Over 170 million people are infected with hepatitis C virus (HCV), a major cause of acute and chronic liver disease that can lead to cirrhosis and hepatocellular carcinoma (Alter and Seeff 2000). The success of this virus is largely due to its ability to cause persistent infections – often lasting for decades – in over 70% of infected individuals. Thus, HCV infection leads to a dynamic interplay between viral replication, host antiviral responses, and viral countermeasures to evade those responses. Understanding these processes will be crucial for devising effective strategies to combat this virus and alleviate the human suffering it exacts. In this chapter we review the current understanding of HCV genome replication, emphasizing the role of viral and host factors in this process. Where applicable, we will draw comparisons to other viruses within this volume. Nevertheless, due to space limitations this review is not meant to be comprehensive, and we apologize in advance to authors whose work could not be cited.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Introduction
Over 170 million people are infected with hepatitis C virus (HCV), a major cause of acute and chronic liver disease that can lead to cirrhosis and hepatocellular carcinoma (Alter and Seeff 2000). The success of this virus is largely due to its ability to cause persistent infections – often lasting for decades – in over 70% of infected individuals. Thus, HCV infection leads to a dynamic interplay between viral replication, host antiviral responses, and viral countermeasures to evade those responses. Understanding these processes will be crucial for devising effective strategies to combat this virus and alleviate the human suffering it exacts. In this chapter we review the current understanding of HCV genome replication, emphasizing the role of viral and host factors in this process. Where applicable, we will draw comparisons to other viruses within this volume. Nevertheless, due to space limitations this review is not meant to be comprehensive, and we apologize in advance to authors whose work could not be cited.
Overview of the HCV Life Cycle
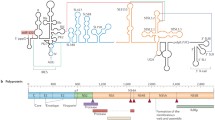
HCV is an enveloped, positive-strand RNA virus classified within the family Flaviviridae. The life cycle of HCV (Fig. 4.1) therefore shares overall similarity to the flaviviruses (Chapter 3) and other positive-strand RNA viruses. Upon infection of a host cell, HCV particles are taken up by receptor-mediated endocytosis and trafficked to endosomes, where the low pH of this compartment induces fusion of the viral envelope and bounding endosomal membrane. The nucleocapsid is then uncoated to release the viral genome into the cytoplasm (step 1), where it can be directly translated to produce the viral structural and non-structural (NS) proteins (step 2). Viral NS proteins and host factors assemble into a cytoplasmic, membrane-bound RNA replicase (step 3), which then recruits the HCV genome out of translation and into replication (step 4). After RNA synthesis, new viral genomes can be recycled back into translation and replication or packaged by viral structural proteins into nascent viral particles (step 5).
Until recently, only limited aspects of the HCV life cycle could be studied because efficient viral culture systems did not exist. Once the viral genome was fully sequenced, infectious cDNA clones were constructed and shown to initiate replication upon intrahepatic inoculation into chimpanzees (Kolykhalov et al. 1997; Yanagi et al. 1997). While these reverse genetic systems were functional in vivo, they were obviously limited by the ethical and practical issues of primate research and did not permit viral replication to be studied in cell culture. The first broadly useful system for studying HCV RNA replication came when Lohmann et al. (1999) engineered bicistronic “subgenomic” replicons to express the selectable marker gene Neo, and selected for rare HCV replication events after transfecting this RNA replicon into the human hepatoma line, Huh-7 (Nakabayashi et al. 1982). Further growth of Neo-resistant cells selected for mutant replicons with increased RNA replication (Blight et al. 2000; Lohmann et al. 2001, 2003). Thus, cell culture-adapted replicons allowed the intracellular aspects of the viral life cycle to be studied and provided much-needed cell-based assays to screen for HCV-specific antivirals. More recently, Takaji Wakita and other investigators showed that the HCV strain JFH-1 was capable of producing infectious virus in cell culture (Lindenbach et al. 2005; Wakita et al. 2005; Zhong et al. 2005), and additional HCV cell culture systems have recently become available (reviewed in (Tellinghuisen et al. 2007)). Thus, the tools to study the complete life cycle of HCV are now in hand.
HCV Genomes
The HCV genome is a monopartite, single-stranded RNA, 9.6 kb in length (Choo et al. 1991). Unlike most cellular mRNAs, the HCV genome lacks a 5′ cap and does not encode a 3′ polyadenosine tail (Tanaka et al. 1995; Kolykhalov et al. 1996). Instead, the HCV genome encodes several cis-acting RNA elements (CREs) that regulate genome translation, RNA replication, and most likely, packaging. Known CREs include sequences and secondary structures within the 5′ and 3′ noncoding regions (NCRs) and the large open reading frame (Fig. 4.2). Unlike the picornaviruses (Chapter 1), HCV does not appear to encode a viral genome-linked protein.
Features of the HCV genome. This map of the HCV genome highlights important secondary structures within the HCV genomic RNA, as described in the text. The large open reading frame is indicated as an open bar. Loop regions involved in the kissing interaction between 5BSL-3.2 and 3′X SL2 are indicated by hearts. AUG, start codon; UGA, stop codon; VR, variable region.
As for all positive-strand RNA viruses, which encode polymerases that lack proof reading activity, HCV exhibits a high degree of genetic variability. Given the size of the HCV genome and an estimated mutation rate of ≈10–4 misincorporations/nt (Crotty et al. 2001), one can calculate that mutants will quickly accumulate and predominate in HCV populations of even modest size. It has been estimated that a chronically infected person makes 1012 virions/day (Perelson et al. 2005). Thus, the sequence diversity present even within a single person is huge. This swarm of genetically related viruses, with no real “wild-type” master sequence, is often referred to as a “quasi-species”. On the global scale, with over 108 infected people, the amount of sequence diversity available to HCV is astronomical. Of course evolutionary fitness varies greatly among these populations. As a result, HCV has evolved into six metastable genotypes, which differ by more than 30% at the nucleotide level (Simmonds et al. 2005).
The HCV 5′ NCR is 341 nt in length, well conserved, and highly structured (Fig. 4.2); it has at least two major functions in the viral life cycle. First, it encodes an internal ribosome entry site (IRES) that allows for cap-independent translation of the viral genome, described below. Second, the 5′ NCR includes one or more overlapping CREs necessary for genome replication. While efficient RNA replication requires nearly the entire 5′ NCR, the minimal 5′ replication element is encoded within the first 120 nt, which includes stem-loops I and II (Friebe et al. 2001; Kim et al. 2002b). Intriguingly, the liver-specific cellular micro RNA (miRNA)-122 binds to the unstructured spacer between these stem-loops, and this interaction is required for an early step in HCV replication (Jopling et al. 2005). Furthermore, by making intergenotypic recombinants with reduced rates of positive- and negative-strand synthesis, Binder and colleagues have confirmed that the 5′ NCR (or more likely, its reverse complement in the negative strand) plays an important role in positive-strand synthesis (Binder et al. 2007b).
The 3′ NCR consists of three subdomains: (1) a short (≈40 nt) variable region located immediately downstream of the termination codon; (2) a polyuridine/polypyrimidine (U/UC) tract of variable length; and (3) a highly conserved, 98 nt region termed the 3′X domain (Fig. 4.2) (Tanaka et al. 1995; Kolykhalov et al. 1996). The poly (U/UC) tract and 3′X domain are required for RNA replication, while the variable region seems to influence replication efficiency (Yanagi et al. 1999; Kolykhalov et al. 2000; Friebe and Bartenschlager 2002; Yi and Lemon 2003). The minimal poly (U/UC) tract appears to be between 26 and 52 nt in length and requires an uninterrupted homopolyuridine tract, suggesting that it is recognized by a trans-acting factor (Friebe and Bartenschlager 2002; Yi and Lemon 2003; You and Rice 2007). The 3′X domain consists of a highly stable 3′ terminal stem-loop, 3′ SL1, and two metastable stem-loops, 3′ SL2 and 3′ SL3 (Tanaka et al. 1996; Blight and Rice 1997). Again, by using intergenotypic chimeras Binder and colleagues confirmed that the 3′X domain serves as an important CRE for negative-strand synthesis (Binder et al. 2007b).
Given that the large open reading frame encompasses over 94% of the HCV genome, it is no surprise that HCV coding region also encodes CREs. Sequences with dual overlapping functions place tight constraints on codon usage, which can be used to help identify such internal CREs (reviewed in (Branch et al. 2005)). As an example, anomalous codon usage was originally thought to reflect a selective pressure to retain an alternative reading frame within the core gene (reviewed in (Branch et al. 2005)). Subsequent genetic analysis determined that this alternative reading frame was dispensable for virus replication, but identified an important CRE, stem-loop VI, embedded within the core gene (Fig. 4.2) (McMullan et al. 2007). While this region is not required for RNA replication in the context of subgenomic RNA replicons, full-length HCV genomes containing mutations in this structure are highly attenuated in vitro and in vivo, and selectively recover the wild-type sequence. Interestingly, one leg of stem-loop VI can base pair with the 5′ NCR, overlapping the miRNA-122 binding site, and thereby down-regulate IRES-mediated translation (Kim et al. 2003).
Embedded within the NS5B gene is another important CRE, 5BSL3.2 (Fig. 4.2) (Lee et al. 2004; You et al. 2004). A key feature of 5BSL3.2 is that one of the loop regions can base pair with the loop region of SL2 in the 3′X region of the genome (Friebe et al. 2005). Mutations in either loop region destroy HCV RNA replication, while compensatory mutations restore it (Friebe et al. 2005; You and Rice 2007). It is not yet known how this long-distance “kissing” interaction regulates HCV RNA replication, but it is interesting to note that base pairing between the coding region and both HCV NCRs is functionally important. This may reflect a general strategy to maintain genome integrity.
In addition to several CREs, the HCV genome exhibits a number of interesting features, such as a low frequency of UA and UU dinucleotides, which may reflect a selection for viral genomes that are poor substrates for the interferon-inducible RNase L (Han and Barton 2002). The HCV genome is also predicted to contain an unusually high rate of internal base pairing, a feature that correlates with a high rate of persistent infection among positive-strand RNA viruses (Simmonds et al. 2004). Thus, these higher-order structures may help to circumvent innate antiviral defenses.
Translation and Polyprotein Processing
As mentioned, the HCV 5′ NCR encodes an IRES that directs cap-independent translation of the viral genome (recently reviewed in (Fraser and Doudna 2007)). The minimal HCV IRES is encoded by sequences within stem-loops II through IV (Fig. 4.2). The central part of this structure, stem-loop III, can directly bind the ribosomal 40S subunit, positioning the start codon within the ribosomal P site (Pestova et al. 1998). Importantly, this interaction appears to bypass the need for canonical eukaryotic translation initiation factors (eIFs), and functionally resembles the mechanism of prokaryotic initiation via the Shine-Delgarno sequence. Structural studies indicate that stem-loops II and III induce a conformational change in the 40S subunit, allowing the RNA-binding cleft to open (Spahn et al. 2001). Subsequently, the IRES-40S complex recruits eIF3 and the eIF2•GTP•Met-tRNA ternary complex, forming a 48S intermediate complex (Ji et al. 2004; Otto and Puglisi 2004). Interestingly, stem-loop IIIb of the HCV IRES functionally and structurally mimics the binding of the 5′ cap binding complex eIF4F (Siridechadilok et al. 2005). Following GTP hydrolysis, these initiation factors are released and the ribosomal 60S subunit is recruited into an IRES-80S complex, which is then capable of initiating protein synthesis (Ji et al. 2004; Otto and Puglisi 2004).
