTPs antibiotics have been shown to be present in the range of 43.6–142 mg/kg (Ekpeghere et al., 2017). Dispersal of ARGs in the environment is highlighted as a global concern as the adulterated water is supplied for various agricultural practices.
Sewage received by WWTPs, thus, contains residual antibiotics from different sources along with the corresponding ARB, including human pathogens (Borjesson et al., 2009). The designing of WWTPs aimed for the removal of solids, organic matter, and nutrients but not for the removal of antibiotics and ARGs. The presence and persistence of antibiotics and ARGs in WWTPs are posing a serious hazard to the environment and public health (LaPara et al., 2012). The existence of antibiotics even in trace amounts in WWTPs allows frequent horizontal gene transfer (HGT) between different bacterial species, and thus, several pathogens are developing resistance particularly against broad-spectrum antibiotics (Rahal et al., 1997). In developing countries of the world, the situation is even more difficult to control because of a breakdown in health services and sanitation which leads to the rapid dissemination of resistant pathogens (Dodge, 1990).
WWTPs significantly contribute toward the problem of antibiotic resistance due to issues associated with their treatment processes. The activated sludge process is the most used wastewater treatment process in the world intended to reduce and eliminate some antibiotics and other pharmaceutical compounds from wastewater. The biodegradation/sorption process is commonly used for the removal of non-volatile compounds such as pharmaceuticals. The sorption process cannot be used for the removal of all types of antibiotics because of the selective specificity of antibiotics toward biosolids (Chander et al., 2005). Chlorination and ultraviolet (UV) treatment are the other methods used for the removal of pathogens and also contribute to the elimination of some pharmaceuticals in wastewater. However, the findings of several studies suggested the inefficiency of disinfection processes in the removal of antibiotics (Batt et al., 2007; Jones et al., 2005). Volatilization (resulting from aeration) or photodegradation (triggered by sunlight) are recognized as effective removal mechanisms; however, they have limited or no impact on antibiotics due to their negligible or non-existent efficacy in breaking down these compounds. In WWTPs the transformation of antibiotics into compounds with increased hydrophilicity or hydrophobicity, as well as their conversion into carbon dioxide that adheres to the activated sludge, poses significant worldwide challenges. These processes make it more challenging to effectively degrade antibiotics in wastewater (Halling-Sørensen et al., 1998; Kümmerer, 2003). Further, finding the absence of the parent pharmaceutical from the WWTP effluent can mislead treatment strategies, the compound can be transformed and persist in the effluent. Identification of the antibacterial potential of the transformed compounds is another issue that needs consideration and immediate scientific attention. It is not enough to have an idea about the number of antibiotics discharged in the wastewater however, it is also important to identify the nature and quantities of degradation products because the transformation of a biologically active compound does not necessarily equate to detoxification. Owing to the above context it is challenging to identify the realistic load of antibiotics in WWTPs. Overall, the methods for ARB and ARGs removal in wastewater and sludge often fall short of complete elimination due to various factors. These include the resilience of certain ARB and ARG, variations in treatment conditions, potential shielding of contaminants, and the specificity of removal techniques. Biological treatment may not entirely eradicate these elements, and chemical treatments may have varying effectiveness. Adsorption methods may not be universally suitable, and membrane filtration might not fully remove smaller molecules. UV disinfection may face limitations, while land application of biosolids carries environmental exposure risks. Comprehensive ARB and ARG removal typically necessitates a multifaceted approach, coupled with responsible antibiotic use to manage antibiotic resistance in the environment. In a study, metagenomic sequencing was used to assess microbial community diversity and the fate ARGs, antibiotic biosynthesis genes (ABSGs), and virulence factor genes (VFGs) in sewage samples from four major WWTPs in the United States. The study found that the diversity and composition of these genes were linked to the treatment processes. While overall ARG and VFG abundances decreased by over 20% after treatment, the activated sludge process (ASP) selectively enriched multidrug resistance ARGs and certain VFGs. Sub-groups of ABSGs also saw substantial increases during ASP. These insights shed light on how conventional wastewater treatment processes impact ARGs and VFGs, aiding in understanding their dissemination through sewage and suggesting strategies to mitigate antibiotic resistance spread via sewage discharge (Le et al., 2022).
2 Antibiotic-Resistant Genes
Maximum antibiotics, ARGs and ARB enter the environment via WWTPs into irrigation, rivers, and biosolids. As a result of anthropogenic activities, aquatic ecosystems contaminated by antibiotic compounds are the chief sites for the occurrence of ARGs. These are transferred to other microbial pathogens via horizontal gene transfer, rendering antibiotics ineffective for the treatment later (Zhou et al., 2016). Hospital wastewater has been shown to host a multitude of Antibiotic Resistance Genes (ARGs). Early studies, dating back to the 1970s, indicated a higher prevalence of transferable resistance in coliform bacteria from hospital wastewater compared to municipal wastewater (Grabow and Prozesky, 1973). Recent research in Romania identified ARGs for tetracyclines, aminoglycosides, chloramphenicol, β-lactams, sulphonamides, quaternary ammonium, and macrolide-lincosamide-streptogramin B antibiotics in hospital wastewater (Szekeres et al., 2017). A review of 37 studies found that hospital wastewater consistently carries more ARGs than community wastewater and serves as a significant source of antibiotic resistance in the environment (Hassoun-Kheir et al., 2020). A group of researchers evaluated the ARGs like tet (X, W, G), sul-1, and intI-1 present in municipal WWTP that is comprised of anaerobic/aerobic/anoxic bioreactors. In anaerobic and anoxic units, a decrease in ARGs succeeding with an increase in ARGs of aerobic units was observed. The overall result demonstrated the effectiveness of anaerobic and anoxic treatments was more superior to aerobic ones (Du et al., 2015). Antimicrobial compounds act by targeting various bacterial cell sites, exhibiting bactericidal or bacteriostatic effects. Bacteria, in response to environmental stress, employ defense mechanisms to counteract these antimicrobials. Common resistance mechanisms involve altering antibiotic target sites, reducing drug affinity, decreasing drug accumulation, employing inactivating enzymes, and acquiring alternative metabolic pathways. Genes encoding the antibiotics are usually located on mobile genetic elements like plasmids, transposons as well as integrons that take part in the dissemination of ARGs and emergence. More specifically integrons include resistance cassettes that code for different ARGs (aminoglycoside, sulfonamide) and efflux pump (qacEΔ1) resistance genes (Xu et al., 2017). In certain conditions with a shared promoter, multigene cassettes may contain various genes, including antibiotic resistance genes (ARGs). These cassettes can exhibit different ARGs' expression in response to selection pressure from specific antibiotics. For example, in the case of MRSA (methicillin-resistant Staphylococcus aureus), a cassette may carry the mecA gene, which imparts resistance to methicillin and related antibiotics (Di Cesare et al., 2016; Sharma et al., 2016).
