Arenaviruses constitute one of the most widespread and diverse groups of rodent-borne viruses, which merit significant attention both as tractable model systems to study acute and persistent viral infections (Oldstone, 2002; Zinkernagel, 2002) and as clinically important human pathogens including several causative agents of severe hemorrhagic fever (HF), chiefly Lassa fever virus (LASV) (Geisbert and Jahrling, 2004; McCormick and Fisher-Hoch, 2002; Peters, 2002). Moreover, evidence indicates that the prototypic Arenavirus lymphocytic choriomeningitis virus (LCMV) is a neglected human pathogen of clinical significance, especially in cases of congenital infection (Barton and Mets, 1999, 2001; Barton et al., 2002; Jahrling and Peters, 1992; Mets et al., 2000; Peters, 2006). No licensed anti-arenavirus vaccines are available, and current anti-arenavirus therapies are limited to the use of the nucleoside analog ribavirin, which is only partially effective and often associated with significant secondary effects including anemia and birth defects. Therefore, it is important to develop novel antiviral strategies to combat arenaviruses. This task will benefit from an improved knowledge about the arenavirus molecular biology, which is the focus of this chapter.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Transcription Termination
- Virus Polymerase
- Ring Finger Protein
- Reverse Genetic System
- Transcription Termination Signal
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Introduction
Arenaviruses constitute one of the most widespread and diverse groups of rodent-borne viruses, which merit significant attention both as tractable model systems to study acute and persistent viral infections (Oldstone, 2002; Zinkernagel, 2002) and as clinically important human pathogens including several causative agents of severe hemorrhagic fever (HF), chiefly Lassa fever virus (LASV) (Geisbert and Jahrling, 2004; McCormick and Fisher-Hoch, 2002; Peters, 2002). Moreover, evidence indicates that the prototypic Arenavirus lymphocytic choriomeningitis virus (LCMV) is a neglected human pathogen of clinical significance, especially in cases of congenital infection (Barton and Mets, 1999, 2001; Barton et al., 2002; Jahrling and Peters, 1992; Mets et al., 2000; Peters, 2006). No licensed anti-arenavirus vaccines are available, and current anti-arenavirus therapies are limited to the use of the nucleoside analog ribavirin, which is only partially effective and often associated with significant secondary effects including anemia and birth defects. Therefore, it is important to develop novel antiviral strategies to combat arenaviruses. This task will benefit from an improved knowledge about the arenavirus molecular biology, which is the focus of this chapter.
The inability to genetically manipulate the Arenavirus genome has hampered studies aimed at understanding its molecular and cell biology, as well as the role played by each viral gene product in virus–host interactions during both acute and persistent infections and associated diseases. The recent development of reverse genetics systems for several arenaviruses including LCMV (Flatz et al., 2006; Lee et al., 2000; Sanchez and de la Torre, 2006), as well as LASV (Hass et al., 2004) and Tacaribe virus (TCRV) (Lopez et al., 2001), has provided investigators with a novel and powerful approach for the investigation of the cis-acting sequences and trans-acting factors that control arenavirus replication and gene expression, as well as assembly and budding. Moreover, the ability to rescue recombinant LCM viruses from cloned cDNAs with predetermined specific mutations and analyze their phenotypic expression in its natural host, the mouse, has created unique opportunities to investigate arenavirus–host interactions that influence a variable infection outcome ranging from virus control and clearance by the host defenses to long-term chronic infection associated with subclinical disease, and severe acute disease including HF. Likewise these new developments are facilitating a detailed understanding of the arenavirus molecular biology. This new knowledge, in turn, is opening new avenues for the development and evaluation of novel antiviral strategies to combat arenaviral infections.
Arenaviruses Important as Both Model Systems and Human Pathogens
LCMV as a Model to Study Virus–Host Interactions Associated with Both Acute and Chronic Viral Infections
The LCMV system provides a primary workhorse in the fields of immunology and viral pathogenesis and has contributed to the development of multiple key concepts in both disciplines including (1) antiviral tolerance (Oldstone, 2002; Zinkernagel, 2002), (2) virus-induced immunosuppression (Oldstone, 2002; Zinkernagel, 2002), and (3) the ability of noncytolytic persistent riboviruses to avoid elimination by the host immune responses and to induce disease by interfering with specialized functions of infected cells, revealing a new way by which viruses do harm in the absence of the classic hallmarks of cytolysis and inflammation (de la Torre and Oldstone, 1996; Oldstone, 2002). Moreover, LCMV represents an excellent model to unravel the mechanisms of virally induced meningitis and foster the development of novel interventions to ameliorate the symptoms and consequences of this clinically important pathogenic process.