There has been some debate about how downstream sequences influence HCV IRES activity, and it now appears that secondary structures are disfavored in the proximal region of the core gene (Rijnbrand et al. 2001). Although originally controversial (reviewed in (Tellinghuisen et al. 2007)), it is also now clear that the HCV 3′ NCR can enhance HCV IRES-mediated translation in hepatic cells, perhaps by promoting ribosomal recycling analogous to the function of the polyA tail in cellular mRNAs (Ito et al. 1998; McCaffrey et al. 2002; Bradrick et al. 2006; Song et al. 2006). In addition, a number of cellular factors have been shown to influence HCV IRES activity in trans including the La autoantigen, which may assist in recruiting the 40S ribosomal subunit to the AUG start codon (reviewed in (Lindenbach et al. 2007b)).
Translation of the HCV genome produces a large polyprotein that is co-translationally and post-translationally cleaved by cellular and viral proteases into at least ten discrete products (Fig. 4.3). These include signal peptidase cleavage at the core/E1, E1/E2, E2/p7, and p7/NS2 junctions. In addition, the mature form of core is generated via an intramembrane cleavage of the C-terminal anchor by signal peptide peptidase. The remaining polyprotein processing steps are catalyzed by two HCV-encoded proteases. The C-terminal domain of NS2 encodes a cysteine protease that is responsible for cleaving the NS2/3 junction (Grakoui et al. 1993; Hijikata et al. 1993). The crystal structure of this domain revealed that it forms an unusual homodimeric protease with twin composite active sites (Lorenz et al. 2006). As described below, all downstream cleavages are mediated by the NS3-4A serine protease.
Features of the HCV proteins. The top illustration indicates the order of the HCV gene products as they are translated in the polyprotein. Open bullet, signal peptide peptidase cleavage; closed bullet, signal peptidase cleavage; open arrowhead, NS2-3 cysteine protease cleavage; closed arrowhead, NS3-4A serine protease cleavage. The bottom illustration indicates the topology of HCV proteins. Where available, atomic coordinates were used to render the illustration to approximate scale. Since NS2 and NS5A were crystallized as dimers, one monomer of each is colored gray.
The core protein and E1 and E2 glycoproteins are structural components of HCV virions, while p7 and NS2 proteins appear to be involved in virus assembly (Jones et al. 2007; Steinmann et al. 2007). The remaining NS proteins are responsible for modulating the intracellular aspects of the HCV life cycle, including RNA replication. Since the core, E1, E2, p7, and NS2 are dispensable for the replication of subgenomic replicons (Lohmann et al. 1999), we therefore turn our attention to the NS proteins involved in RNA replication.
Replicase Components
NS3-4A
NS3 (70 kDa) is a key component of the HCV replicase, encoding an N-terminal serine protease domain and a C-terminal RNA helicase/nucleoside triphosphatase (NTPase) domain. The activities of both enzymes are essential for viral replication. The serine protease domain of NS3 can cleave NS3/4A in cis and then interact with NS4A, which contributes one ß-strand to the chymotrypsin-like fold and activates the serine protease activity (reviewed in (Penin et al. 2004b)). In turn, NS4A anchors the NS3-4A complex to cellular membranes via an N-terminal membrane anchor. The NS3-4A serine protease is responsible for cleaving the viral polyprotein at the NS4A/B, NS4B/5A, and NS5A/5B junctions. In addition, NS3-4A can cleave the cellular proteins IPS-1 and TRIF, which normally transduce signals to activate gene expression in response to viral infection (Li et al. 2005a, b; Meylan et al. 2005). By cleaving these substrates, NS3-4A helps to circumvent cellular antiviral defenses, as described below.
NS3-4A is a member of the superfamily 2 RNA helicases, which use the energy from ATP hydrolysis to power double-stranded RNA unwinding. Recent enzymatic studies have revealed that NS3-4A unwinds 18-bp segments via several discrete 3 bp steps, each of which uses a spring-loaded mechanism to coordinate ATP hydrolysis with smaller, 1 bp advances along the substrate (Serebrov and Pyle 2004; Dumont et al. 2006; Myong et al. 2007). Although the NS3 helicase domain is functional on its own, full helicase activity requires full-length NS3 and NS4A, which contribute to substrate binding (Pang et al. 2002; Beran et al. 2007). Furthermore, NS3-4A helicase appears to function as a dimer or other higher-order multimer (Serebrov and Pyle 2004; Mackintosh et al. 2006). NS3-4A preferentially binds polyuridine, which stimulates NTPase activity in vitro (Suzich et al. 1993; Kanai et al. 1995); it is interesting to speculate that the poly (U/UC) may perform a similar function in vivo, targeting NS3-4A helicase activity to the 3′ NCR. Despite these details, the precise function of the NS3-4A helicase during viral replication remains unknown. One important clue is that the NS3-4A helicase is genetically linked to NS5A, NS5B, and the 5′ NCR for efficient positive-strand synthesis (Binder et al. 2007b). Thus, perhaps the HCV helicase is responsible for fraying the double-stranded product of negative-strand synthesis, revealing a CRE that directs positive-strand synthesis.
At 54-aa, NS4A (8 kDa) is the smallest NS protein. As mentioned, NS4A serves to anchor the NS3-4A complex to cellular membranes, contributes to the folding of the serine protease, and stimulates RNA helicase activity. In addition, NS4A plays an important albeit unclear role in NS5A hyperphosphorylation (described below) (Kaneko et al. 1994; Koch and Bartenschlager 1999; Lindenbach et al. 2007a). Mutagenesis of the C-terminal acidic region in NS4A revealed that the efficiency of RNA replication correlates with NS4A’s ability to mediate NS5A phosphorylation (Lindenbach et al. 2007a). Some of these NS4A-mediated replication defects were suppressed by second-site changes in NS3, indicating additional functional interactions between these two proteins.
NS4B
NS4B is a small (27 kDa) hydrophobic integral membrane protein that co-translationally associates with ER membranes via an internal signal sequence and at least four central transmembrane spanning helices (Hügle et al. 2001; Lundin et al. 2003; Elazar et al. 2004). Despite the relative hydrophobicity of the protein, the bulk of NS4B appears to be on the cytoplasmic face of the ER membrane, particularly regions at the N- and C-termini, although the topology of the N-terminus remains an area of debate, and may involve other HCV non-structural proteins, such as NS5A (Lundin et al. 2003; Elazar et al. 2004; Lundin et al. 2006). NS4B may also be palmitoylated, which can apparently facilitate its oligomerization (Yu et al. 2006). For some HCV strains, the central cytoplasmic loop of NS4B apparently encodes a nucleotide-binding motif and an in vitro GTPase activity has been demonstrated for this protein (Einav et al. 2004). However the relevance of these observations remains unclear, as at least one cell culture-adaptive mutation disrupts this motif yet leads to increased RNA replication (Bartenschlager et al. 2004).
Several studies have implicated NS4B in RNA replication. A number of cell culture-adaptive mutations have been mapped to NS4B (reviewed in (Bartenschlager et al. 2004)), and allelic variation within NS4B correlates with RNA replication efficiency (Blight 2007). One specific role for NS4B may be to serve as a scaffold for viral replicase assembly. Overexpression of NS4B induces the rearrangement of cellular membranes into structures that resemble the sites of RNA replication (Egger et al. 2002; Gosert et al. 2003) and can trigger ER stress leading to an unfolded protein response (Zheng et al. 2005). NS4B has recently been shown to interact with Rab5, a regulator of membrane fusion, as well as other components of the early endosomal compartment (Stone et al. 2007). NS4B also appears to regulate the activity of sterol regulatory element binding proteins (SREBPs), major regulators of lipid metabolism, perhaps for the purpose of membrane synthesis and reorganization (Lundin et al. 2006).
NS5A
NS5A is a large (56–58 kDa), hydrophilic, RNA-binding phosphoprotein of unknown function. Yet recent biochemical and genetic experiments have provided new insights into this enigmatic protein. NS5A is peripherally anchored to intracellular membranes via an N-terminal amphipathic helix (Brass et al. 2002; Penin et al. 2004a; Sapay et al. 2006; Brass et al. 2007). Deletion or alteration of this helix leads to a diffuse cytoplasmic localization of NS5A and is lethal for RNA replication (Elazar et al. 2003; Penin et al. 2004a). Furthermore, other amphipathic viral membrane anchors cannot substitute for the NS5A membrane anchor, suggesting that this region likely interacts with other HCV replicase components (Lee et al. 2006; Teterina et al. 2006).
Limited proteolysis of purified NS5A suggests that the remainder of NS5A folds into three domains separated by two flexible, low complexity sequence blocks (Tellinghuisen et al. 2004). Domain I contains four conserved cysteines that coordinate a single zinc atom (Tellinghuisen et al. 2004). These residues are essential for RNA replication, presumably via their structural role in metal ion coordination. The structure of domain I has been determined by X-ray crystallography, revealing a novel protein fold for NS5A (Tellinghuisen et al. 2005). One obvious feature of domain I is a large, basic groove formed by the interface of monomers in the dimeric NS5A structure. It is tempting to speculate that this groove might represent the site of RNA binding to NS5A, although this remains to be experimentally determined (Huang et al. 2005; Tellinghuisen et al. 2005). Domain I also contains a large, conserved region on its surface that includes residues involved in NS5B binding and inhibiting polymerase activity (Shirota et al. 2002). NS5A domains II and III are poorly conserved and not well characterized. Domain II appears to contain some alpha helical content but attempts at determining the structure of this domain indicate that it may be natively unfolded (Liang et al. 2006, 2007). Domain III appears even more plastic than domain II, as this region can tolerate large deletions and insertions without disrupting RNA replication (Moradpour et al. 2004b; Appel et al. 2005b; Liu et al. 2006; McCormick et al. 2006).
NS5A clearly has an important role in HCV RNA replication. First, a number of cell culture-adaptive mutations that greatly enhance RNA replication have been mapped to NS5A (Blight et al. 2000; Lohmann et al. 2001). Conversely, RNA replication is ablated by a number of mutations in NS5A (Elazar et al. 2003; Penin et al. 2004a; Tellinghuisen et al. 2004; Appel et al. 2005b). Furthermore, NS5A colocalizes with other replicase components at the site of active RNA synthesis (Moradpour et al. 2004b). NS5A is the only HCV NS protein that can be complemented in trans, suggesting that this protein may be a dynamic component of the replicase that can enter and exit the replicase throughout the HCV life cycle (Appel et al. 2005a; Tong and Malcolm 2006).
One of the most striking aspects of NS5A is the correlation between NS5A phosphorylation and RNA replication. NS5A exists in basally phosphorylated (56 kDa) and hyperphosphorylated (58 kDa) forms, based on their mobility in SDS-PAGE (Kaneko et al. 1994; Tanji et al. 1995). Soon after cell culture-adaptive mutations were discovered, it was apparent that NS5A mutations that dramatically increase RNA replication also tend to decrease NS5A hyperphosphorylation, suggesting that these phosphorylation events may regulate the level of replication (Blight et al. 2000). The relevant basal and hyperphosphorylation acceptor sites have not been fully defined, but the predominant sites of phosphorylation are likely to be serine residues (Tanji et al. 1995; Reed et al. 1997; Reed and Rice 1999; Katze et al. 2000; Appel et al. 2005b). Nonetheless, accumulating evidence suggests that basal phosphorylation primarily targets residues in domains II and III, whereas hyperphosphorylation sites cluster in domain I and the low complexity linker between domains I and II. Residues implicated in basal phosphorylation are not required for RNA replication (Appel et al. 2005b). Hyperphosphorylation of NS5A requires the expression of NS3, NS4A, NS4B, and NS5A in cis (Koch and Bartenschlager 1999; Neddermann et al. 1999). While the mechanisms linking NS5A hyperphosphorylation and RNA replication remain to be determined, one important clue is that the NS5A phosphoforms differentially interact with hVAP-A, a SNARE-like vesicle sorting protein that has been implicated in HCV replication (Evans et al. 2004; Randall et al. 2007).