Wetland sediments are a crucial source of aquatic ARGs because of the regular release of microbe into water. The level of ARGs mainly depends on the environmental factors of the wetland with domestic sewage as the major source of ARGs. A group of researchers demonstrated the presence of ARB, sul (1&3), tet (A, B, C, E), and qnr (S) genes in surface flow wetland (Fang et al., 2017). In wastewater associated with the microbial community, a large number of ARGs has been demonstrated and by the release of wastewater into the aquatic environment these can move along the water cycle. Notably, the prime sites for occurrence and dispersal of ARGS are the aquatic systems that get polluted with antibiotics mainly from various human activities (Rodriguez-Mozaz et al., 2015). Various qnr genes on plasmids encode proteins that reduce the effectiveness of DNA gyrase and topoisomerase IV, resulting in low-level fluoroquinolone resistance. qnrB genes, originating from different Citrobacter species, and qnrA genes from Shewanella algae are prevalent. These genes are often found on class 1 integrons, co-carried with other resistance determinants. qnr genes are frequently detected in aqueous environments, including WWTPs, their effluents, soil irrigated with wastewater, and wetlands along urban coasts, indicating potential environmental dissemination of antibiotic resistance determinants (Mutuku et al., 2022).
3 Methods for Analyzing Antibiotic Resistance Genes
As research on antibiotics is mainly focused on microbes for the past 70 years, wherein the foremost method employed is pure culture isolation. Testing of antibiotic susceptibility is reasonably affordable and provides fundamental data required for the treatment of patients related to resistance. Worldwide, the clinical breakpoints committees such as Clinical and Laboratory Standards Institute (CLSI) and EUCAST, www.eucast.org help in interpreting antibiotic resistance. But these clinical breakpoints are not applicable for wastewater bacteria. In this case, epidemiological cut-off values i.e., ECOFFs have been introduced for determining the wastewater. The term ECOFF is used to define the Minimum Inhibitory Concentration (MIC) threshold at which bacterial isolates exhibit phenotypically detectable acquired resistance mechanisms (Kahlmeter and Turnidge, 2022). Pure culture isolation is a daunting task and may not work for organisms difficult to culture and therefore, for determining antibiotic-resistant genes, various molecular biology tools have been used like polymerase chain reaction (PCR), quantitative PCR, digital PCR and genome sequencing (Martínez et al., 2014).
3.1 PCR and its Variants
PCR is a basic genetic method and powerful detection tool with significant diagnostic values employed in numerous scientific areas. This method is used in the identification of resistance, virulence factors, and other properties of microorganisms. Based on amplification of specific target genes or application of probes, detection of antibiotic-resistant genes along with their expression was observed. Real-time PCR as compared to conventional PCR requires less duration of time and therefore yields products in a very short period (March Rosselló & Bratos Pérez, 2016). Various real-time PCR is known that can detect the pathogens and the genes responsible for antibiotic resistance of different specimens. Such kind of PCR is automated and can amplify the target within few minutes. Verigenesystem developed by nanosphere is used for identification of antimicrobial resistance marker. It has detected 9 species 3 genera of bacteria and 2 resistance genes (mecA & vanA/B) with high specificity (100%) and sensitivity in gram-positive bacteria. In the case of gram-negative bacteria, 5 species and 4 genera of bacteria and resistance genes (CTX-M, ESBLs, KPC) were detected within 3 h of time exhibiting 93% specificity and sensitivity (Ledeboer et al., 2015).
The Xpert Carba-Rcatridge from GeneXpert can detect genes for KPC, NDM, VIM, IMP, and OXA-48 within an hour using PCR amplification. Eazyplex system carries out DNA amplification via LAMP (loop-mediated isothermal amplification). Amplification in this technique is done through chain displacement without a change in temperature and the amplicon formed is detected in real-time. Numerous such kits in different formats and different amplifuication methods that can determine the genes responsible for antibiotic resistance are in market (Hinic et al., 2015; Bloemberg et al., 2014).
Analysis of antibiotics resistant genes from environmental DNA can be achieved by PCR and quantitative PCR techniques. The kits available for clinical samples can be repurposed to detect the ARB or ARGs in WWTPs. The initial processing of the samples and nucleic acid isolation needs optimization. To achieve optimization, spiking and marker use are considered. For instance, for SARS-CoV2 process optimization, Phi6 bacteriophage was employed as a surrogate for enveloped viruses. In the analysis of Antibiotic Resistance (AR), sample processing is essential. This involves actions like concentrating Antibiotic Resistance Genes (ARGs), eliminating impurities, and ensuring sample uniformity. Common processing methods involve filtration and/or centrifugation. Filtration is typically employed for liquid samples, while centrifugation is preferred for samples with a significant solid content, such as activated sludge. Hundreds of ARGs as well as other genes can be detected simultaneously in one run by qPCR array (Zhu et al., 2013). Using qPCR, a comparative analysis of 10 genes related to tetracycline resistance and three sulfonamide resistance genes was conducted in two Polish WWTPs. The study revealed an augmentation of selected ARGs following wastewater treatment processes highlighting the pivotal role of WWTPs in amplifying the distribution of antibiotic resistance determinants in the environment (Pazda et al., 2020). Droplet Digital PCR (ddPCR™) has been used to analyze Antibiotic Resistance Genes (ARGs). ddPCR offers precise quantification by counting nucleic acid molecules enclosed in defined water-in-oil droplets. The study assessed one mobile element integrase gene (intI1) and seven ARGs, including four tet genes (tetA, tetC, tetQ, tetW), one macrolide resistance gene (ermB), and one sulphonamide resistance gene (sul2). The impact of ultrasonic treatment on these ARGs was examined in different types of sludge from Wastewater Treatment Plants (WWTPs). The results indicate that ultrasonic pre-treatment had limited effects on the absolute concentrations of total ARGs, with some fluctuations observed in specific ARGs and intI1 abundance (Rumky et al. 2022). Thus, PCR based techniques provide a better opportunity for the detection of ARGs, sequences of mobile genetic elements, and target genes of bacterial species present in WWTPs or in the environement.