Arenaviruses and the Diseases That They Cause
Arenaviruses cause chronic infections of rodents with a worldwide distribution (Buchmeier et al., 2001). Asymptomatically infected animals move freely in their natural habitat and may invade human habitation. Humans are infected through mucosal exposure to aerosols, or by direct contact of abrade skin with infectious materials.
Several arenaviruses cause severe HF disease in humans and pose a serious public health problem (Buchmeier et al., 2001; McCormick and Fisher-Hoch, 2002; Peters, 2002). In recent years, increased traveling to and from endemic regions has led to the importation of LASV into non-endemic regions including the United States (Freedman and Woodall, 1999; Holmes et al., 1990; Isaacson, 2001). On the other hand, it is worth noting that compelling evidence indicates that LCMV is a neglected human pathogen of clinical significance, especially in cases of congenital infection leading to hydrocephalus, mental retardation, and chorioretinitis in infants (Barton et al., 2002; Jahrling and Peters, 1992; Mets et al., 2000). In addition, LCMV poses special threat to immuno-compromised individuals, as illustrated by recent cases of transplant-associated infections by LCMV with a fatal outcome in the United States (Fischer et al., 2006; Peters, 2006).
Arenavirus Genome Organization
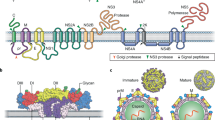
Arenaviruses are enveloped viruses with a bi-segmented negative single-stranded RNA genome and a life cycle restricted to the cell cytoplasm (Buchmeier et al., 2007; Meyer et al., 2002). Individual arenaviruses exhibit some variability in the lengths of the two genomic RNA segments, L (ca. 7.2 kb) and S (ca. 3.5 kb), but their overall organization is well conserved across the virus family. As with other negative strand (NS) RNA viruses, arenaviruses are characterized by a lack of infectivity of their purified genome RNA species and the presence of a virion-associated RNA-dependent-RNA polymerase (RdRp). However, the Arenavirus coding strategy has unique features compared to prototypical NS RNA viruses. Each Arenavirus genome segment uses an ambisense coding strategy to direct the synthesis of two polypeptides in opposite orientation, separated by a non-coding intergenic region (IGR) with a predicted folding of a stable hairpin structure (Buchmeier et al., 2007; Meyer et al., 2002). The S RNA encodes the viral glycoprotein precursor, GPC (ca. 75 kDa), and the nucleoprotein, NP (ca. 63 kDa), whereas the L RNA encodes the viral RNA-dependent-RNA polymerase (RdRp, or L polymerase) (ca. 200 kDa) and a small RING finger protein Z (ca. 11 kDa). The NP and L coding regions are transcribed into a genomic complementary mRNA, whereas the GPC and Z coding regions are not translated directly from genomic RNA, but rather from genomic sense mRNAs that are transcribed using as templates the corresponding antigenome RNA species, which also function as replicative intermediates (Fig. 10.1). The term ambisense refers to this situation in which regions located down- and upstream of the IGR of the S and L genome RNA species are of negative and positive sense, respectively. However, genomic S and L RNAs cannot function as mRNAs and be directly translated into GPC and Z proteins, respectively.
Virions contain the L and S genomic RNAs as helical nucleocapsid structures that are organized into circular configurations, with lengths ranging from 400 to 1300 nm (Young and Howard, 1983). The L and S genomic RNA species are not present in equimolar amounts within virions (L:S ratios ∼1:2), and low levels of both L and S antigenomic RNA species are also present within virions. In addition, it has been documented that host ribosomes can be incorporated into virions, but the biological implications of this remain to be determined (Buchmeier et al., 2007; Muller et al., 1983). Likewise, significant levels of the Z mRNA appear to be incorporated into virions (Salvato et al., 1992), but it is unknown whether this reflects a functional requirement or an imprecise encapsidation process.
Terminal Nucleotide Sequences
Arenaviruses exhibit high degree of sequence conservation at the 3′-end of the L and S RNA segments (17 out of 19 nucleotides [nt] are identical), suggesting that this conserved terminal sequence element constitutes the virus promoter for polymerase entry. Arenaviruses, similar to other NS RNA viruses, also exhibit complementarity between the 5′- and 3′-ends of their genomes and antigenomes. The almost exact inverted complement of the 3 ′-end 19 nt is found at the 5′-termini of genomes and antigenomes of arenaviruses. Thus, the 5′- and 3′-ends of both L and S genome segments are predicted to form panhandle structures. This prediction is supported by electron microscopy data showing the existence of circular ribonucleoprotein (RNP) complexes within arenavirus virion particles. This terminal complementarity may reflect the presence at the 5′-ends of cis-acting signals sequences that provide a nucleation site for RNA encapsidation, required to generate the nucleocapsid (NC) templates recognized by the virus polymerase. Terminal complementarity may also be a consequence of strong similarities between the genome and antigenome promoters used by the virus polymerases. This terminal complementarity has been proposed to favor the formation of both intra- and inter-molecular L and S duplexes that might be part of the replication initiation complex (Salvato, 1993b). For several arenaviruses, an additional non-templated G residue has been detected on the 5′-end of their genome RNAs (Garcin and Kolakofsky, 1992; Raju et al., 1990).