Perhaps the most compelling data in the area of NS5A phosphorylation and replication comes from the study of NS5A kinases. One large-scale compound library screen for small molecules that decrease the hyperphosphorylation of NS5A turned up molecules that allow RNA replication to proceed efficiently even in the absence of adaptive mutations (Neddermann et al. 2004). Some of these compounds likely block casein kinase 1α (CKIα), and genetic silencing of CKIα produces results similar to treatment of cells with these inhibitors (Quintavalle et al. 2006). Phosphorylation of NS5A by CKIα requires prior phosphorylation of nearby residues in NS5A, presumably by one or more additional kinases, indicating that NS5A phosphorylation is likely to be a cascade of events (Quintavalle et al. 2007). Indeed, a number of other potential NS5A kinases have been identified, including AKT, casein kinase II, p70s6K, MEK, and MKK1 (see (Huang et al. 2007) for a recent review). Perhaps the best-characterized potential NS5A kinase activity, after that of CKIα, is casein kinase II (CKII). A number of biochemical experiments have implicated CKII, or a related CMGC kinase family member, as a kinase that phosphorylates multiple sites in NS5A domain III (Reed et al. 1997; Kim et al. 1999; Huang et al. 2004). Despite progress in identifying NS5A kinases, more work is needed to understand the network of NS5A phosphorylation events and their role in regulating HCV genome replication.
NS5A has been reported to interact with a large constellation of host proteins involved in innate immunity, signaling, apoptosis, and lipid trafficking (Macdonald and Harris 2004). Many of these interactions, although interesting, have no demonstrated effect on HCV biology. For instance, a recent siRNA screen targeting a number of published NS5A-interaction partners confirmed only a few essential interactions (Randall et al. 2007). Nevertheless, some NS5A-interacting host factors do have an essential role in HCV genome replication. Three recent examples include FBL-2, FKBP8, and TBC120. The interaction of NS5A with the geranylgeranylated F-box protein FBL-2 is required for replication of a genotype 1b replicon, and small molecule inhibitors of geranylgeranylation or siRNA silencing of FBL-2 inhibit(s) this replicon (Wang et al. 2005). FBL-2, like other F-box proteins, is believed to target proteins for degradation, although its specific substrates remain to be identified. NS5A also interacts with FKBP8, an immunophilin that shares similarity with the cyclophilin family of peptidyl-prolyl cis–trans isomerases (PPIs), although FKBP8 appears to lack PPI activity (Okamoto et al. 2006). Interestingly, FKBP8 binds to NS5A as a trimeric complex with the chaperone HSP90 to modulate HCV RNA replication. The significance of these interactions for HCV protein folding or post-translational modification is not understood. TBC120 is an NS5A-binding host protein that is essential for HCV RNA replication (Sklan et al. 2007a, b). This protein appears to be similar to the Rab GTPase-activating proteins, and like VAP-A, may play a role in membrane trafficking and reorganization during the HCV life cycle.
NS5B
NS5B (68 kDa) is a central component of the HCV replicase, the RNA-dependent RNA polymerase (RdRp) that synthesizes all viral RNAs. NS5B was initially predicted to function as an RNA polymerase based on the presence of the conserved GDD motif common to the active site of other polymerases (Choo et al. 1989). Mutation of this GDD motif abolishes infectivity of HCV transcripts in chimpanzees and blocks RNA replication in cell culture (Lohmann et al. 1999; Kolykhalov et al. 2000). NS5B is a hydrophilic protein that associates with ER-derived membranes via a C-terminal hydrophobic tail-anchor that can post-translationally insert into membranes (Schmidt-Mende et al. 2001; Ivashkina et al. 2002). The NS5B tail-anchor is required for HCV RNA replication, and its removal leads to nuclear localization of NS5B (Moradpour et al. 2004a), although it can be functionally replaced by a similar tail-anchor sequence from a poliovirus protein (Lee et al. 2006). Nevertheless, removal of this sequence allows the expression and purification of soluble NS5B that retains polymerase activity, which has allowed extensive structural and enzymatic analyses on NS5B (Lohmann et al. 1997; Ferrari et al. 1999).
A number of crystal structures of the soluble, tail-anchor deleted form of NS5B have been generated (Ago et al. 1999; Bressanelli et al. 1999; Lesburg et al. 1999; Bressanelli et al. 2002; O'Farrell et al. 2003). These structures have been reviewed elsewhere in great detail (De Francesco et al. 2003). The overall fold of NS5B is similar to that of other single chain polymerases, with a classic right-hand topology containing distinct palm, finger, and thumb domains. Like other polymerases, the palm domain of NS5B contains the residues responsible for catalysis, nucleotide binding, and RNA template coordination. Unlike other polymerases, extensive interactions exist between the finger and thumb domains in NS5B, resulting in a fully enclosed, preformed active site capable of binding nucleotides without further conformational changes. It is thought that this closed form of NS5B may represent the structure of the polymerase during strand initiation, and that further conformational changes are required for elongation. More recent structural efforts have captured an open form of the polymerase in which thumb domain movements disrupt contact with the finger domain, which may represent a processive form of the polymerase (Biswal et al. 2005). Another unique feature of the NS5B polymerase is the presence of a ß-hairpin loop near the active site, which may position the 3′-end of the template in the proper orientation relative to the active site (Hong et al. 2001; O'Farrell et al. 2003; Ranjith-Kumar et al. 2003; Kim et al. 2005). Once the template is properly positioned, the ß-hairpin may be displaced from the active site to allow the large double-stranded RNA product to exit the active site region. Another unusual feature of NS5B is the presence of a GTP-binding allosteric regulatory site in the thumb domain (Bressanelli et al. 2002). A structure of NS5B complexed with non-nucleoside inhibitors suggests the importance of the region of the thumb subdomain near this allosteric site in conformational changes required for the transition of NS5B from the initiation state to an elongation state (O'Farrell et al. 2003). The residues comprising the GTP-binding site are not required for the in vitro polymerase activity of NS5B but are essential for RNA replication in the replicon system (Ranjith-Kumar et al. 2003; Cai et al. 2005). Another unusual regulatory feature observed in the structure of NS5B is a long C-terminal loop that encircles the thumb domain and inserts in the region of the active site (Lévêque et al. 2003). This loop decreases the RNA-binding and polymerase activities of NS5B in vitro (Lévêque et al. 2003; Ranjith-Kumar et al. 2003). It is clear NS5B possesses many features that regulate its activity, what remains to be understood is how these features function dynamically in the context of replication.
The enzymatic activity of NS5B has been extensively studied (see (Lohmann et al. 2000) for review). Although NS5B is capable of extending annealed RNA and DNA primers or self-primed “copy back” templates (Behrens et al. 1996; Lohmann et al. 1997; Al et al. 1998; Yamashita et al. 1998), NS5B most likely uses primer-independent “de novo” initiation during authentic RNA replication (Luo et al. 2000; Zhong et al. 2000). A crystal structure of NS5B with bound nucleotides strongly supports this model, as this structure is similar to the de novo initiation complex of the bacteriophage phi 6 polymerase (Bressanelli et al. 2002). Furthermore, the aforementioned ß-hairpin and C-terminal regulatory loop likely favor de novo initiation over copy back and primer extension activities by excluding double-stranded templates (Cheney et al. 2002; Lévêque et al. 2003; Ranjith-Kumar et al. 2003). NS5B prefers to initiate with a purine residue templated by a free 3′-end, but surprisingly, initiation can also occur on circular RNA templates, indicating that a free 3′-end is not absolutely required for de novo initiation (Kao et al. 2000; Ranjith-Kumar and Kao 2006). Early work with NS5B indicated that it is also capable of adding non-templated residues to the 3′-end of templates via a terminal transferase-like activity (Behrens et al. 1996; Ranjith-Kumar et al. 2001; Shim et al. 2002). These findings are controversial, as disparity exists in observing this activity among different research groups, and it has been suggested that terminal transferase activity may be a copurifying enzyme from the expression host (Lohmann et al. 1997; Yamashita et al. 1998; Oh et al. 1999; Kashiwagi et al. 2002). Nevertheless, one group reported that NS5B terminal transferase activity is dependent on active site residues within NS5B (Ranjith-Kumar et al. 2001).
In addition to the complexities of regulation that became apparent from NS5B structures, additional levels of regulation of NS5B exist. NS5B forms oligomers and exhibits cooperativity in RNA synthesis (Wang et al. 2002), suggesting that polymerase activity is regulated by homotypic intermolecular interactions. Similar findings have been made for the poliovirus RdRP (Chapter 1). In addition NS5B polymerase activity is modulated by a number of HCV replicase proteins, including NS3, a positive stimulator, and NS4A and NS5A, both negative regulators (Piccininni et al. 2002; Shirota et al. 2002). Clearly, the activity of NS5B in the viral replicase might be quite different from that observed in vitro using purified NS5B. Additionally, a number of host cell proteins have been shown to interact with and modify the activity of NS5B. NS5B can be phosphorylated by the cellular kinase PRK2, and this modification increases HCV RNA replication (Kim et al. 2004). In addition, NS5B, like NS5A, interacts with vesicle sorting proteins like hVAP-A and B. The interaction of NS5B with hVAP-B appears to increase polymerase stability, and therefore RNA replication (Tu et al. 1999; Gao et al. 2004). Perhaps the most exciting observation in this area in recent years is the interaction of NS5B with cyclophilins, another class of PPIs. This interaction was uncovered when cyclosporin A (CsA) was found to suppress HCV RNA replication in a dose-dependent manner (Watashi et al. 2003; Nakagawa et al. 2004). It was later found that CsA disrupts the interaction of NS5B with cyclophilin B, which is required for efficient recruitment and replication of HCV RNA (Watashi et al. 2005). HCV replication is reduced by knockdown of cyclophilin B, or by the non-immunosuppressive CsA derivative, DEBI0-025 (Nakagawa et al. 2005; Paeshuyse et al. 2006). Thus, the immunosuppressive activity of CsA is not required for inhibition of replication. A number of HCV mutants resistant to CsA have been selected, and mapping studies suggest both NS5B and NS5A may play a role in the inhibitory activities of CsA (Fernandes et al. 2007; Robida et al. 2007). CsA is not likely to become a widely used HCV antiviral in the immediate future as this drug is only highly effective in inhibiting genotype 1b replicons (Ishii et al. 2006).
Membrane interactions of HCV. A. An illustration of HCV-induced membrane rearrangements, as interpreted by the authors from an electron micrograph published by Gosert et al. (2003). N, nucleus; ER, endoplasmic reticulum; MW, membranous web; M, mitochondrion. B. A model for the HCV replicase within a spherule.