3.2 Microarrays
This method is known for the detection of target molecules hybridized to a specific probe on a solid base using image analysis. It can analyze vast number of resistance genes in a single assay. Many of the microarrays like Check MDR (CT102, CT103, CT103XL) can detect several genes that code for different types of β-lactamases (ESBLs, AmpCs, carbapenemases) have been marketed (Bogaerts et al., 2016; Cuzon et al., 2012; Stuart et al., 2012). Pathogen identification and microbial source tracking (MST) enhance water quality evaluation, health risk assessment, and pollution source remediation. A microarray, used with dead-end ultrafiltration and whole-genome amplification, detected various pathogens, viruses, and antibiotic resistance genes in different water types, including sewage-contaminated samples. Sensitivity for sewage-related gene targets was around 51–57%, lower than specificity (79–81%) (Li et al., 2016).
3.3 Metagenomics
With some limitations of qPCR targeted analysis and failure to design primers for new or unknown genes, the most suitable approach is Metagenomics. It is based on the sequencing of the whole metagenome present in the environmental sample. It is becoming a widely used and affordable approach for analyzing the ARB and ARGs present in the waste water treatment plants (Thirunavukarasou et al., 2022). It is not only restricted to the detection of species already known genes but also has the potential to determine the new ARGs. A shotgun metagenomic approach was used to analyze ARGs and mobile genetic elements (MGEs) in activated sludge samples from two hospital wastewater treatment plants in Daegu, South Korea. Microbial community diversity was assessed through 16S rRNA metagenome sequencing. Cloacibacterium caeni and Lewinella nigricans dominated in domestic sewage wastewater (SWW) effluents, while Bacillus subtilis and Staphylococcus epidermidis were prevalent in hospital wastewater (HWW) effluents. Notably, HWW had higher ARG abundance, including multidrug resistance, macrolide-lincosamide-streptogramin, beta-lactam, bacitracin, and tetracycline, indicating antibiotic use in human medicine. Higher levels of MGEs in HWW raised concerns about horizontal gene transfer (Manoharan et al., 2021).
A group of researchers determined the ARGs present in WWTP of 8 activated sludges collected twice in both seasons. As a result, a wide spectrum of ARGs was detected with approximately 200 subtypes, many of which were reported for the first time. The ARGs identified were mainly aminoglycoside, tetracycline, sulfonamide, and chloramphenicol. It is quite evident from the above study that wastewater influent possessed the highest ARGs subsequently followed by affluent, anaerobic digestion and activated sludge. This approach has also been implicated for the detection of ARGs in different environmental samples (Yang et al., 2014). A study performed by Nesme et al. revealed the different ARGs present in 71 environment specimens. The other study also investigated the occurrence patterns of ARGs from 10 different samples including WWTPs. A total of 260 subtypes of ARGs were determined within a range of 5.4 × 10–6 to 2.2 × 10–1 copy of ARG/16s rRNA. The total ARG in a different environment was the same as the level of anthropogenic impact. It was clear from the results WWTPs contribute to the major hotspot of abundant ARG dissemination (Li et al., 2015). To detect low-abundance ARGs, a high-throughput amplicon sequencing method was developed, targeting 251 ARGs, 8 mobile genetic element genes, and 19 metal resistance genes. The new method outperformed traditional shotgun sequencing, offering significantly improved sensitivity, diversity, and cost-effectiveness. The approach was applied to environmental and clinical samples, demonstrating its efficiency in ARG surveillance and evolution assessment. By enhancing our understanding of resistome dynamics, this method provides valuable insights into the global challenge of antibiotic resistance (Li et al., 2022). Amplicon sequencing panels are available commercially as well such as The AmpliSeq for Illumina Antimicrobial Resistance Research Panel. The panel offers a fast, precise, and economical solution for the detection of 28 distinct antibiotic classes. It consists of two pools comprising 815 amplicons designed to evaluate the presence of 478 antimicrobial resistance (AMR) genes. This collaborative effort involved experts from Lawrence Livermore National Laboratory, University of Chicago, Argonne National Laboratory, Los Alamos National Laboratory, and Naval Research Laboratory.
4 Impact of Antibiotic Resistance on Public Health
The rapid emergence of antibiotic resistance development through wastewater and WWTPs is a serious concern and negatively affects the environment and public health. Heavily populated cities drinking-water aquifers are usually polluted with wastewater effluent (Sedlak et al., 2000). Through such practices, antibiotics and resistant bacteria are introduced into the drinking-water systems and increasing the potential risk of human exposure. There are different possible routes by which antibiotics and ARGs from WWTPs and agricultural runoffs can reach the consumers of treated water. The presence of discharge points of municipal WWTPs upstream of a pumping station, seepage and runoff of lagoon wastewater from animal feeding operations, and farm application of the lagoon sediments are few potentially risky sources that allow the introduction of antibiotics and antibiotic-resistant bacteria in the water supplies. The amount of antibiotics and ARB in the water supplies is highly influenced by the disinfection process employed and its frequency of use. Chlorine is a generally effective disinfection process, a significant increase in ARB was observed in chlorinated swine lagoons. The plausible reason behind that is the presence of chlorine-resistant bacteria may reduce bacterial removal efficiency during chlorination due to the development of resistance against disinfectants (Macauley et al., 2006). WHO has categorized 12 bacterial species and their accompanying AMR profiles that are found highly hazardous to human health (WHO, 2019). Majorly the Gram-negative bacteria are included in the list, the most common etiologic agents and mainly found in hospital- and/or community-acquired infections. Table 1 shows the categorization of these bacterial species and drugs for which they possess resistance.