Intergenic Regions
Arenavirus IGR are predicted to fold into a stable hairpin structure. Transcription termination of the S-derived NP and GP occurs at multiple sites within the predicted stem of the IGR, suggesting that a structural motif rather than a sequence-specific signal promotes the release of the Arenavirus polymerase from the template RNA. Some arenaviruses including LCMV contain one single predicted stem loop within the S IGR, whereas the S IGR of others, e.g., TCRV, is predicted to contain two distinct IGRs downstream to the translation termination codons from NP and GPC (Buchmeier et al., 2007; Meyer et al., 2002).
Sequence Heterogeneity
The family Arenaviridae comprises two distinct complexes: the LCMV-Lassa complex, which includes the Old World arenaviruses, and the Tacaribe virus (TCRV) complex, which includes all known New World arenaviruses. Early sequence analysis of laboratory-adapted arenaviruses revealed a significant degree of genetic stability with amino acid sequence homologies of 90%–95% among different strains of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV), whereas significant higher levels of genetic diversity (37%–56%) were observed for homologous proteins of different arenaviruses species (Buchmeier et al., 2007; Southern, 1996). More recent genetic studies on arenavirus field isolates, including Lassa (Bowen et al., 2000), Junin (Garcia et al., 2000), Guanarito (GTO) (Weaver et al., 2000), Pirital (PIR) (Fulhorst et al., 1999), and Whitewater Arroyo (Fulhorst et al., 2001), have revealed a high degree of genetic variation among geographical and temporal isolates of the same virus species. Notably, a remarkably high level of genetic divergence (26% and 16% at the nt and amino acid level, respectively) has been documented among PIR isolates within very small geographic regions (Weaver et al., 2001). The substantial degree of inter- and intra-species genetic variation among arenaviruses appears to have important biological correlates, as suggested by the significant variation in biological properties observed among LCMV strains (Sevilla et al., 2002). Thus, dramatic phenotypic differences have been documented among genetically very closely related LCMV isolates exhibiting only a few amino acid differences in their proteins.
Arenavirus Proteins
The NP is the most abundant viral polypeptide in both infected cells and virions. NP is the main structural element of the viral RNP and plays an essential role in viral RNA synthesis. We have shown that NP exhibits also an IFN counteracting activity (Martinez-Sobrido et al., 2006). The viral glycoprotein precursor GPC is posttranslationally proteolytically processed by the S1P cellular protease to yield the two mature virion glycoproteins GP-1 (40–46 kDa) and GP-2 (35 kDa) (Beyer et al., 2003; Pinschewer et al., 2003b). GPC contains a 58-amino-acid signal peptide (SSP) that is expressed as a stable polypeptide in infected cells that remains stably associated to the GP complex (GPcx). Besides its role in targeting the nascent polypeptide to the endoplasmic reticulum, this SSP likely serves additional roles in the trafficking and function of the viral envelope glycoproteins (Eichler et al., 2003a, 2004, 2003b; Froeschke et al., 2003; York et al., 2004). GP-1 mediates virus interaction with host cell surface receptors and is held in place at the top of the spike by ionic interactions with the N-terminus of the transmembrane GP-2 (Buchmeier et al., 2001; Neuman et al., 2005).
The Arenavirus L protein has the characteristic sequence motifs conserved among the RdRp (L proteins), of negative strand (NS) RNA viruses (Salvato et al., 1989; Sanchez and de la Torre, 2005; Tordo et al., 1992; Vieth et al., 2004). Detailed sequence analysis and secondary structure predictions have been documented for the LFV L polymerase (Vieth et al., 2004). These studies identified several regions of strong alpha-helical content and a putative coiled-coil domain at the N-terminus, whose functional roles remain to be determined.