Membrane Alterations
Like all positive-strand RNA viruses, HCV genome replication is membrane associated. HCV replication occurs within a dense cluster of perinuclear vesicles often referred to as the “membranous web” (Fig. 4.4A) (Egger et al. 2002). The source of these membranes is likely to be the ER or a closely related compartment (reviewed in (Bartenschlager et al. 2004)) and can be induced by expressing NS4B (Egger et al. 2002). The membranous web has been positively identified as the site of HCV synthesis RNA through metabolic labeling of nascent RNAs and colocalization of viral replicase components (Gosert et al. 2003; Moradpour et al. 2004b). Biochemical analysis of membrane fractions from HCV replicon-bearing cells indicate that RdRP activity is protected from nuclease and protease digestion within a detergent-sensitive compartment (Miyanari et al. 2003; Aizaki et al. 2004; Yang et al. 2004; Quinkert et al. 2005). Thus, it is thought that HCV replication occurs within these vesicles (Fig. 4.4B). This model is in agreement with studies on numerous other positive-strand RNA viruses. Recent biophysical studies on the Flock House virus replicase indicated that these “spherules” are invaginations lined with viral replicase proteins, contain a single negative-strand intermediate and only a few positive strands, and retain communication with the cytosol via a thin neck (Kopek et al. 2007). It has been further suggested that these structures bear structural and functional homology to incompletely budded retrovirus particles or icosahedral, double-stranded RNA virus particles (Schwartz et al. 2002).
HCV replication induces genes involved in lipid metabolism, including ATP citrate lyase and acetyl-CoA synthetase (Su et al. 2002; Kapadia and Chisari 2005). Thus, membrane proliferation is likely to be required for membranous web formation and replicase assembly. Indeed, HCV replication is stimulated by saturated and monounsaturated fatty acids and inhibited by polyunsaturated fatty acids or inhibitors of lipid synthesis (Kapadia and Chisari 2005). The role of cholesterol in this process remains obscured by the fact that for genotype 1b replicons, RNA replication is dependent on the mevalonate biosynthetic pathway, most likely for the geranylgeranylation of the NS5A-interacting protein FBL2 (Ye et al. 2003; Kapadia and Chisari 2005; Wang et al. 2005). Nevertheless, extraction of cellular cholesterol with methyl-ß-cyclodextrin – a crude method to be sure – had only a modest effect on HCV genome replication (Aizaki et al. 2004; Kapadia et al. 2007). Thus, the role of cholesterol in HCV replicase function remains unclear, and it would be interesting to reassess this in the context of a replicon that is not reliant on FBL2.
Mechanisms of RNA Replication
As for all positive-strand viruses, the flow of genetic information is relatively straightforward: the positive-strand genomic RNA is used to make a negative-strand RNA intermediate, which then serves as a template for synthesizing new positive strands (Fig. 4.5). However this model is deceptively simple, as there are multiple levels of regulation controlling this process, and the mechanisms of HCV RNA replication are only beginning to be understood. The complexity of intracellular events associated with HCV infection is staggering, with the viral genome serving as an mRNA for translation of viral proteins, as a template for RNA replication, and as carrier of genetic information within progeny virions. Clearly, the trafficking of viral RNA between these processes must be regulated to avoid conflicts. For instance, it seems unlikely that the viral genome can be simultaneously used as a substrate for translation, with ribosomes moving down the genome in the 5′ to 3′ direction, and as a template for negative-strand synthesis, with the viral replicase copying the RNA in the 3′ to 5′ direction. Thus, it is clear that all of the steps in the viral life cycle are highly coordinated.
By simple and elegant methods of quantitation, Quinkert et al. (2005) determined that each HCV replicon-bearing cell contains about 1,000 positive-strand RNAs, 100 negative strands, and about 1,000,000 copies of each viral protein. Thus, the viral genome serves as a template for translation far more often than as a template for RNA replication. An excess of viral structural proteins makes sense, as the formation of nascent virus particles will likely require at least 180 copies of each structural protein for each positive-strand RNA that is packaged (assuming a T=3 particle). Given that they are derived from a single polyprotein precursor, HCV NS proteins must be generated in a roughly equimolar amount as the viral structural proteins. The function of excess NS proteins is not yet clear, as only a small fraction of them are sequestered within the membrane-bound replicase at any given time (Miyanari et al. 2003; Quinkert et al. 2005). As described below, there is increasing evidence that some of the HCV NS proteins have gained additional, non-replicative activities such as to manipulate the innate antiviral response and cell-signaling pathways.
As alluded to earlier, the HCV genome must be recruited out of translation and into a membrane-bound replicase. The signals controlling this transition are not yet fully understood, although a number of important clues have recently emerged. Based on what is known about the picornaviruses (Chapter 1), this switch likely involves the 5′ and 3′ NCRs, which function in both translation and replication. As mentioned previously, HCV RNA replication requires miR-122 binding to the 5′ NCR (Jopling et al. 2005). This is particularly interesting given that (1) miRNAs can reduce translation of cellular mRNAs by sequestering them within specialized cytoplasmic processing “P” bodies (reviewed in (Parker and Sheth 2007)); and (2) P-bodies appear to recruit the genome of brome mosaic virus, another positive-strand RNA virus (Chapter 5), out of translation and into replication (Beckham et al. 2007). Thus, it is tempting to speculate that the binding of miRNA-122 to the HCV genome may regulate the switch from genome translation to RNA replication. In line with this model, cellular mRNAs targeted by miRNA-122 are translationally silenced and targeted to P-bodies; this silencing is derepressed by the cellular protein HuR (Bhattacharyya et al. 2006). Interestingly, HuR binds to the positive- and negative-stranded forms of the HCV genome, and is required for the replication cycle of HCV (Spångberg et al. 2000; Randall et al. 2007). It will be interesting indeed to see whether miRNA-122 and HuR are relevant to the utilization of HCV genomes in translation vs. replication. Another seemingly important clue is that the polypyrimidine tract binding protein (PTB) binds to both the 5′ NCR and core-coding region of the HCV RNA, where it modulates translation from the viral IRES, and to the 3′ NCR where it may suppress RNA replication (Tsuchihara et al. 1997; Ito and Lai 1999; Anwar et al. 2000; Tischendorf et al. 2004). In addition, it is interesting to note that the polycytidine-binding protein 2 (PCBP-2) binds to the HCV 5′ NCR (Spångberg and Schwartz 1999; Fukushi et al. 2001); during poliovirus replication, this protein interacts with the viral 5′ NCR and RdRP to control the switch between translation and replication (Chapter 1).
Once HCV RNA has been recruited into the replicase, RNA synthesis presumably begins (Fig. 4.5). Aside from what has been learned regarding strand initiation and elongation with purified NS5B polymerase in vitro, little is known about this process. Membrane extracts from HCV replicon-bearing cells have been used to study replicase-associated RdRP activity, but these reactions only appear to involve strand elongation and not initiation on new templates (Ali et al. 2002; Hardy et al. 2003; Lai et al. 2003; Aizaki et al. 2004; Yang et al. 2004; Quinkert et al. 2005). Since all of the viral genetic material must be copied during each replication cycle, it is thought that negative-strand synthesis must begin via a primer-independent de novo initiation event. As mentioned earlier, the 3′-end of the HCV genome is folded in a stable stem-loop structure. When the authentic 3′-end of the HCV genome is used as a template for de novo initiation in an in vitro reaction, only internal initiation products (i.e., 5′ truncated negative strands) are generated (Kao et al. 2000; Oh et al. 2000; Sun et al. 2000; Kim et al. 2002a). However the addition of a few unpaired nts to the 3′-end allows template-length negative strands to be synthesized (Oh et al. 2000). Thus, perhaps NS3-4A RNA helicase is needed to unwind the 3′X SL1 to allow authentic initiation. Alternatively, it has been suggested that the NS5B terminal transferase activity provides these unpaired 3′-ends (Ranjith-Kumar et al. 2001). It remains unclear how these extensions would be subsequently resolved.
Once the negative-strand RNA is synthesized it remains associated with the positive-strand RNA, either in partially double-stranded replicative intermediates (RI) or fully double-stranded replicate forms (RF) (Fig. 4.5) (Ali et al. 2002). The negative-strand RNA then serves as a template to direct the synthesis of multiple positive strands, leading to asymmetry in RNA synthesis, with approximately ten positive strands generated for each minus strand (Lanford et al. 1995; Lohmann et al. 1999; Miyanari et al. 2003; Aizaki et al. 2004; Ranjith-Kumar et al. 2004). How this asymmetry is regulated remains unknown, but likely involves CREs, the composition of the viral replicase, or the differential processivity of the replicase on different templates. Recent studies with chimeric replicons have suggested that genotype-specific contacts required for efficient negative-strand synthesis are made between the 3′X tail and the NS5B polymerase, whereas genotype-specific positive-strand synthesis likely utilizes a CRE on the 3′-end of the negative-strand RNA (i.e., the reverse complement of the 5′ NCR) and requires NS3, NS5A, and NS5B (Binder et al. 2007b). These data provide the first evidence of differential requirements of replicase proteins for replication at the 5′- and 3′-end of the genome, and as such, may be important clues for the regulation of strand synthesis asymmetry.
Cellular Response to Infection
As with all viruses that are capable of establishing persistent infections, HCV must face the innate antiviral response of the host cell. It is thought that cells recognize and respond to the unusual features of viral genomes, such as double-stranded RNAs or RNAs lacking a 5′-methylated cap (Hornung et al. 2006: Pichlmair, 2006 #1699). Identifying how HCV manages to deal with this response has been an area of intense investigation for many years.
Much of the early work in this area was focused on the NS5A protein and its role in interferon resistance via interaction with PKR, a double-stranded RNA sensor (see (Tan and Katze 2001) for review). A region in NS5A, termed the interferon sensitivity-determining region (ISDR), was found to possess a high mutation rate in clinical samples, which weakly correlated with sensitivity to IFN therapy (Enomoto et al. 1995, 1996). Furthermore, the NS5A ISDR was shown to bind PKR and inhibit the IFN-induced activity of PKR on downstream targets, most notably eIF2α, thereby counteracting the antiviral effects of PKR (Gale et al. 1997, 1998a, b). Yet the significance of the ISDR is an area of debate, with some groups showing no relationship between ISDR and IFN response in patients (Pawlotsky 1999). Furthermore, deletion or mutation of the ISDR does not affect the IFN sensitivity of HCV replicons, suggesting this sequence does not play a direct role in IFN response in cell culture (Blight et al. 2000; Liu et al. 2006). In addition, NS5A has been shown to induce interleukin-8, which antagonizes the antiviral effects of IFN (Polyak et al. 2001). Although none of these reported NS5A activities seem to be the major mechanism by which HCV escapes antiviral defenses, it is possible that they act synergistically in the context of an authentic infection and modulate IFN sensitivity. Indeed, the ability of NS5A to manipulate many host-signaling pathways and interact with a diverse range of host-signaling molecules, few of which have a dramatic effect on virus replication, may collectively represent clues toward the overall manipulation of the host cell by HCV (see (Macdonald and Harris 2004) for review).