The presence of even traces of antibiotics in WWTPs and the availability of high bacterial species make the environment highly favorable for the development of resistant strains. The presence of antibiotics works as a selection pressure and this stress induces mutations in bacteria and thus they acquire resistance not only against a single drug but also against multiple antibiotics. Further, these resistance strains rapidly transfer their ARGs to the available high bacterial population, and thus, resistance spread exponentially through HGT methods (Korzeniewska et al., 2013). Waste generated by hospitals is considered a major source of ARB which is further transferred to WWTPs and contaminates surface water/groundwater and agricultural soil. Proper safety measures should be acquired before the discharge of hospital waste and reliable testing of discharge samples must be employed to analyze a load of resistant bacterial strains and the number of antimicrobial residue/agents in the hospital waste to adopt necessary control measures for the same. To prevent the dissemination of ARB, the use of effective disinfection processes with their correct doses is essentially required (Exner et al., 2017). The use of sub-lethal doses can trigger resistance. Identification of effective disinfectants and measurement of their appropriate doses are very crucial and essential steps used to eliminate ARB from waste discharge. The discharge of antibiotics by living beings into the environment through their feces and urine is a serious matter of concern. The humans and animals generated waste contains a mixture of partially metabolized bioactive compounds and xenobiotic compounds. The discharged antibiotics by this waste are further incorporated into the municipal sewers, sewage sludge, and the soil by different routes. As municipal wastewater contains high levels of inorganic and organic matter that accelerate the growth of microorganisms present in the waste including commensal, pathogenic, and non-pathogenic bacteria, further emergence of antibiotics in this waste causes the rapid development of resistance through HGT (Silva et al., 2006). Thus, the WWTPs are known as potential hot spots for promoting the spread of antibiotic resistance and overall increase ARB and ARGs in the environment (Moura et al., 2011). Research findings showed the presence of higher percentages of MDR bacteria in the effluent of treated wastewater than in the affluent, confirming the increased impact of remnants of antibiotics in the aquatic environment. Thus, there is an emerging need for the development of effective strategies of antibiotics removal/degradation from WWTPs as the persistence of microbial contaminants is a major threat to public health (Harnisz, 2013). Different factors like the design and operation of WWTPs affect the dynamics of ARB and ARGs in wastewater. Even after treatment of water, complete removal of microbes cannot be ensured, and their potential risks cannot be overlooked. In distribution system pipe proliferation of bacteria in drinking has been observed even after chlorination (Marathe et al., 2017).
Different factors like an increase of antibiotic-resistant bacteria, decrease in the count of antibiotic-sensitive bacteria, type of antibiotic resistance acquired, and dose of disinfectant, all affect the increase in the ARB content in the effluent of WWTPs. Pathogenic bacterial strains which acquire resistance against one or more antibiotics might be transferred from the environment to humans and pose a significant threat (Blasco et al., 2008). The chances of acquiring resistance by human pathogens are also increasing and affecting their population dynamics. The presence of multi-resistance bacteria and genes in drinking water has been reported. Multi-resistant Salmonella is identified in water used for spraying vegetables (Parvathi et al., 2011). The development of multidrug resistance phenotypes imposes further limitations on the available therapeutic options. Thus, the selection of an effective process for wastewater treatment can play an important role in the reduction of ARB and ARG contaminants from the effluent which will further reduce their possible risks to the environment and public health. Different factors can control the choice of wastewater treatment method to be used. Improvement in treatment technologies can reduce the chances of ARB infections.
Transfer of ARGs from non-pathogenic strains to pathogenic strains that may infect humans and cause severe health risks is a serious issue. Pomati et al. (2006) reported inhibition in the proliferation of human embryonic cells by a complex mixture of therapeutic drugs (including four antibiotics) and these pharmaceuticals might potentially affect aquatic life as well. Inhibitory effects of low levels of pharmaceuticals were observed on the catalytic activities of different xenobiotic-metabolizing enzymes present in carp liver (Thibaut et al., 2006; Shah Maulin, 2020, 2021a, 2021b). In a study conducted by Wilson et al. (2003) eco-toxicological effects of ciprofloxacin antibiotic along with triclosan (antiseptic) and tergito NP10 (surfactant) were reported in the aquatic environment. The mixture of pharmaceutical products significantly affected the rate of algal biomass production and algal community structure. The authors used different concentrations of ciprofloxacin antibiotics and studied its impact on the algal community structure in the upstream and downstream samples of a WWTP. Findings suggested that that continuous exposure of ciprofloxacin influenced the algal community structure and later it shifts the food web structure of streams. Halling-Sørensen (2001) evaluated the EC50 value of certain antibiotics to activated sludge bacteria and Nitrosomonas europaea and found maximum toxic effects were exerted by chlortetracycline and oxolinic acid. The assessment of human health risks associated with ARB and ARGs in aquatic environments remains a challenge due to the lack of specific data, such as dose-response relationships and exposure assessments. Amarasiri et al. outlines recent studies on human health risk evaluations related to ARB and ARGs, emphasizing the need for additional data to conduct a refined quantitative microbial risk assessment (QMRA) for various scenarios. QMRA, which combines information on occurrence, exposure, and dose-response, has been applied to assess health risks related to pathogens like E. coli, Campylobacter, and Legionella. While QMRA is considered a suitable method for assessing the additional human health risks posed by ARB, more research is needed. Recent studies have evaluated exposure to antibiotic-resistant E. coli through drinking water and irrigation, indicating potential risks. The transfer of ARGs varies based on bacterial counts, and even a low dose of ARB can pose health risks, particularly for individuals with compromised immune systems. However, limited data on exposure and dose-response for ARB and ARGs hinder the quantitative assessment of these risks, necessitating further research to better understand their implications in various scenarios (Amarasiri et al., 2020).
The antibiotic resistance ultimately decreases societal productivity and increases the number of side effects (WHO report, 2019). The major risk groups who are highly susceptible and can be devastatingly affected by antibiotic resistance are.
-
Infants, especially premature babies, due to their less developed and weak immune systems.
-
Seniors, who are living in long-term care facilities or seniors’ residences for a long period. As they have more chances of infection exposure, they are in close contact with many other people and have weakened immune systems due to prolonged illness or extended use of medicines.
-
Homeless people or those who are living in crowded or unhygienic conditions. They have a higher risk of infection exposure.
-
People with weak immune systems due to prolonged illness or injury.
-
People are associated with healthcare facilities and working in daycare centers or other settings where chances of spreading infections are very high, especially when proper infection prevention and control measures are not followed.