The arenavirus RING finger protein Z has no homologue among other known NS RNA viruses. Z is a structural component of the virion (Salvato et al., 1992). In LCMV-infected cells Z has been shown to interact with several cellular proteins including the promyelocytic leukemia (PML) protein (Borden et al., 1998) and the eukaryote translation initiation factor 4E (eIF4E) (Campbell Dwyer et al., 2000; Kentsis et al., 2001), which have been proposed to contribute to the noncytolytic nature of LCMV infection and repression of cap-dependent translation, respectively. Biochemical studies suggested that Z might be the arenavirus counterpart of the matrix (M) protein found in other negative strand RNA viruses (Salvato, 1993a; Salvato et al., 1992). Consistent with this proposal, more recent evidence has shown that Z is the driving force of arenavirus budding (Perez et al., 2003; Strecker et al., 2003; Urata et al., 2006). The expression of Z during the progression from early to late phases of the LCMV life cycle appears to be highly regulated, and thereby Z might play different roles during the life cycle of LCMV.
Arenavirus Life Cycle
Cell Attachment and Entry
Consistent with a broad host range and cell type tropism, a highly conserved and widely expressed cell surface protein, alpha-dystroglycan (aDG) has been identified as a main receptor for LCMV, LASV and Clade C NW arenaviruses (Kunz et al., 2002). However, several other arenaviruses appear to use an alternative receptor (Kunz et al., 2004; Spiropoulou et al., 2002), and recently transferring receptor 1 was identified as a cellular receptor used for entry of the NW HF arenaviruses Machupo and Junin (Radoshitzky et al., 2007). Upon receptor binding, arenavirus virions are internalized by uncoated vesicles and released into the cytoplasm by a pH-dependent membrane fusion step that is mediated by GP-2 (Borrow and Oldstone, 1994; Di Simone and Buchmeier, 1995; Di Simone et al., 1994). This fusion event is mediated by GP-2, which is structurally similar to the fusion active membrane proximal portions of the GP of other enveloped viruses (Gallaher et al., 2001).
RNA Replication and Transcription
The fusion between viral and cellular membranes releases the viral RNP into the cytoplasm, which is ensued by viral RNA synthesis. LCMV mRNAs have extra non-templated nt and a cap structure at their 5′-ends, but the origin of both the cap and 5′-non-templated nt extensions remains to be determined. Transcription termination of subgenomic non-polyadenylated viral mRNAs was mapped to multiple sites within the distal side of the IGR (Meyer and Southern, 1994; Tortorici et al., 2001), suggesting that the IGR acts as a bona fide transcription termination signal for the virus polymerase. The basic steps of Arenavirus RNA replication and gene transcription are illustrated in Fig. 10.2 using the S segment as example; the same scheme applies also to the L segment. NP and L are transcribed into a genomic complementary mRNA, whereas the GPC and Z are not translated directly from genomic RNA, but rather from genomic sense mRNAs that are transcribed using as templates the corresponding antigenome RNA species, which also function as replicative intermediates.
Viral Trans-Acting Factors Required for RNA Replication and Transcription
Reverse genetic studies using an LCMV minigenome rescue system identified NP and L as the minimal viral trans-acting factors required for efficient RNA synthesis mediated by the virus polymerase (Lee et al., 2000). Similar findings have been now documented for LASV (Hass et al., 2004) and the New World Arenavirus TCRV (Lopez et al., 2001). Notably, both genetic and biochemical evidence have indicated that oligomerization of L is required for the activity of the LCMV L polymerase (Sanchez and de la Torre, 2005), a finding similar to that previously documented for the paramyxoviruses Sendai (Smallwood et al., 2002) and parainfluenza virus 3 (PIV3) (Smallwood and Moyer, 2004).
Cis-Acting Signals Involved in the Regulation of LCMV RNA Synthesis
Sequence Specificity and Structure Define the Functional Genome Promoter
All arenavirus genomes examined to date have a highly conserved sequence element at their 3′-termini (17/19 nt are identical), and the inverted complement of this sequence is found at the genome 5′-termini. Terminal complementarity in L and S RNAs predicts the formation of a conserved and thermodynamically stable panhandle structure that, similar to influenza- and bunya-viruses, was proposed to contribute to the control of RNA synthesis.
Mutation-function analysis of the genome 3′/5′-termini in the control of viral RNA synthesis using a transcription and replication competent LCMV minigenome (MG) rescue system (Perez and de la Torre, 2002) revealed the minimal LCMV genomic promoter to be contained within the 3′-terminal 19 nt. Moreover, deletions and nt substitutions within the MG 5′-end that disrupted terminal complementarity abolished also genome promoter activity. Notably, compensatory mutations that restore paring between the 3′- and 5′-termini did not result in restoration of genome promoter activity. Likewise, these studies did not identify mutations within the promoter sequences that affected independently either RNA replication or transcription. A detailed mutation-function analysis of the virus genome promoter has been documented for LASV (Hass et al., 2006), revealing that the LASV genome promoter also regulates transcription and replication in a coordinated manner. These studies also showed that the LASV genome promoter is composed of two functional elements, a sequence-specific region from residues 1 to 12 and a variable complementary region from residues 13 to 19. The first region appears to interact with the replication complex mainly via base-specific interactions, while in the second region solely base pairing between 3′ and 5′ promoter ends is important for promoter function.