More recently the focus has shifted to the role of the NS3-4A complex in circumventing innate antiviral defenses. NS3-4A antagonizes at least two key innate antiviral defenses by short-circuiting the transduction of the viral RNA-sensing signals of the retinoic acid inducible gene-I (RIG-I) helicase and the Toll-like receptor-3 (TLR3) systems (see (Johnson and Gale 2006) for review). In the manipulation of both pathways, the NS3-4A protease cleaves key adapter proteins that are required for effective signal transduction to IRF-3 and NF-κB, important transcription factors for the innate immune response. NS3-4A manipulates the TLR3 pathway by cleaving an adapter molecule, TRIF, required for the activation of IRF-3 and NF-κB in response to extracellular forms of double-stranded RNA (Ferreon et al. 2005; Li et al. 2005a). In the case of the RIG-I pathway, the NS3-4A protease cleaves a CARD domain-containing adapter protein designated IPS-1 (also known as Cardif, MAVS, and VISA) (Li et al. 2005b; Meylan et al. 2005). IPS-1 is an outer mitochondrial protein responsible for transducing signals from both RIG-I and another viral RNA-sensing helicase, MDA5, to IRF-3 and NF-κB. Cleavage of IPS-1 blocks the induction of gene transcription in response to cytoplasmic RNAs, allowing the virus to escape from this potent antiviral pathway (Breiman et al. 2005; Foy et al. 2005; Karayiannis 2005). Further evidence that RIG-I limits HCV replication is the observation that the Huh-7.5 hepatoma cell line, a clone of Huh-7 cells that is highly permissive for HCV RNA replication, expresses a dominant negative form of RIG-I (Sumpter et al. 2005). Nevertheless, the relative importance of the RIG-I pathway in mediating Huh-7.5 permissiveness has been questioned by at least one other group (Binder et al. 2007a), so it seems likely that the picture is not yet complete.
References
Ago, H., Adachi, T., Yoshida, A., Yamamoto, M., Habuka, N., Yatsunami, K. and Miyano, M. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure Fold Des 7: 1417–26.
Aizaki, H., Lee, K. J., Sung, V. M., Ishiko, H. and Lai, M. M. 2004. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324:450–61.
Al, R. H., Xie, Y., Wang, Y. and Hagedorn, C. H. 1998. Expression of recombinant hepatitis C virus non-structural protein 5B in Escherichia coli. Virus Res 53: 141–9.
Ali, N., Tardif, K. D. and Siddiqui, A. 2002. Cell-free replication of the hepatitis C virus subgenomic replicon. J Virol 76: 12001–7.
Alter, H. J. and Seeff, L. B. 2000. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin Liver Dis 20: 17–35.
Anwar, A., Ali, N., Tanveer, R. and Siddiqui, A. 2000. Demonstration of functional requirement of polypyrimidine tract-binding protein by SELEX RNA during hepatitis C virus internal ribosome entry site-mediated translation initiation. J Biol Chem 275: 34231–5.
Appel, N., Herian, U. and Bartenschlager, R. 2005a. Efficient rescue of hepatitis C virus RNA replication by trans-complementation with nonstructural protein 5A. J Virol 79: 896–909.
Appel, N., Pietschmann, T. and Bartenschlager, R. 2005b. Mutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J Virol 79: 3187–94.
Bartenschlager, R., Frese, M. and Pietschmann, T. 2004. Novel insights into hepatitis C virus replication and persistence. Adv Virus Res 63: 71–180.
Beckham, C. J., Light, H. R., Nissan, T. A., Ahlquist, P., Parker, R. and Noueiry, A. 2007. Interactions between brome mosaic virus RNAs and cytoplasmic processing bodies. J Virol 81: 9759–68.
Behrens, S. E., Tomei, L. and De Francesco, R. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J 15: 12–22.
Beran, R. K., Serebrov, V. and Pyle, A. M. 2007. The serine protease domain of hepatitis C viral NS3 activates RNA helicase activity by promoting the binding of RNA substrate. J Biol Chem 282: 34913–20.
Bhattacharyya, S. N., Habermacher, R., Martine, U., Closs, E. I. and Filipowicz, W. 2006. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb Symp Quant Biol 71: 513–21.
Binder, M., Kochs, G., Bartenschlager, R. and Lohmann, V. 2007a. Hepatitis C virus escape from the interferon regulatory factor 3 pathway by a passive and active evasion strategy. Hepatology 46: 1365–74.
Binder, M., Quinkert, D., Bochkarova, O., Klein, R., Kezmic, N., Bartenschlager, R. and Lohmann, V. 2007b. Identification of determinants involved in initiation of hepatitis C virus RNA synthesis by using intergenotypic replicase chimeras. J Virol 81: 5270–83.
Biswal, B. K., Cherney, M. M., Wang, M., Chan, L., Yannopoulos, C. G., Bilimoria, D., Nicolas, O., Bedard, J. and James, M. N. 2005. Crystal structures of the RNA-dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J Biol Chem 280: 18202–10.
Blight, K. J. 2007. Allelic variation in the hepatitis C virus NS4B protein dramatically influences RNA replication. J Virol 81: 5724–36.
Blight, K. J., Kolykhalov, A. A. and Rice, C. M. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290: 1972–4.
Blight, K. J. and Rice, C. M. 1997. Secondary structure determination of the conserved 98-base sequence at the 3' terminus of hepatitis C virus genome RNA. J Virol 71: 7345–52.
Bradrick, S. S., Walters, R. W. and Gromeier, M. 2006. The hepatitis C virus 3'-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acids Res 34: 1293–303.
Branch, A. D., Stump, D. D., Gutierrez, J. A., Eng, F. and Walewski, J. L. 2005. The hepatitis C virus alternate reading frame (ARF) and its family of novel products: the alternate reading frame protein/F-protein, the double-frameshift protein, and others. Semin Liver Dis 25: 105–17.
Brass, V., Bieck, E., Montserret, R., Wolk, B., Hellings, J. A., Blum, H. E., Penin, F. and Moradpour, D. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J Biol Chem 277: 8130–9.
Brass, V., Pal, Z., Sapay, N., Deleage, G., Blum, H. E., Penin, F. and Moradpour, D. 2007. Conserved determinants for membrane association of nonstructural protein 5A from hepatitis C virus and related viruses. J Virol 81: 2745–57.
Breiman, A., Grandvaux, N., Lin, R., Ottone, C., Akira, S., Yoneyama, M., Fujita, T., Hiscott, J. and Meurs, E. F. 2005. Inhibition of RIG-I-dependent signaling to the interferon pathway during hepatitis C virus expression and restoration of signaling by IKKepsilon. J Virol 79: 3969–78.
Bressanelli, S., Tomei, L., Rey, F. A. and De Francesco, R. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J Virol 76: 3482–92.
Bressanelli, S., Tomei, L., Roussel, A., Incitti, I., Vitale, R. L., Mathieu, M., De Francesco, R. and Rey, F. A: 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc Natl Acad Sci USA 96: 13034–9.
Cai, Z., Yi, M., Zhang, C. and Luo, G. 2005. Mutagenesis analysis of the rGTP-specific binding site of hepatitis C virus RNA-dependent RNA polymerase. J Virol 79: 11607–17.
Cheney, I. W., Naim, S., Lai, V. C., Dempsey, S., Bellows, D., Walker, M. P., Shim, J. H., Horscroft, N., Hong, Z. and Zhong, W. 2002. Mutations in NS5B polymerase of hepatitis C virus: impacts on in vitro enzymatic activity and viral RNA replication in the subgenomic replicon cell culture. Virology 297: 298–306.
Choo, Q.-L., Kuo, G., Weiner, A. J., Overby, L. R., Bradley, D. W. and Houghton, M: 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244: 359–62.
Choo, Q.-L., Richman, K. H., Han, J. H., Berger, K., Lee, C., Dong, C., Gallegos, C., Coit, D., Medina-Selby, A., Barr, P. J., Weiner, A. J., Bradley, D. W., Kuo, G. and Houghton, M: 1991. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA 88: 2451–5.
Crotty, S., Cameron, C. E. and Andino, R. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc Natl Acad Sci USA 98: 6895–900.
De Francesco, R., Tomei, L., Altamura, S., Summa, V. and Migliaccio, G. 2003. Approaching a new era for hepatitis C virus therapy: inhibitors of the NS3-4A serine protease and the NS5B RNA-dependent RNA polymerase. Antiviral Res 58: 1–16.
Dumont, S., Cheng, W., Serebrov, V., Beran, R. K., Tinoco, I., Jr., Pyle, A. M. and Bustamante, C. 2006. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature 439: 105–8.
Egger, D., Wolk, B., Gosert, R., Bianchi, L., Blum, H. E., Moradpour, D. and Bienz, K. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol 76: 5974–84.
Einav, S., Elazar, M., Danieli, T. and Glenn, J. S. 2004. A nucleotide binding motif in hepatitis C virus (HCV) NS4B mediates HCV RNA replication. J Virol 78: 11288–95.
Elazar, M., Cheong, K. H., Liu, P., Greenberg, H. B., Rice, C. M. and Glenn, J. S. 2003. Amphipathic helix-dependent localization of NS5A mediates hepatitis C virus RNA replication. J Virol 77: 6055–61.
Elazar, M., Liu, P., Rice, C. M. and Glenn, J. S. 2004. An N-terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication. J Virol 78: 11393–400.
Enomoto, N., Sakuma, I., Asahina, Y., Kurosaki, M., Murakami, T., Yamamoto, C., Izumi, N., Marumo, F. and Sato, C: 1995. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J Clin Invest 96: 224–30.
Enomoto, N., Sakuma, I., Asahina, Y., Kurosaki, M., Murakami, T., Yamamoto, C., Ogura, Y., Izumi, N., Marumo, F. and Sato, C: 1996. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med 334: 77–81.
Evans, M. J., Rice, C. M. and Goff, S. P. 2004. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc Natl Acad Sci USA 101: 13038–43.
Fernandes, F., Poole, D. S., Hoover, S., Middleton, R., Andrei, A. C., Gerstner, J. and Striker, R. 2007. Sensitivity of hepatitis C virus to cyclosporine A depends on nonstructural proteins NS5A and NS5B. Hepatology 46: 1026–33.
Ferrari, E., Wright-Minogue, J., Fang, J. W., Baroudy, B. M., Lau, J. Y. and Hong, Z: 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J Virol 73: 1649–54.
Ferreon, J. C., Ferreon, A. C., Li, K. and Lemon, S. M. 2005. Molecular determinants of TRIF proteolysis mediated by the hepatitis C virus NS3/4A protease. J Biol Chem 280: 20483–92.
Foy, E., Li, K., Sumpter, R., Jr., Loo, Y. M., Johnson, C. L., Wang, C., Fish, P. M., Yoneyama, M., Fujita, T., Lemon, S. M. and Gale, M., Jr. 2005. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci USA 102: 2986–91.
Fraser, C. S. and Doudna, J. A. 2007. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat Rev Microbiol 5: 29–38.
Friebe, P. and Bartenschlager, R. 2002. Genetic analysis of sequences in the 3' nontranslated region of hepatitis C virus that are important for RNA replication. J Virol 76: 5326–38.
Friebe, P., Boudet, J., Simorre, J. P. and Bartenschlager, R. 2005. Kissing-loop interaction in the 3' end of the hepatitis C virus genome essential for RNA replication. J Virol 79: 380–92.
Friebe, P., Lohmann, V., Krieger, N. and Bartenschlager, R. 2001. Sequences in the 5' nontranslated region of hepatitis C virus required for RNA replication. J Virol 75: 12047–57.
Fukushi, S., Okada, M., Kageyama, T., Hoshino, F. B., Nagai, K. and Katayama, K. 2001. Interaction of poly(rC)-binding protein 2 with the 5'-terminal stem loop of the hepatitis C-virus genome. Virus Res 73: 67–79.
Gale, M., Jr., Blakely, C. M., Kwieciszewski, B., Tan, S. L., Dossett, M., Tang, N. M., Korth, M. J., Polyak, S. J., Gretch, D. R. and Katze, M. G: 1998a. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol 18: 5208–18.