Antibiotic resistance is an emerging threat for society. To avoid the environmental spreading of ARB and ARGs, specific assessment tools and methods are required to identify the antibiotic residue and the antibiotic-resistant determinants present in the wastewater. Intensive and result-oriented research is required for developing effective treatment and disinfection methods for the complete elimination or degradation of ARB in WWTPs as their implications on public health and the environment are challenging and difficult to handle. The WWTPs are best suited for the removal of solids, organic matter, and nutrients, but unfortunately, their design and operation do not support the removal of antibiotics and ARB. However, advanced technologies are being used in WWTPs for water treatment but identification of effective treatment technology and complete removal of ARB and ARG from the effluent of WWTPs is still very challenging. Mathematical modeling appropaches can also help us understand how bacteria and antibiotics interact in water treatment plants. These models have been useful for wastewater treatment but haven't focused much on antibiotic resistance. Some models have explored resistance in specific environments. Baker et al., developed a mathematical model to quantify the spread of antimicrobial resistance in stored agricultural waste (Baker et al., 2016). More data is required to fine-tune such models and include resistance in everyday wastewater treatment plant operations. Combining different modelling approaches, from population-level models to individual-based models, could give a clearer picture of how antibiotic resistance spreads. The main challenge is the lack of data to validate and improve these models. Developing models that consider resistance, bacteria, gene transfer, and antibiotic levels could help us understand and control antibiotic resistance better.
5 Conclusion and Future Perspectives
Different studies suggest the presence of high microbial densities, residual antibiotics, and high microbial, growth rates are fundamental issues of conventional WWTP design that may represent the perfect niche for promoting rapid transfer of ARGs and spread of multi antibiotic resistance among residents bacteria. Wastewater treatment plants are well designed for the treatment of wastewater but to improve their antibiotic removal efficiencies their designs and operation should be optimized. The advanced treatment technologies used in drinking-water production such as activated carbon, ozonation, and membrane technologies can efficiently reduce the concentration of micropollutants, however; these are not affordable for many municipal WWTP facilities. Using the combined physical and biological treatment process and optimizing the operational condition of WWTPs can help in the effective removal of pharmaceuticals. It is difficult to identify the actual load of antibiotics in WWTPs as many of them transformed into another form and reactivate. Furthermore, in the complex environment of WWTPs, the assessment of effective doses of antibiotics which can raise risks of resistance development is also very difficult. For evaluation of the complete risk of pharmaceuticals in the environment, it is important to identify the types, abundance of transformed products, and their biological activities that contribute to adverse ecological and health effects. The advance and sensitive analytical instrumentation can be used for the identification of traces of metabolites present in the WWTPs. Toxicity assessment of metabolites and mixtures of microcontaminants is essentially required for a complete and reliable assessment of their adverse ecological and human health consequences. There is an emerging need to update the current water quality standards to identify the quality of water in terms of the acceptable levels of micropollutants present in it before its discharge into the environment. Critical quality assessment of wastewater, recycled into WWTPs, is important to reduce the significant hazards posed on the environment and public health.
The absence of standardized regulations for monitoring microcontaminants has led to a rise in environmental antibiotic resistance, endangering public health and ecological stability. Wastewater treatment plant effluents, containing ARGs, significantly contribute to the spread of resistance among various bacteria through horizontal gene transfer. This poses a risk to humans and animals, challenging the “One Health” initiative endorsed by the World Health Organization and hindering the achievement of the United Nations Sustainable Development Goals. To mitigate this, advanced technologies are essential for removing antibiotics and ARGs from wastewater. Setting stringent limits on antibiotic release from sources like hospitals and agriculture, while also controlling other drugs and biocides driving resistance, is crucial.
The effluent of WWTPs is used for many purposes without knowing the potential hazards accompanied by it. Critical assessment of actual microbial load, amount of residual antibiotics or their transformed compounds, and their biological activities must be checked before the recyled water is used. Antibiotic resistance and water sustainability are global grand challenges that need immediate attention and must be addressed. There is a need for continuous investment and innovation in research, technology, and policy development, particularly where the two challenges intersect. Very few epidemiological studies have been conducted to evaluate evidence of recycled water as a source of microbial illness. Next-generation sequencing, metagenomics, and other new molecular epidemiological approaches are providing insights into the relative contributions of various sources to antibiotic-resistant diseases in humans. Assessment of their impact on the environment and public health is required. However, quantification of risks with the effect of other related sources is quite challenging. Identification of low-cost mitigation strategies that work with existing infrastructure or upgrade plans, and can also provide additional benefits of water treatment, nutrient recycling, and watershed protection is the need of the hour. Further research on environmental antibiotic contamination and its link to resistance development is needed to enhance intervention measures. Although the occurrence of antibiotic ARGs in the One-Health cycle may vary, it is believed that some ARGs create connections between different niches and environmental areas. Integrated monitoring programs are necessary to track the spread of ARB and ARGs across the One-Health framework. Challenges include aligning methods and sites, deciding what biological entities to target (bacteria, mobile genetic elements, or ARGs), and ensuring method sensitivity and accuracy. The ultimate goal is to provide data for authorities and policymakers to develop guidelines for preventing resistance spread, but translating scientific data into practical information remains a significant challenge.
The above-mentioned areas need susbtancial scientific inputs to address the issue of the rapid emergence of antibiotic resistance and its potential hazards to the environment and public health. To conclude, addressing the issue of ARGs and ARBs and their potential impact on public health, several strategies should be considered. These strategies involve implementing more robust surveillance and monitoring systems to track the trends and presence of ARGs and ARBs in WWTPs, researching and developing advanced wastewater treatment technologies that specifically target and reduce these contaminants, and establishing regulatory measures and guidelines to control their release into the environment. Promoting public awareness, fostering collaboration among research institutions, wastewater authorities, and healthcare organizations, and adopting a holistic “One Health” approach that recognizes the interconnectedness of human, animal, and environmental health are all pivotal. Furthermore, encouraging antibiotic stewardship within healthcare settings, promoting international cooperation, and continuing to research innovative technologies for ARG and ARB removal are essential components. Long-term health impact studies are also necessary to gain insights into the consequences of the increased presence of ARGs and ARBs in the environment. These comprehensive strategies are designed to mitigate the risks associated with ARGs and ARBs in wastewater and protect public health from the growing menace of antibiotic resistance.
References
Amarasiri, M., Sano, D., & Suzuki, S. (2020). Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: Current knowledge and questions to be answered. Critical Reviews in Environmental Science and Technology, 50(19), 2016–2059.