These findings support the view that arenavirus genome promoters regulate transcription and replication in a coordinated manner and that both sequence specificity within the 3′-terminal 19 nt and the integrity of the predicted panhandle structure are required for the activity of the genome promoter. Moreover, the two biosynthetic processes, RNA replication and transcription, directed by the virus polymerase complex appeared to be coordinated by the same cis-acting regulatory sequences. Initiation of RNA synthesis by the arenavirus polymerase has been proposed to employ a prime and realign mechanism, which would account for the presence of a non-templated G at the 5′-ends of the arenavirus genomic and antigenomic RNAs (Garcin and Kolakofsky, 1990, 1992). Results obtained using MGs with a variety of 5′-end sequences have provided evidence in support of this model (Perez and de la Torre, 2002).
The IGR Present Within Each Arenavirus Genome Segment is a Bona Fide Transcription Termination Signal, but Plays also a Critical Role in Assembly of Infectious Particles
Arenavirus mRNAs have extra non-templated nucleotides (nt) and a cap structure at their 5′-ends, but the origin of both the cap and 5′-non-templated nt extensions remains to be determined. The 3′-termini of the subgenomic non-polyadenylated viral mRNAs have been mapped to multiple sites within the distal side of the IGR (Meyer et al., 2002; Southern, 1996). All arenavirus IGR sequences are predicted to fold into single or double stem-loop structures (Buchmeier et al., 2001; Meyer et al., 2002; Southern, 1996), suggesting a structure-dependent transcription termination mechanism reminiscent of rho-independent termination in prokaryotes (Yarnell and Roberts, 1999).
Studies using the LCMV MG rescue system where a variety of RNA analogues of the S genome segment, containing or not an IGR, served as a template for synthesis of full-length anti-MG (aMG) replicate and subgenomic size mRNA species for reporter gene expression showed that a MG without IGR was amplified by the virus polymerase with equal efficiency but subgenomic mRNA species were undetectable (Pinschewer et al., 2005). Intriguingly, however, reporter gene expression from IGR-deficient aMG CAT-sense RNA of genomic length was found to be only about 5-fold less efficient than from subgenomic CAT mRNA derived from an IGR-containing MG, but at least 100-fold more efficient than a T7 RNA polymerase transcript with the same sequence. These results validated the IGR as a bona fide transcription termination signal, but revealed also that in the absence of IGR-mediated transcription termination, a fraction of full-length aMG RNA behaves as bona fide mRNA. Conceptually similar findings have been also documented for the NW arena TCRV (Lopez and Franze-Fernandez, 2007). Likewise studies using a TCRV MG rescue system demonstrated that the transcription termination signal provided by the IGR is structure, but not sequence, dependent (Lopez and Franze-Fernandez, 2007).
Unexpectedly, LCMV MGs without IGR were dramatically impaired in their ability to passage reporter gene activity via infectious VLP (Pinschewer et al., 2005), suggesting that in addition to its role in the control of RNA synthesis, the arenavirus IGR plays a role in virus assembly or budding, or both, required for the efficient virus propagation. Whether this role of the IGR depends on sequence specificity or structure, or both, remains to be determined.
Intracellular levels of NP determine levels of viral RNA synthesis but do not regulate the balance between RNA replication and transcription. For arenaviruses it has been proposed, and widely accepted, that intracellular NP levels modulate the balance between RNA replication and transcription. Intracellular NP levels increase during the course of the infection and unfold secondary RNA structures within the IGR. This results in attenuation of structure-dependent transcription termination at the IGR, which promotes replication of genome and antigenome RNA species. However, more recent studies using the LCMV MG rescue system revealed that both RNA replication and transcription were equally enhanced by incrementally increasing amounts of NP up to levels in the range of LCMV-infected cells (Pinschewer et al., 2003a). These data, similar to those described for the paramyxovirus RSV (Fearns et al., 1997), are consistent with a central role for NP in transcription and RNA replication of the LCMV genome, but they do not support a central role of NP levels in balancing the two biosynthetic processes.