Gale, M. J., Jr., Korth, M. J. and Katze, M. G: 1998b. Repression of the PKR protein kinase by the hepatitis C virus NS5A protein: a potential mechanism of interferon resistance. Clin Diagn Virol 10: 157–62.
Gale, M. J., Jr., Korth, M. J., Tang, N. M., Tan, S. L., Hopkins, D. A., Dever, T. E., Polyak, S. J., Gretch, D. R. and Katze, M. G: 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230: 217–27.
Gao, L., Aizaki, H., He, J. W. and Lai, M. M. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J Virol 78: 3480–8.
Gosert, R., Egger, D., Lohmann, V., Bartenschlager, R., Blum, H. E., Bienz, K. and Moradpour, D. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol 77: 5487–92.
Grakoui, A., McCourt, D. W., Wychowski, C., Feinstone, S. M. and Rice, C. M: 1993. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci USA 90: 10583–7.
Han, J. Q. and Barton, D. J. 2002. Activation and evasion of the antiviral 2'-5' oligoadenylate synthetase/ribonuclease L pathway by hepatitis C virus mRNA. RNA 8: 512–25.
Hardy, R. W., Marcotrigiano, J., Blight, K. J., Majors, J. E. and Rice, C. M. 2003. Hepatitis C virus RNA synthesis in a cell-free system isolated from replicon-containing hepatoma cells. J Virol 77: 2029–37.
Hijikata, M., Mizushima, H., Akagi, T., Mori, S., Kakiuchi, N., Kato, N., Tanaka, T., Kimura, K. and Shimotohno, K: 1993. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J Virol 67: 4665–75.
Hong, Z., Cameron, C. E., Walker, M. P., Castro, C., Yao, N., Lau, J. Y. and Zhong, W. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285: 6–11.
Hornung, V., Ellegast, J., Kim, S., Brzozka, K., Jung, A., Kato, H., Poeck, H., Akira, S., Conzelmann, K. K., Schlee, M., Endres, S. and Hartmann, G. 2006. 5'-Triphosphate RNA is the ligand for RIG-I. Science 314: 994–7.
Huang, L., Hwang, J., Sharma, S. D., Hargittai, M. R., Chen, Y., Arnold, J. J., Raney, K. D. and Cameron, C. E. 2005. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J Biol Chem 280: 36417–28.
Huang, L., Sineva, E. V., Hargittai, M. R., Sharma, S. D., Suthar, M., Raney, K. D. and Cameron, C. E. 2004. Purification and characterization of hepatitis C virus non-structural protein 5A expressed in Escherichia coli. Protein Expr Purif 37: 144–53.
Huang, Y., Staschke, K., De Francesco, R. and Tan, S. L. 2007. Phosphorylation of hepatitis C virus NS5A nonstructural protein: a new paradigm for phosphorylation-dependent viral RNA replication? Virology 364: 1–9.
Hügle, T., Fehrmann, F., Bieck, E., Kohara, M., Krausslich, H. G., Rice, C. M., Blum, H. E. and Moradpour, D. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 284: 70–81.
Ishii, N., Watashi, K., Hishiki, T., Goto, K., Inoue, D., Hijikata, M., Wakita, T., Kato, N. and Shimotohno, K. 2006. Diverse effects of cyclosporine on hepatitis C virus strain replication. J Virol 80: 4510–20.
Ito, T. and Lai, M. M: 1999. An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3'-untranslated sequence. Virology 254: 288–96.
Ito, T., Tahara, S. M. and Lai, M. M: 1998. The 3'-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J Virol 72: 8789–96.
Ivashkina, N., Wolk, B., Lohmann, V., Bartenschlager, R., Blum, H. E., Penin, F. and Moradpour, D. 2002. The hepatitis C virus RNA-dependent RNA polymerase membrane insertion sequence is a transmembrane segment. J Virol 76: 13088–93.
Ji, H., Fraser, C. S., Yu, Y., Leary, J. and Doudna, J. A. 2004. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci USA 101: 16990–5.
Johnson, C. L. and Gale, M., Jr. 2006. CARD games between virus and host get a new player. Trends Immunol 27: 1–4.
Jones, C. T., Murray, C. L., Eastman, D. K., Tassello, J. and Rice, C. M. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J Virol 81: 8374–83.
Jopling, C. L., Yi, M., Lancaster, A. M., Lemon, S. M. and Sarnow, P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309: 1577–81.
Kanai, A., Tanabe, K. and Kohara, M: 1995. Poly(U) binding activity of hepatitis C virus NS3 protein, a putative RNA helicase. FEBS Lett 376: 221–4.
Kaneko, T., Tanji, Y., Satoh, S., Hijikata, M., Asabe, S., Kimura, K. and Shimotohno, K: 1994. Production of two phosphoproteins from the NS5A region of the hepatitis C viral genome. Biochem Biophys Res Commun 205: 320–6.
Kao, C. C., Yang, X., Kline, A., Wang, Q. M., Barket, D. and Heinz, B. A. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J Virol 74: 11121–8.
Kapadia, S. B., Barth, H., Baumert, T., McKeating, J. A. and Chisari, F. V. 2007. Initiation of hepatitis C virus infection is dependent on cholesterol and cooperativity between CD81 and scavenger receptor B type I. J Virol 81: 374–83.
Kapadia, S. B. and Chisari, F. V. 2005. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc Natl Acad Sci USA 102: 2561–6.
Karayiannis, P. 2005. The hepatitis C virus NS3/4A protease complex interferes with pathways of the innate immune response. J Hepatol 43: 743–5.
Kashiwagi, T., Hara, K., Kohara, M., Kohara, K., Iwahashi, J., Hamada, N., Yoshino, H. and Toyoda, T. 2002. Kinetic analysis of C-terminally truncated RNA-dependent RNA polymerase of hepatitis C virus. Biochem Biophys Res Commun 290: 1188–94.
Katze, M. G., Kwieciszewski, B., Goodlett, D. R., Blakely, C. M., Neddermann, P., Tan, S. L. and Aebersold, R. 2000. Ser(2194) is a highly conserved major phosphorylation site of the hepatitis C virus nonstructural protein NS5A. Virology 278: 501–13.
Kim, M., Kim, H., Cho, S. P. and Min, M. K. 2002a. Template requirements for de novo RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase on the viral X RNA. J Virol 76: 6944–56.
Kim, S. J., Kim, J. H., Kim, Y. G., Lim, H. S. and Oh, J. W. 2004. Protein kinase C-related kinase 2 regulates hepatitis C virus RNA polymerase function by phosphorylation. J Biol Chem 279: 50031–41.
Kim, Y. K., Kim, C. S., Lee, S. H. and Jang, S. K. 2002b. Domains I and II in the 5' nontranslated region of the HCV genome are required for RNA replication. Biochem Biophys Res Commun 290: 105–12.
Kim, J., Lee, D. and Choe, J: 1999. Hepatitis C virus NS5A protein is phosphorylated by casein kinase II. Biochem Biophys Res Commun 257: 777–81.
Kim, Y. K., Lee, S. H., Kim, C. S., Seol, S. K. and Jang, S. K. 2003. Long-range RNA-RNA interaction between the 5' nontranslated region and the core-coding sequences of hepatitis C virus modulates the IRES-dependent translation. RNA 9: 599–606.
Kim, Y. C., Russell, W. K., Ranjith-Kumar, C. T., Thomson, M., Russell, D. H. and Kao, C. C. 2005. Functional analysis of RNA binding by the hepatitis C virus RNA-dependent RNA polymerase. J Biol Chem 280: 38011–9.
Koch, J. O. and Bartenschlager, R: 1999. Modulation of hepatitis C virus NS5A hyperphosphorylation by nonstructural proteins NS3, NS4A, and NS4B. J Virol 73: 7138–46.
Kolykhalov, A. A., Agapov, E. V., Blight, K. J., Mihalik, K., Feinstone, S. M. and Rice, C. M: 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277: 570–4.
Kolykhalov, A. A., Feinstone, S. M. and Rice, C. M: 1996. Identification of a highly conserved sequence element at the 3' terminus of hepatitis C virus genome RNA. J Virol 70: 3363–71.
Kolykhalov, A. A., Mihalik, K., Feinstone, S. M. and Rice, C. M. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3' nontranslated region are essential for virus replication in vivo. J Virol 74: 2046–51.
Kopek, B. G., Perkins, G., Miller, D. J., Ellisman, M. H. and Ahlquist, P. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol 5: e220.
Lai, V. C., Dempsey, S., Lau, J. Y., Hong, Z. and Zhong, W. 2003. In vitro RNA replication directed by replicase complexes isolated from the subgenomic replicon cells of hepatitis C virus. J Virol 77: 2295–300.
Lanford, R. E., Chavez, D., Chisari, F. V. and Sureau, C: 1995. Lack of detection of negative-strand hepatitis C virus RNA in peripheral blood mononuclear cells and other extrahepatic tissues by the highly strand-specific rTth reverse transcriptase PCR. J Virol 69: 8079–83.
Lee, H., Liu, Y., Mejia, E., Paul, A. V. and Wimmer, E. 2006. The C-terminal hydrophobic domain of hepatitis C virus RNA polymerase NS5B can be replaced with a heterologous domain of poliovirus protein 3A. J Virol 80: 11343–54.
Lee, H., Shin, H., Wimmer, E. and Paul, A. V. 2004. cis-acting RNA signals in the NS5B C-terminal coding sequence of the hepatitis C virus genome. J Virol 78: 10865–77.
Lesburg, C. A., Cable, M. B., Ferrari, E., Hong, Z., Mannarino, A. F. and Weber, P. C: 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat Struct Biol 6: 937–43.
Lévêque, V. J., Johnson, R. B., Parsons, S., Ren, J., Xie, C., Zhang, F. and Wang, Q. M. 2003. Identification of a C-terminal regulatory motif in hepatitis C virus RNA-dependent RNA polymerase: structural and biochemical analysis. J Virol 77: 9020–8.
Li, K., Foy, E., Ferreon, J. C., Nakamura, M., Ferreon, A. C., Ikeda, M., Ray, S. C., Gale, M., Jr. and Lemon, S. M. 2005a. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci USA 102: 2992–7.
Li, X. D., Sun, L., Seth, R. B., Pineda, G. and Chen, Z. J. 2005b. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci USA 102: 17717–22.
Liang, Y., Kang, C. B. and Yoon, H. S. 2006. Molecular and structural characterization of the domain 2 of hepatitis C virus non-structural protein 5A. Mol Cells 22: 13–20.
Liang, Y., Ye, H., Kang, C. B. and Yoon, H. S. 2007. Domain 2 of nonstructural protein 5A (NS5A) of hepatitis C virus is natively unfolded. Biochemistry 46: 11550–8.
Lindenbach, B. D., Evans, M. J., Syder, A. J., Wolk, B., Tellinghuisen, T. L., Liu, C. C., Maruyama, T., Hynes, R. O., Burton, D. R., McKeating, J. A. and Rice, C. M. 2005. Complete replication of hepatitis C virus in cell culture. Science 309: 623–6.
Lindenbach, B. D., Pragai, B. M., Montserret, R., Beran, R. K., Pyle, A. M., Penin, F. and Rice, C. M. 2007a. The C terminus of hepatitis C virus NS4A encodes an electrostatic switch that regulates NS5A hyperphosphorylation and viral replication. J Virol 81: 8905–18.
Liu, S., Ansari, I. H., Das, S. C. and Pattnaik, A. K. 2006. Insertion and deletion analyses identify regions of non-structural protein 5A of Hepatitis C virus that are dispensable for viral genome replication. J Gen Virol 87: 323–7.