Baker, M., Hobman, J. L., Dodd, C. E., Ramsden, S. J., & Stekel, D.J. (2016). Mathematical modelling of antimicrobial resistance in agricultural waste highlights importance of gene transfer rate. FEMS Microbiology Ecology, 92(4), fiw040.
Batt, A. L., Bruce, I. B., & Aga, D. S. (2006). Evaluating the vulnerability of surface waters to antibiotic contamination from varying wastewater treatment plant discharges. Environmental Pollution, 142, 295–302.
Batt, A., Kim, S., & Aga, D. (2007). Comparison of the occurrence of antibiotics in four full-scale wastewater treatment plants with varying designs and operations. Chemosphere, 68, 428–435.
Blasco, M. D., Esteve, C., & Alcaide, E. (2008). Multiresistant waterborne pathogens isolated from water reservoirs and cooling systems. Journal of Applied Microbiology, 105, 469–475.
Bloemberg, G. V., Polsfuss, S., Meyer, V., Bottger, E. C., & Hombach, M. (2014). Evaluation ofthe AID ESBL line probe assay for rapid detection of extended-spectrumbeta-lactamase (ESBL) and KPC carbapenemase genes in Enterobacteriaceae. Jantimicrob Chemotherapy, 69, 85–90.
Bogaerts, P., Cuzon, G., Evrard, S., Hoebeke, M., Naas, T., & Glupczynski, Y. (2016). Evaluation of a DNA microarray for rapid detection of the most prevalent extended-spectrumbeta-lactamases, plasmid-mediated cephalosporinases and carbapenemases inEnterobacteriaceae, Pseudomonas and Acinetobacter. International Journal of Antimicrobial Agents, 48, 189–193.
Borjesson, S., Melin, S., Matussek, A., & Lindgren, P. E. (2009). A seasonal study of the mecA gene and Staphylococcus aureus including methicillinresistant S. aureus in a municipal wastewater treatment plant. Water Research, 43, 925−932.
Cassini, A., Högberg, L. D., Plachouras, D., Quattrocchi, A., Hoxha, A., Simonsen, G. S., Colomb-Cotinat, M., Kretzschmar, M.E ., Devleesschauwer, B., & Cecchini, M. et al. (2015). Faculty opinions recommendation of attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modeling analysis. Fac Opin Post-Publ Peer Rev Biomed Lit, 2019(19), 56–66.
CDC report. (2019). The biggest antibiotic-resistant threats in the U.S. Centers for Disease Control and Prevention. https://www.cdc.gov/drugresistance/biggest-threats.html.
Chander, Y., Kumar, K., Goyal, S. M., & Gupta, S. C. (2005). Antibacterial activity of soil-bound antibiotics. Journal of Environmental Quality, 34, 1952–1957.
Cuzon, G., Naas, T., Bogaerts, P., Glupczynski, Y., & Nordmann, P. (2012). Evaluation of a DNAmicroarray for the rapid detection of extended-spectrum beta-lactamases (TEM, SHV and CTX-M), plasmid-mediated cephalosporinases (CMY-2-like, DHA, FOX, ACC-1, ACT/MIR and CMY-1-like/MOX) and carbapenemases (KPC, OXA-48, VIM, IMP and NDM). Journal of Antimicrobial Chemotherapy, 67, 1865–1869.
Di Cesare, A., Eckert, E. M., D’Urso, S., Bertoni, R., Gillan, D. C., Wattiez, R., et al. (2016). Co-occurrence of integrase 1, antibiotic and heavy metal resistance genes in municipal wastewater treatment plants. Water Research, 94, 208–214. https://doi.org/10.1016/j.watres.2016.02.049.
Dodge, C. P. (1990). Health implications of war in Uganda and Sudan. Social Science & Medicine, 3(1), 691–698.
Du, J., Geng, J., Ren, H., Ding, L., Xu, K., & Zhang, Y. (2015). Variation of antibiotic resistance genes in municipal wastewater treatment plant with A2O-MBR system. Environmental Science and Pollution Research., 22(5), 3715–3726.
Ekpeghere, K. I., Lee, J. W., Kim, H. Y., Shin, S. K., & Oh, J. E. (2017). Determination and characterization of pharmaceuticals in sludge from municipal and livestock wastewater treatment plants. Chemosphere, 168, 1211–1221.
Exner, M. et al. (2017). Antibiotic resistance: What is so special about multidrug- resistant gram-negative bacteria? GMS Hygiene and Infection Control, 12, Doc05. PMC. Web. October 8, 2017.
Fang, H., Zhang, Q., Nie, X., Chen, B., Xiao, Y., Zhou, Q., et al. (2017). Occurrence and elimination of antibiotic resistance genes in a long-term operation integrated surface flow constructed wetland. Chemosphere, 173(99–10), 6. https://doi.org/10.1016/j.chemosphere.2017.01.027.
Grabow, W. O., & Prozesky, O. W. (1973). Drug resistance of coliform bacteria in hospital and city sewage. Antimicrobial Agents Chemotherapy, 3, 175–180. https://doi.org/10.1128/AAC.3.2.175.
Halling-Sørensen, B. (2001). Inhibition of aerobic growth and nitrification of bacteria in sewage sludge by antibacterial agents. Archives of Environmental Contamination and Toxicology, 40, 451–460.
Halling-Sørensen, B., Nors Nielsen, S., Lanzky, P. F., Ingerslev, F., Holten Lützhøft, H. C., & Jorgensen, S. E. (1998). Occurrence, fate and effects of pharmaceutical substances in the environment—A review. Chemosphere, 36, 357–393.
Harnisz, M. (2013). Total resistance of native bacteria as an indicator of changes in the water environment. Environmental Pollution, 174, 85–92.
Hassoun-Kheir, N., Stabholtz, Y., Kreft, J.-U., de la Cruz, R., Romalde, J. L., Nesme, J., et al. (2020). Comparison of antibiotic-resistant bacteria and antibiotic resistance genes abundance in hospital and community wastewater: a systematic review. Science of the Total Environment, 5, 140804. https://doi.org/10.1016/j.scitotenv.2020.140804.
Hinic, V., Ziegler, J., Straub, C., Goldenberger, D., & Frei, R. (2015). Extended-spectrumbeta-lactamase (ESBL) detection directly from urine samples with the rapidisothermal amplification-based eazyplex(R) SuperBug CRE assay: proof of concept. Journal of Microbiology Methods, 119, 203–205.