Role of the Z Protein in the Control of Arenavirus RNA Synthesis
Z Exhibits a Dose-Dependent Inhibitory Effect on RNA Replication and Transcription of the LCMV MG
Z was not required for intracellular transcription and replication of an LCMV MG, but rather Z exhibited a dose-dependent inhibitory effect on both transcription and replication of LCMV MG (Cornu and de la Torre, 2001, 2002; Cornu et al., 2004; Lee et al., 2000) (see below). Similar findings have been also reported for TV (Lopez et al., 2001) and LASV (Hass et al., 2004).
Mutation-function studies identified regions and specific amino acid residues within Z contributing to its inhibitory activity on RNA synthesis mediated by the LCMV polymerase (Cornu and de la Torre, 2002). Serial deletion mutants of the N- and C-termini of Z showed that the N-terminus (residues 1–16) and C-terminus (residues 79–90) do not contribute to the Z inhibitory activity. Moreover, results from the use of chimera proteins between Z and Xenopus Neuralized, a nonviral RING finger protein, indicated that the structural integrity of the Z ring domain (RD) was required but not sufficient for the inhibitory activity of Z. Likewise, a highly conserved tryptophan (W) residue located at position 36 in ARM-Z, next to the second conserved cysteine (C) of the Z RD, had a major contribution to the Z inhibitory activity. The inhibitory activity of Z on virus RNA synthesis appeared to be related to the degree of genetic proximity between Z and the viral trans-acting factors L and NP. Thus, Z proteins from different LCMV strains had similar inhibitory activities on the expression of LCMV MG, whereas the Z protein of the genetically more distantly related TCRV had about 10-fold lower inhibitory activity on LCMV MG expression (Cornu and de la Torre, 2002).
Homotypic viral interference can be readily demonstrated with several arenaviruses including LCMV (Welsh and Pfau, 1972). This phenomenon is not strictly strain specific as illustrated by the existence of interference among different pairs of LCMV strains. Heterotypic interference between arenaviruses has been occasionally reported, and its degree appears to correlate with the genetic relationship of the viruses (Damonte et al., 1983; Welsh and Pfau, 1972). It is therefore plausible that increased expression of Z protein during the virus life cycle might contribute to block replication of an additional infection by a genetically closely related arenavirus. Superinfection exclusion could influence arenavirus evolution and contribute to explain the observed population partitioning in the field, resulting in the maintenance of independent evolutionary lineages of the same strain within a small geographic range.
Cells Expressing Z Become Highly Resistant to Virus Infection Due to a Blockade in Virus RNA Synthesis
Cells transduced with recombinant, replication-deficient adenoviruses expressing Z (rAd-Z) from either LCMV or LASV became highly resistant to infection with LCMV or LASV, respectively (Cornu et al., 2004), whereas cells transduced with a control rAd expressing GFP remained fully susceptible to both LCMV and LASV. This resistance was specific as the rAd-Z transduced cells remained fully susceptible to measles virus (MV) (Cornu et al., 2004). These findings indicated that the Z-mediated inhibitory activity operates also during the course of the natural cycle of virus infection, and it is not a property observed only in the context of a MG system. Cells transduced with rAd-Z remained susceptible to infection with a recombinant VSV where the LCMV G substituted for the VSV G (Cornu et al., 2004), suggesting that Z-mediated resistance to infection was not due to a blockade of virus entry, but rather to a strong inhibitory effect of Z on LCMV RNA replication and transcription.
The TCRV Z protein was reported to interact with the virus L polymerase, and this interaction was proposed to be responsible for TCRV Z-mediated inhibition of RNA replication and expression of a TCRV MG (Jacamo et al., 2003). Intriguingly, for LCMV the use of either biochemical (Co-IP) or genetic (mammalian-TH) approaches have failed to provide evidence of a Z–L interaction. This could be due to intrinsic differences between the biology of TCRV and LCMV, or differences in the two experimental systems. Further studies would be necessary to elucidate the mechanisms by which Z exerts its inhibitory activity on RNA synthesis by the virus polymerase, as well as to determine the biological implications of this Z activity. The recent observation that the activity of the LCMV L polymerase requires an L–L interaction (Sanchez and de la Torre, 2005) raises the possibility that Z might interfere with L–L interaction and thereby affect the virus polymerase activity. An alternative way whereby Z could mediate inhibition of viral RNA synthesis stems from our finding that Z interacts with N. As with the M proteins of several other NS RNA viruses, Z–NP interaction could inhibit the biosynthetic activity of the virus RNP.