Lohmann, V., Hoffmann, S., Herian, U., Penin, F. and Bartenschlager, R. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77: 3007–19.
Lohmann, V., Korner, F., Dobierzewska, A. and Bartenschlager, R. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J Virol 75: 1437–49.
Lohmann, V., Korner, F., Herian, U. and Bartenschlager, R: 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol 71: 8416–28.
Lohmann, V., Korner, F., Koch, J. O., Herian, U., Theilmann, L. and Bartenschlager, R: 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285: 110–3.
Lohmann, V., Roos, A., Korner, F., Koch, J. O. and Bartenschlager, R. 2000. Biochemical and structural analysis of the NS5B RNA-dependent RNA polymerase of the hepatitis C virus. J Viral Hepat 7: 167–74.
Lorenz, I. C., Marcotrigiano, J., Dentzer, T. G. and Rice, C. M. 2006. Structure of the catalytic domain of the hepatitis C virus NS2-3 protease. Nature 442: 831–5.
Lundin, M., Lindstrom, H., Gronwall, C. and Persson, M. A. 2006. Dual topology of the processed hepatitis C virus protein NS4B is influenced by the NS5A protein. J Gen Virol 87: 3263–72.
Lundin, M., Monne, M., Widell, A., Von Heijne, G. and Persson, M. A. 2003. Topology of the membrane-associated hepatitis C virus protein NS4B. J Virol 77: 5428–38.
Luo, G., Hamatake, R. K., Mathis, D. M., Racela, J., Rigat, K. L., Lemm, J. and Colonno, R. J. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J Virol 74: 851–63.
Macdonald, A. and Harris, M. 2004. Hepatitis C virus NS5A: tales of a promiscuous protein. J Gen Virol 85: 2485–502.
Mackintosh, S. G., Lu, J. Z., Jordan, J. B., Harrison, M. K., Sikora, B., Sharma, S. D., Cameron, C. E., Raney, K. D. and Sakon, J. 2006. Structural and biological identification of residues on the surface of NS3 helicase required for optimal replication of the hepatitis C virus. J Biol Chem 281: 3528–35.
McCaffrey, A. P., Ohashi, K., Meuse, L., Shen, S., Lancaster, A. M., Lukavsky, P. J., Sarnow, P. and Kay, M. A. 2002. Determinants of hepatitis C translational initiation in vitro, in cultured cells and mice. Mol Ther 5: 676–84.
McCormick, C. J., Maucourant, S., Griffin, S., Rowlands, D. J. and Harris, M. 2006. Tagging of NS5A expressed from a functional hepatitis C virus replicon. J Gen Virol 87: 635–40.
McMullan, L. K., Grakoui, A., Evans, M. J., Mihalik, K., Puig, M., Branch, A. D., Feinstone, S. M. and Rice, C. M. 2007. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc Natl Acad Sci USA 104: 2879–84.
Meylan, E., Curran, J., Hofmann, K., Moradpour, D., Binder, M., Bartenschlager, R. and Tschopp, J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437: 1167–72.
Miyanari, Y., Hijikata, M., Yamaji, M., Hosaka, M., Takahashi, H. and Shimotohno, K. 2003. Hepatitis C virus non-structural proteins in the probable membranous compartment function in viral genome replication. J Biol Chem 278: 50301–8.
Moradpour, D., Brass, V., Bieck, E., Friebe, P., Gosert, R., Blum, H. E., Bartenschlager, R., Penin, F. and Lohmann, V. 2004a. Membrane association of the RNA-dependent RNA polymerase is essential for hepatitis C virus RNA replication. J Virol 78: 13278–84.
Moradpour, D., Evans, M. J., Gosert, R., Yuan, Z., Blum, H. E., Goff, S. P., Lindenbach, B. D. and Rice, C. M. 2004b. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J Virol 78: 7400–9.
Myong, S., Bruno, M. M., Pyle, A. M. and Ha, T. 2007. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science 317: 513–6.
Nakabayashi, H., Taketa, K., Miyano, K., Yamane, T. and Sato, J: 1982. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res 42: 3858–63.
Nakagawa, M., Sakamoto, N., Enomoto, N., Tanabe, Y., Kanazawa, N., Koyama, T., Kurosaki, M., Maekawa, S., Yamashiro, T., Chen, C. H., Itsui, Y., Kakinuma, S. and Watanabe, M. 2004. Specific inhibition of hepatitis C virus replication by cyclosporin A. Biochem Biophys Res Commun 313: 42–7.
Nakagawa, M., Sakamoto, N., Tanabe, Y., Koyama, T., Itsui, Y., Takeda, Y., Chen, C. H., Kakinuma, S., Oooka, S., Maekawa, S., Enomoto, N. and Watanabe, M. 2005. Suppression of hepatitis C virus replication by cyclosporin a is mediated by blockade of cyclophilins. Gastroenterology 129: 1031–41.
Neddermann, P., Clementi, A. and De Francesco, R: 1999. Hyperphosphorylation of the hepatitis C virus NS5A protein requires an active NS3 protease, NS4A, NS4B, and NS5A encoded on the same polyprotein. J Virol 73: 9984–91.
Neddermann, P., Quintavalle, M., Di Pietro, C., Clementi, A., Cerretani, M., Altamura, S., Bartholomew, L. and De Francesco, R. 2004. Reduction of hepatitis C virus NS5A hyperphosphorylation by selective inhibition of cellular kinases activates viral RNA replication in cell culture. J Virol 78: 13306–14.
O'Farrell, D., Trowbridge, R., Rowlands, D. and Jäger, J. 2003. Substrate complexes of hepatitis C virus RNA polymerase (HC-J4): structural evidence for nucleotide import and de-novo initiation. J Mol Biol 326: 1025–35.
Oh, J. W., Ito, T. and Lai, M. M: 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J Virol 73: 7694–702.
Oh, J. W., Sheu, G. T. and Lai, M. M. 2000. Template requirement and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template. J Biol Chem 275: 17710–7.
Okamoto, T., Nishimura, Y., Ichimura, T., Suzuki, K., Miyamura, T., Suzuki, T., Moriishi, K. and Matsuura, Y. 2006. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. Embo J 25: 5015–25.
Otto, G. A. and Puglisi, J. D. 2004. The pathway of HCV IRES-mediated translation initiation. Cell 119: 369–80.
Paeshuyse, J., Kaul, A., De Clercq, E., Rosenwirth, B., Dumont, J. M., Scalfaro, P., Bartenschlager, R. and Neyts, J. 2006. The non-immunosuppressive cyclosporin DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology 43: 761–70.
Pang, P. S., Jankowsky, E., Planet, P. J. and Pyle, A. M. 2002. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J 21: 1168–76.
Parker, R. and Sheth, U. 2007. P bodies and the control of mRNA translation and degradation. Mol Cell 25: 635–46.
Pawlotsky, J. M: 1999. Hepatitis C virus (HCV) NS5A protein: role in HCV replication and resistance to interferon-alpha. J Viral Hepat 6 Suppl 1: 47–8.
Penin, F., Brass, V., Appel, N., Ramboarina, S., Montserret, R., Ficheux, D., Blum, H. E., Bartenschlager, R. and Moradpour, D. 2004a. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J Biol Chem 279: 40835–43.
Penin, F., Dubuisson, J., Rey, F. A., Moradpour, D. and Pawlotsky, J. M. 2004b. Structural biology of hepatitis C virus. Hepatology 39: 5–19.
Perelson, A. S., Herrmann, E., Micol, F. and Zeuzem, S. 2005. New kinetic models for the hepatitis C virus. Hepatology 42: 749–54.
Pestova, T. V., Shatsky, I. N., Fletcher, S. P., Jackson, R. J. and Hellen, C. U: 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev 12: 67–83.
Piccininni, S., Varaklioti, A., Nardelli, M., Dave, B., Raney, K. D. and McCarthy, J. E. 2002. Modulation of the hepatitis C virus RNA-dependent RNA polymerase activity by the non-structural (NS) 3 helicase and the NS4B membrane protein. J Biol Chem 277: 45670–9.
Polyak, S. J., Khabar, K. S., Paschal, D. M., Ezelle, H. J., Duverlie, G., Barber, G. N., Levy, D. E., Mukaida, N. and Gretch, D. R. 2001. Hepatitis C virus nonstructural 5A protein induces interleukin-8, leading to partial inhibition of the interferon-induced antiviral response. J Virol 75: 6095–106.
Quinkert, D., Bartenschlager, R. and Lohmann, V. 2005. Quantitative analysis of the hepatitis C virus replication complex. J Virol 79: 13594–605.
Quintavalle, M., Sambucini, S., Di Pietro, C., De Francesco, R. and Neddermann, P. 2006. The alpha isoform of protein kinase CKI is responsible for hepatitis C virus NS5A hyperphosphorylation. J Virol 80: 11305–12.
Quintavalle, M., Sambucini, S., Summa, V., Orsatti, L., Talamo, F., De Francesco, R. and Neddermann, P. 2007. Hepatitis C virus NS5A is a direct substrate of casein kinase I-alpha, a cellular kinase identified by inhibitor affinity chromatography using specific NS5A hyperphosphorylation inhibitors. J Biol Chem 282: 5536–44.
Randall, G., Panis, M., Cooper, J. D., Tellinghuisen, T. L., Sukhodolets, K. E., Pfeffer, S., Landthaler, M., Landgraf, P., Kan, S., Lindenbach, B. D., Chien, M., Weir, D. B., Russo, J. J., Ju, J., Brownstein, M. J., Sheridan, R., Sander, C., Zavolan, M., Tuschl, T. and Rice, C. M. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci USA 104: 12884–9.
Ranjith-Kumar, C. T., Gajewski, J., Gutshall, L., Maley, D., Sarisky, R. T. and Kao, C. C. 2001. Terminal nucleotidyl transferase activity of recombinant Flaviviridae RNA-dependent RNA polymerases: implication for viral RNA synthesis. J Virol 75: 8615–23.
Ranjith-Kumar, C. T., Gutshall, L., Sarisky, R. T. and Kao, C. C. 2003. Multiple interactions within the hepatitis C virus RNA polymerase repress primer-dependent RNA synthesis. J Mol Biol 330: 675–85.
Ranjith-Kumar, C. T. and Kao, C. C. 2006. Recombinant viral RdRps can initiate RNA synthesis from circular templates. RNA 12: 303–12.
Ranjith-Kumar, C. T., Sarisky, R. T., Gutshall, L., Thomson, M. and Kao, C. C. 2004. De novo initiation pocket mutations have multiple effects on hepatitis C virus RNA-dependent RNA polymerase activities. J Virol 78: 12207–17.
Reed, K. E. and Rice, C. M: 1999. Identification of the major phosphorylation site of the hepatitis C virus H strain NS5A protein as serine 2321. J Biol Chem 274: 28011–8.
Reed, K. E., Xu, J. and Rice, C. M: 1997. Phosphorylation of the hepatitis C virus NS5A protein in vitro and in vivo: properties of the NS5A-associated kinase. J Virol 71: 7187–97.
Rijnbrand, R., Bredenbeek, P. J., Haasnoot, P. C., Kieft, J. S., Spaan, W. J. and Lemon, S. M. 2001. The influence of downstream protein-coding sequence on internal ribosome entry on hepatitis C virus and other flavivirus RNAs. Rna 7: 585–97.
Robida, J. M., Nelson, H. B., Liu, Z. and Tang, H. 2007. Characterization of hepatitis C virus subgenomic replicon resistance to cyclosporine in vitro. J Virol 81: 5829–40.