Jones, O. A., Lester, J. N., & Voulvoulis, N. (2005). Pharmaceuticals: A threat to drinking water? Trends in Biotechnology, 23, 163–167.
Kahlmeter, G., & Turnidge, J. (2022). How to: eCOFFs—the why, the how, and the don'ts of EUCAST epidemiological cutoff values. Clinical Microbiology and Infection, 28(7), 952–954.
Korzeniewska, E., Korzeniewska, A., & Harnisz, M. (2013). Antibiotic resistant Escherichia coli in hospital and municipal sewage and their emission to the environment. Ecotoxicology and environmental Safety, 91, 96–102.
Kümmerer, K. (2003). Significance of antibiotics in the environment. Journal of Antimicrobial Chemotherapy, 52, 5–7.
LaPara, T. M., Burch, T. R., McNamara, P. J., Tan, D. T., Yan, M., & Eichmiller, J. J. (2011). Tertiary-treated municipal wastewater is a significant point source of antibiotic resistance genes into duluth-superior harbor. Environmental Science and Technology, 45, 9543–9549.
LaPara, T. M., & Burch, T. R. (2012). Municipal wastewater as a reservoir of antibiotic resistance? In P. Keen, & M. Monforts (Eds.), Antimicrobial Resistance in the Environment. Wiley, ISBN 978-0-470-90542-5.
Larson, E. (2007). Community factors in the development of antibiotic resistance. Annual Review of Public Health, 28(1), 435–447.
Laxminarayan, R., Duse, A., Wattal, C., Zaidi, A. K., Wertheim, H. F., Sumpradit, N., Vlieghe, E., Hara, G. L., Gould, I. M., Goossens, H., et al. (2013). Antibiotic resistance-the need for global solutions. The Lancet Infectious Diseases, 13, 1057–1098.
Le, L. T., Huang, Z., Whiteson, K., & Jiang, S. (2022) The occurrence and diversity of antibiotic resistance and virulence factor genes in wastewater from four North American treatment plants. Environmental Science: Water Research & Technology, 8(8), 1650–1664.
Ledeboer, N. A., Lopansri, B. K., Dhiman, N., Cavagnolo, R., Carroll, K. C., Granato, P., et al. (2015). Identification of gram-negative bacteria and genetic resistance determi-nants from positive blood culture broths by use of the Verigene Gram-NegativeBlood culture multiplex microarray-based molecular assay. Journal of Clinical Microbiology, 53, 2460–2472.
Li, X., Harwood, V. J., Nayak, B., & Weidhaas, J. L. (2016). Ultrafiltration and microarray for detection of microbial source tracking marker and pathogen genes in riverine and marine systems. Applied and Environmental Microbiology, 82(5), 1625–1635.
Li, Y., Shi, X., Zuo, Y., Li, T., Liu, L., Shen, Z., Shen, J., Zhang, R., & Wang, S. (2022). Multiplexed target enrichment enables efficient and in-depth analysis of antimicrobial resistome in metagenomes. Microbiology Spectrum, 10(6), e02297–22.
Li, B., Yang, Y., Ma, L. P., Ju, F., Guo, F., Tiedje, J. M., & Zhang, T. (2015). Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME Journal, 9(11), 2490–2502.
Ling, L. L., Schneider, T., Peoples, A. J., Spoering, A. L., Engels, I., Conlon, B. P., Mueller, A., Schäberle, T. F., Hughes, D. E., Epstein, S., Jones, M., Lazarides, L., Steadman, V. A., Cohen, D. R., Felix, C. R., Fetterman, K. A., Millett, W. P., Nitti, A. G., Zullo, A. M., … Lewis, K. (2015). A new antibiotic kills pathogens without detectable resistance. Nature, 517, 455.
Lirermore, D. M. (1995). _lactamases in laboratory and clinical resistance. Clinical Microbiology Reviews, 8(5), 57–84.
Macauley, J. J., Qiang, Z., Adams, C., D., Surampalli, R., & Mormile, M. R. (2006). Disinfection of swine wastewater using chlorine, ultraviolet light and ozone. Water Research, 40, 2017–2026.
Manoharan, R. K., Srinivasan, S., Shanmugam, G., & Ahn, Y. H. (2021). Shotgun metagenomic analysis reveals the prevalence of antibiotic resistance genes and mobile genetic elements in full scale hospital wastewater treatment plants. Journal of Environmental Management, 296, 113270.
Marathe, N. P., Pal, C., Gaikwad, S. S., Jonsson, V., Kristiansson, E., & Larsson, D. G. J. (2017). Untreated urban waste contaminates Indian river sediments with resistance genes to last resort antibiotics. Water Research, 124, 388–397.
March Rosselló, G. A., & Bratos Pérez, M. A. (2016). Antibiograma rápido en MicrobiologíaClínica. Enfermedades Infecciosas y Microbiología Clínica, 34, 61–68.
Martínez, J. L. (2009). Environmental pollution by antibiotics and by antibiotic resistance determinants. Environmental Pollution, 157, 2893–2902.
Martínez, J. L., et al. (2014). What is a resistance gene? Ranking risk in resistomes. Nature Reviews Microbiology, 13, 116–123.
Minh, T. B., Leung, H. W., Loi, I. H., Chan, W. H., So, M. K., Mao, J., Choi, D., Lam, J. C. W., Zheng, G., Martin, M., et al. (2009). Antibiotics in the Hong Kong metropolitan area: Ubiquitous distribution and fate in Victoria Harbour. Marine Pollution Bulletin, 58, 1052–1062.
Moura, A., Carolina Pereira, C., Henriques, I., & Correia, A. (2011). Novel gene cassettes and integrons in antibiotic-resistant bacteria isolated from urban wastewaters. Research Microbiology, 163(2011), 92–100.
Mutuku, C., Gazdag, Z., & Melegh, S. (2022). Occurrence of antibiotics and bacterial resistance genes in wastewater: resistance mechanisms and antimicrobial resistance control approaches. World Journal of Microbiology and Biotechnology, 38(9), 152.
O’neill, J. (2014). Review on antimicrobial resistance: Tackling a crisis for the health and wealth of nations. HM Government.
Parvathi, P., Vijayan, J., Murali, G., & Chandran, P. (2011). Comparative virulence genotyping and antimicrobial susceptibility profiling of environmental and clinical Salmonella enterica from Cochin, India. Current Microbiology, 62, 21–26.