Assembly and Budding
GP and Z are Required for the Generation of Infectious VLP
Production of LCMV occurs by budding at the surface of infected cells. For most enveloped NS RNA viruses, this process is assumed to depend on the interaction between the RNP core and the virus-encoded transmembrane glycoproteins (GP), which is mediated by the matrix (M) protein. Arenaviruses do not code for an obvious counterpart of M, but early cross-linking studies showed complex formation between NP and Z, suggesting a possible role of Z in virion morphogenesis (Salvato, 1993b; Salvato et al., 1992). Studies using the LCMV MG rescue system showed that generation of infectious arenavirus-like particles (VLP) required both Z and GP (Lee et al., 2002). Importantly, the correct processing of GPC was strictly required for the generation of either infectious VLPs (Lee et al., 2002) or retroviral pseudotyped particles (Beyer et al., 2003; Pinschewer et al., 2003b). Moreover, correct processing of GPC necessitates the structural integrity of GP-2 cytoplasmic tail (Kunz et al., 2003).
Z is the Driving Force of Arenavirus Budding
The requirement of GP for the generation of infectious VLP was expected due to its role in receptor recognition and virus entry (Kunz et al., 2002), whereas the need for Z indicated a role of this protein in virus assembly or budding, or both. This, in turn, suggested that consistent with earlier biochemical data (Salvato, 1993b; Salvato et al., 1992) and recent ultrastructural data on arenavirus virions determined by cryo-electron microscopy (Neuman et al., 2005), Z could be the Arenavirus functional counterpart of the M proteins that mediate budding in other NS RNA viruses. Studies using reverse genetics approaches to examine the requirement of LCMV proteins for efficient cell release of VLP containing bona fide viral nucleocapsids (NC) revealed that the production of MG RNA-containing NC was not impaired in the absence of GP, but dramatically diminished in the absence of Z (Perez et al., 2003), indicating that Z was playing a central role in LCMV budding.
Z has Features of Bona Fide Budding Proteins and Contains Canonical Late (L) Domain Motifs That are Functionally Active in Promoting Z-Mediated Budding
Consistent with its role as the driving force of arenavirus budding, Z exhibited self-budding activity in the absence of other viral proteins (Perez et al., 2003; Strecker et al., 2003; Urata et al., 2006). A feature characteristic of viral budding proteins is the flexibility of their L domains; one L domain substitutes for another in promoting virion release (Freed, 2002). Z also exhibited this feature as determined by the budding properties of Z-Gag chimeric proteins where Z was fused to an RSV Gag protein that lacked both its membrane targeting and binding signal (M domain) and L domain (Perez et al., 2003).
Consistent with their features of bona fide budding proteins, arenavirus Z contains canonical late (L) domain motifs similar to those present in Gag and M proteins of several viruses (Freed, 2002). The Z protein of LCMV contains a single PPPY motif, whereas the Z protein of the highly pathogenic arenavirus LFV possesses both PTAP and PPPY motifs separated by nine amino acids (Perez et al., 2003; Strecker et al., 2003; Urata et al., 2006). Mutation-function studies confirmed that these L domain motifs present in Z mediated the budding activity of Z (Perez et al., 2003; Strecker et al., 2003). Ebola VP40 protein contains overlapping PTAP and PPXY L domains, but each one of them was found to be sufficient to promote efficient VP40-mediated budding (Licata et al., 2003). In contrast, in the case of LASV Z both L domains were found to be required for efficient Z-mediated budding (Perez et al., 2003).
Myristoylation of Z is Required for its Budding Activity
Z is devoid of hydrophobic transmembrane domains, but it accumulates near to the inner surface of the plasma membrane and is strongly membrane associated. All known arenavirus Z proteins contain a glycine (G) at position 2 and nearby K and R residues characteristic of a myristoylation motif. Metabolic labeling showed incorporation of [3H]myristic acid by wild-type, but not G2A mutant; Z protein and the mutation G2A abrogated Z-mediated budding without affecting viral RNA replication and transcription (Perez et al., 2004). Likewise, treatment with the myristoylation inhibitor 2-hydroxymyristic acid (2-OHM) inhibited Z-mediated budding, abrogated formation of virus-like particles, and caused a dramatic reduction in virus production in LCMV-infected cells (Perez et al., 2004). Moreover, addition to the N-terminus of Z(G2A) of the myristoylation domain of the tyrosine protein kinase Src restored budding activity in the Z(G2A)G2A (Perez et al., 2004). These findings and similar ones described by others (Strecker et al., 2003, 2006) have also been documented. These findings indicate that myristoylation of Z plays a key role in Arenavirus budding. Similar findings have been also documented for LASV Z.
Z–GP Interact
Based on the roles played by Z and GP in the arenavirus life cycle, it would be predicted that Z and the GP should interact in a manner required for the formation of mature infectious virion particles. Accordingly, recent evidence has shown the subcellular co-localization and biochemical association of Z and GP (Capul et al., 2007). Notably, neither the RING domain nor the L domains were required for this Z–GP interaction (Capul et al., 2007). In contrast myristoylation of Z played a critical role in Z–GP interaction as determined by the failure of a G2A mutant of Z to interact with GP (Capul et al., 2007). These results may reflect that accumulation of Z at certain membranes within the cell might be a limiting factor for its association with GP.