Sapay, N., Montserret, R., Chipot, C., Brass, V., Moradpour, D., Deleage, G. and Penin, F. 2006. NMR structure and molecular dynamics of the in-plane membrane anchor of nonstructural protein 5A from bovine viral diarrhea virus. Biochemistry 45: 2221–33.
Schmidt-Mende, J., Bieck, E., Hugle, T., Penin, F., Rice, C. M., Blum, H. E. and Moradpour, D. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J Biol Chem 276: 44052–63.
Schwartz, M., Chen, J., Janda, M., Sullivan, M., den Boon, J. and Ahlquist, P. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol Cell 9: 505–14.
Serebrov, V. and Pyle, A. M. 2004. Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature 430: 476–80.
Shim, J. H., Larson, G., Wu, J. Z. and Hong, Z. 2002. Selection of 3′-template bases and initiating nucleotides by hepatitis C virus NS5B RNA-dependent RNA polymerase. J Virol 76: 7030–9.
Shirota, Y., Luo, H., Qin, W., Kaneko, S., Yamashita, T., Kobayashi, K. and Murakami, S. 2002. Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA polymerase (RdRP) NS5B and modulates RNA-dependent RNA polymerase activity. J Biol Chem 277: 11149–55.
Simmonds, P., Bukh, J., Combet, C., Deleage, G., Enomoto, N., Feinstone, S., Halfon, P., Inchauspe, G., Kuiken, C., Maertens, G., Mizokami, M., Murphy, D. G., Okamoto, H., Pawlotsky, J. M., Penin, F., Sablon, E., Shin, I. T., Stuyver, L. J., Thiel, H. J., Viazov, S., Weiner, A. J. and Widell, A. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42: 962–73.
Simmonds, P., Tuplin, A. and Evans, D. J. 2004. Detection of genome-scale ordered RNA structure (GORS) in genomes of positive-stranded RNA viruses: Implications for virus evolution and host persistence. RNA 10: 1337–51.
Siridechadilok, B., Fraser, C. S., Hall, R. J., Doudna, J. A. and Nogales, E. 2005. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310: 1513–5.
Sklan, E. H., Serrano, R. L., Einav, S., Pfeffer, S. R., Lambright, D. G. and Glenn, J. S. 2007a. TBC1D20 is a RAB1 GAP that mediates HCV replication. J Biol Chem 282: 36354–61.
Sklan, E. H., Staschke, K., Oakes, T. M., Elazar, M., Winters, M., Aroeti, B., Danieli, T. and Glenn, J. S. 2007b. A Rab-GAP TBC domain protein binds hepatitis C virus NS5A and mediates viral replication. J Virol 81: 11096–105.
Song, Y., Friebe, P., Tzima, E., Junemann, C., Bartenschlager, R. and Niepmann, M. 2006. The hepatitis C virus RNA 3'-untranslated region strongly enhances translation directed by the internal ribosome entry site. J Virol 80: 11579–88.
Spahn, C. M., Kieft, J. S., Grassucci, R. A., Penczek, P. A., Zhou, K., Doudna, J. A. and Frank, J. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40 s ribosomal subunit. Science 291: 1959–62.
Spångberg, K. and Schwartz, S: 1999. Poly(C)-binding protein interacts with the hepatitis C virus 5' untranslated region. J Gen Virol 80 (Pt 6): 1371–6.
Spångberg, K., Wiklund, L. and Schwartz, S. 2000. HuR, a protein implicated in oncogene and growth factor mRNA decay, binds to the 3' ends of hepatitis C virus RNA of both polarities. Virology 274: 378–90.
Steinmann, E., Penin, F., Kallis, S., Patel, A. H., Bartenschlager, R. and Pietschmann, T. 2007. Hepatitis C Virus p7 Protein Is Crucial for Assembly and Release of Infectious Virions. PLoS Pathog 3: e103.
Stone, M., Jia, S., Heo, W. D., Meyer, T. and Konan, K. V. 2007. Participation of rab5, an early endosome protein, in hepatitis C virus RNA replication machinery. J Virol 81: 4551–63.
Su, A. I., Pezacki, J. P., Wodicka, L., Brideau, A. D., Supekova, L., Thimme, R., Wieland, S., Bukh, J., Purcell, R. H., Schultz, P. G. and Chisari, F. V. 2002. Genomic analysis of the host response to hepatitis C virus infection. Proc Natl Acad Sci USA 99: 15669–74.
Sumpter, R., Jr., Loo, Y. M., Foy, E., Li, K., Yoneyama, M., Fujita, T., Lemon, S. M. and Gale, M., Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol 79: 2689–99.
Sun, X. L., Johnson, R. B., Hockman, M. A. and Wang, Q. M. 2000. De novo RNA synthesis catalyzed by HCV RNA-dependent RNA polymerase. Biochem Biophys Res Commun 268: 798–803.
Suzich, J. A., Tamura, J. K., Palmer-Hill, F., Warrener, P., Grakoui, A., Rice, C. M., Feinstone, S. M. and Collett, M. S: 1993. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J Virol 67: 6152–8.
Tan, S. L. and Katze, M. G. 2001. How hepatitis C virus counteracts the interferon response: the jury is still out on NS5A. Virology 284: 1–12.
Tanaka, T., Kato, N., Cho, M. J. and Shimotohno, K: 1995. A novel sequence found at the 3' terminus of hepatitis C virus genome. Biochem Biophys Res Commun 215: 744–9.
Tanaka, T., Kato, N., Cho, M. J., Sugiyama, K. and Shimotohno, K: 1996. Structure of the 3' terminus of the hepatitis C virus genome. J Virol 70: 3307–12.
Tanji, Y., Kaneko, T., Satoh, S. and Shimotohno, K: 1995. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J Virol 69: 3980–6.
Tellinghuisen, T. L., Evans, M. J., von Hahn, T., You, S. and Rice, C. M. 2007. Studying hepatitis C virus: making the best of a bad virus. J Virol 81: 8853–67.
Tellinghuisen, T. L., Marcotrigiano, J., Gorbalenya, A. E. and Rice, C. M. 2004. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J Biol Chem 279: 48576–87.
Tellinghuisen, T. L., Marcotrigiano, J. and Rice, C. M. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435: 374–9.
Teterina, N. L., Gorbalenya, A. E., Egger, D., Bienz, K., Rinaudo, M. S. and Ehrenfeld, E. 2006. Testing the modularity of the N-terminal amphipathic helix conserved in picornavirus 2C proteins and hepatitis C NS5A protein. Virology 344: 453–67.
Tischendorf, J. J., Beger, C., Korf, M., Manns, M. P. and Kruger, M. 2004. Polypyrimidine tract-binding protein (PTB) inhibits Hepatitis C virus internal ribosome entry site (HCV IRES)-mediated translation, but does not affect HCV replication. Arch Virol 149: 1955–70.
Tong, X. and Malcolm, B. A. 2006. Trans-complementation of HCV replication by non-structural protein 5A. Virus Res 115: 122–30.
Tsuchihara, K., Tanaka, T., Hijikata, M., Kuge, S., Toyoda, H., Nomoto, A., Yamamoto, N. and Shimotohno, K: 1997. Specific interaction of polypyrimidine tract-binding protein with the extreme 3'-terminal structure of the hepatitis C virus genome, the 3'X. J Virol 71: 6720–6.
Tu, H., Gao, L., Shi, S. T., Taylor, D. R., Yang, T., Mircheff, A. K., Wen, Y., Gorbalenya, A. E., Hwang, S. B. and Lai, M. M: 1999. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology 263: 30–41.
Wakita, T., Pietschmann, T., Kato, T., Date, T., Miyamoto, M., Zhao, Z., Murthy, K., Habermann, A., Krausslich, H. G., Mizokami, M., Bartenschlager, R. and Liang, T. J. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11: 791–6.
Wang, C., Gale, M., Jr., Keller, B. C., Huang, H., Brown, M. S., Goldstein, J. L. and Ye, J. 2005. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol Cell 18: 425–34.
Wang, Q. M., Hockman, M. A., Staschke, K., Johnson, R. B., Case, K. A., Lu, J., Parsons, S., Zhang, F., Rathnachalam, R., Kirkegaard, K. and Colacino, J. M. 2002. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J Virol 76: 3865–72.
Watashi, K., Hijikata, M., Hosaka, M., Yamaji, M. and Shimotohno, K. 2003. Cyclosporin A suppresses replication of hepatitis C virus genome in cultured hepatocytes. Hepatology 38: 1282–8.
Watashi, K., Ishii, N., Hijikata, M., Inoue, D., Murata, T., Miyanari, Y. and Shimotohno, K. 2005. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol Cell 19: 111–22.
Yamashita, T., Kaneko, S., Shirota, Y., Qin, W., Nomura, T., Kobayashi, K. and Murakami, S: 1998. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J Biol Chem 273: 15479–86.
Yanagi, M., Purcell, R. H., Emerson, S. U. and Bukh, J: 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci USA 94: 8738–43.
Yanagi, M., St Claire, M., Emerson, S. U., Purcell, R. H. and Bukh, J: 1999. In vivo analysis of the 3' untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc Natl Acad Sci USA 96: 2291–5.
Yang, G., Pevear, D. C., Collett, M. S., Chunduru, S., Young, D. C., Benetatos, C. and Jordan, R. 2004. Newly synthesized hepatitis C virus replicon RNA is protected from nuclease activity by a protease-sensitive factor(s). J Virol 78: 10202–5.
Ye, J., Wang, C., Sumpter, R., Jr., Brown, M. S., Goldstein, J. L. and Gale, M., Jr. 2003. Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc Natl Acad Sci USA 100: 15865–70.
Yi, M. and Lemon, S. M. 2003. 3' nontranslated RNA signals required for replication of hepatitis C virus RNA. J Virol 77: 3557–68.
You, S. and Rice, C. M. 2007. 3' RNA elements in hepatitis C virus replication: Kissing partners and long poly (U). J Virol 82: 184–95.
You, S., Stump, D. D., Branch, A. D. and Rice, C. M. 2004. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J Virol 78: 1352–66.
Yu, G. Y., Lee, K. J., Gao, L. and Lai, M. M. 2006. Palmitoylation and polymerization of hepatitis C virus NS4B protein. J Virol 80: 6013–23.
Zheng, Y., Gao, B., Ye, L., Kong, L., Jing, W., Yang, X., Wu, Z. and Ye, L. 2005. Hepatitis C virus non-structural protein NS4B can modulate an unfolded protein response. J Microbiol 43: 529–36.
Zhong, J., Gastaminza, P., Cheng, G., Kapadia, S., Kato, T., Burton, D. R., Wieland, S. F., Uprichard, S. L., Wakita, T. and Chisari, F. V. 2005. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA 102: 9294–9.
Zhong, W., Uss, A. S., Ferrari, E., Lau, J. Y. and Hong, Z. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J Virol 74: 2017–22.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2009 Springer Science+Business Media, LLC
About this chapter
Cite this chapter
Lindenbach, B.D., L. Tellinghuisen, T. (2009). Hepatitis C Virus Genome Replication. In: Raney, K., Gotte, M., Cameron, C. (eds) Viral Genome Replication. Springer, Boston, MA. https://doi.org/10.1007/b135974_4
Download citation
DOI: https://doi.org/10.1007/b135974_4
Published:
Publisher Name: Springer, Boston, MA
Print ISBN: 978-0-387-89425-6
Online ISBN: 978-0-387-89456-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)