Pazda, M., Rybicka, M., Stolte, S., Piotr Bielawski, K., Stepnowski, P., Kumirska, J., Wolecki, D., & Mulkiewicz, E. (2020). Identification of selected antibiotic resistance genes in two different wastewater treatment plant systems in Poland: A preliminary study. Molecules, 25(12), 2851.
Peng, X., Tan, J., Tang, C., Yu, Y., & Wang, Z. (2008). Multiresidue determination of fluoroquinolone, sulfonamide, trimethoprim, and chloramphenicol antibiotics in urban waters in China. Environmental Toxicology and Chemistry, 27, 73.
Pomati, F., Castiglioni, S., Zuccato, E., Fanelli, R., Vigetti, D., Rossetti, C., & Calamari, D. (2006). Effects of a complex mixture of therapeutic drugs at environmental levels on human embryotic cells. Environmental Science and Technology, 40, 2442–2447.
Pruden, A., Arabi, M., & Storteboom, H. N. (2012). Correlation between upstream human activities and riverine antibiotic resistance genes. Environmental Science and Technology, 46, 11541–11549.
Rahal, K., Wang, F., & Schindler, J. et al. (1997). Reports on surveillance of and microbial resistance on individual countries. Clinical Infectious Diseases, 24(Suppl 1), S169-75.
Rodriguez-Mozaz, S., Chamorro, S., Marti, E., Huerta, B., Gros, M., Sànchez- Melsió, A., et al. (2015). Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Research, 69, 234–242. https://doi.org/10.1016/j.watres.2014.11.021.
Rumky, J., Kruglova, A., & Repo, E. (2022). Fate of antibiotic resistance genes (ARGs) in wastewater treatment plant: Preliminary study on identification before and after ultrasonication. Environmental Research, 215, 114281.
Sedlak, D. L., Gray, J. L., & Pinkston, K. E. (2000). Understanding microcontaminants in recycled water. Environmental Science and Technology, 34, 508A-515A.
Shah, M. P. (2020). Microbial bioremediation & biodegradation. Springer.
Shah, M. P. (2021a). Removal of refractory pollutants from wastewater treatment plants. CRC Press.
Shah, M. P. (2021b). Removal of emerging contaminants through microbial processes. Springer.
Sharma, V. K., Johnson, N., Cizmas, L., McDonald, T. J., & Kim, H. (2016). A review of the influence of treatment strategies on antibiotic resistant bacteria and antibiotic resistance genes. Chemosphere, 150, 702–714. https://doi.org/10.1016/j.chemosphere.2015.12.084.
Silva, J., Castillo, G., Callejas, L., Lopez, H., & Olmos, J. (2006). Frequency of transferable multiple antibiotic resistance amongst coliform bacteria isolated from a treated sewage effluent in Antofagasta, Chile, Elect. Journal of Biotechnology, 9, 533–540.
Stuart, J. C., Voets, G., Scharringa, J., Fluit, A. C., & Leverstein-van Hall, M. A. (2012). Detec-tion of carbapenemase-producing Enterobacteriaceae with a commercial DNAmicroarray. Journal of Medical Microbiology, 61, 809–812.
Szekeres, E., Baricz, A., Chiriac, C. M., Farkas, A., Opris, O., Soran, M. L., et al. (2017). Abundance of antibiotics, antibiotic resistance genes and bacterial community composition in wastewater effluents from different Romanian hospitals. Environment and Pollution, 225, 304–315. https://doi.org/10.1016/j.envpol.2017.01.054.
Szczepanowski, R., Link e, B., Krahn, I., Gartemann, K.-H., Gützkow, T., Eichler, W., Pühler, A., & Schlüter, A. (2009). Detection of 140 clinically relevant antibiotic-resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiology, 2009(155), 2306–2319.
Thibaut, R., Schnell, S., & Porte, C. (2006). The interference of pharmaceuticals with endogenous and xenobiotic metabolizing enzymes in cap liver: An in-vitro study. Environmental Science & Technology.
Thirunavukarasou, A., Kaur, S., & Khera, H. K. (2022). Metagenomics for studying microbes in wastewater treatment plants. In Microbial Community Studies in Industrial Wastewater Treatment, (pp. 171–183). CRC Press.
WHO. (2017). Antibacterial agents in clinical development: An analysis of the antibacterial clinical development pipeline, including tuberculosis. World Health Organization. WHO/EMP/IAU/2017.11.
Wilson, B. A., Smith, V. H., deNoyelles, F., Jr., & Larive, C. K. (2003). Effects of three pharmaceutical and personal care products on natural freshwater algal assemblages. Environmental Science Technology, 37, 1713–1719.
World Health Organization. (2019). Report on global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics.
Xu, Y. B., Hou, M. Y., Li, Y. F., Huang, L., Ruan, J. J., Zheng, L., et al. (2017). Distribution of tetracycline resistance genes and AmpC b- lactamase genes in representative non-urban sewage plants and correlations with treatment processes and heavy metals. Chemosphere, 170, 274–281. https://doi.org/10.1016/j.chemosphere.2016.12.027.
Yang, Y., Li, B., Zou, S. C., Fang, H. H. P., & Zhang, T. (2014). Fate of antibioticresistance genes in sewage treatment plant revealed bymetagenomic approach. Water Research, 62, 97–106.
Zhou, B., Wang, C., Zhao, Q., Wang, Y., Huo, M., Wang, J., et al. (2016). Prevalence and dissemination of antibiotic resistance genes and coselection of heavy metals in Chinese dairy farms. Journal of Hazardous Materials, 320, 10–17. https://doi.org/10.1016/j.jhazmat.2016.08.007.
Zhu, Y.-G., et al. (2013). Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proceedings of the National Academy of Sciences of the United States of America, 110, 3435–3440.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Kour, N., Singh, J., Khera, H.K. (2023). Increasing Prevalence of Antibiotic-Resistant Genes in Wastewater: Impact on Public Health. In: Shah, M.P. (eds) Genomics of Antibiotic Resistant Bacteria in Industrial Waste Water Treatment. Springer, Cham. https://doi.org/10.1007/978-3-031-44618-4_5
Download citation
DOI: https://doi.org/10.1007/978-3-031-44618-4_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-44617-7
Online ISBN: 978-3-031-44618-4
eBook Packages: Earth and Environmental ScienceEarth and Environmental Science (R0)