Production of Infectious LCMV from Cloned cDNAs
The ability to generate predetermined specific mutations within the LCMV genome, and analyze their phenotypic expression in appropriate cell culture systems and the virus natural host, the mouse, has represented a major step forward for the elucidation of the molecular and cellular mechanisms underlying LCMV–host interactions, including the bases of LCMV persistence and associated disease. In addition, the procedures developed for the rescue of rLCMV should allow for the rescue of LFV and other HF arenaviruses, which may accelerate the development and fine-tuning of live-attenuated arenavirus vaccines (Lukashevich et al., 2005), and facilitate their safe production for use in endemic areas where they are urgently needed (Geisbert and Jahrling, 2004; Geisbert et al., 2005; McCormick and Fisher-Hoch, 2002).
Rescue of Infectious rLCMV from Cloned cDNAs
Prior to the successful rescue of LCMV entirely from cloned cDNAs, a helper virus-based system was documented that allowed for the rescue of LCMV carrying a recombinant S segment (rS) (Pinschewer et al., 2003b). This system was based on intracellular reconstitution of a recombinant LCMV S (rS) where the glycoprotein of vesicular stomatitis virus (VSVG) was substituted for the glycoprotein of LCMV and produced intracellularly from cDNA under control of a polymerase I promotor. Coexpression of the LCMV proteins NP and L allowed expression of VSVG from rS, and infection of transfected cells with wild-type (wt) LCMV resulted in reassortment of the L segment of wt LCMV with the rS at low frequency. Selection of the rLCMV over the LCMVwt used as helper virus was facilitated by the use of a cell line (SRD-12B) deficient in the S1P protease. The rationale for this approach was based on the fact that LCMV infectivity, but not that of rLCMV/VSVG, requires correct processing of LCMV-GPC by the cellular protease S1P.
The rLCMV/VSVG provided investigators with a powerful tool to rescue rLCM viruses containing engineered rS segments. For this, cells instructed to express the rS RNP of interest are infected with rLCMV/VSVG, and the virus progeny is subjected to selection with a neutralizing antibody to VSV G to eliminate the helper rLCMV/VSVG. This approach however required several rounds of selection and was limited to the rescue of LCMV carrying recombinant S segments. These limitations were circumvented with the development of reverse genetics system to allow for the rescue of infectious LCMV entirely from cloned cDNAs, without the need of using a helper virus. Both a T7 RNA polymerase (T7RP) (Sanchez and de la Torre, 2006) and RNA polymerase I (pol-I) (Flatz et al., 2006) systems have been developed to direct intracellular synthesis to recombinant L and S genome, or antigenome, RNA species. Both the T7RP and pol-I-based rescue systems used pol-II-based expression plasmids to provide the viral trans-acting factors L and NP. Both systems exhibited similar efficiencies. The pol-I-based system offers the advantages that (1) the generation of the correct 3′-end of the Sag and Lag RNA species does not depend on the efficiencies of self-cleavable ribozymes and (2) there is no need for a plasmid expressing T7RP. On the other hand this system has the limitation of the species specificity of the pol-I promoters, which determines the need of generating different vectors for efficient intracellular synthesis of virus genome RNA species in cell types from different species.
Production of rLCMV was readily detected 48 h after transfection, which was followed by a rapid increase in virus production reaching titers of 107 PFU/ml. These rLCMV exhibited growth and biological properties predicted for LCMV. Notably, similar rescue efficiencies were obtained using genome of antigenome L and S expressing plasmids (Sanchez and de la Torre, 2006), indicating that annealing between viral mRNAs and genome, or antigenome, RNA species does not pose a significant problem for the rescue of arenaviruses.
Use of rLCMV to Address Biological Questions
The rLCMV/VSVG has been used to examine a variety of biological questions, which illustrate the tremendous impetus for arenavirus research derived from the ability to generate rLCMV from cloned cDNAs (Bergthaler et al., 2006; Merkler et al., 2006; Pinschewer et al., 2004). The development and use of reverse genetics approaches have revolutionized the analysis of the cis-acting signals and trans-acting proteins required for RNA replication, transcription, maturation, and budding of other negative strand RNA viruses (Conzelmann, 2004; Kawaoka, 2004; Neumann et al., 2002). These approaches are now applicable to arenaviruses and will permit to dissect the role, and underlying mechanisms, of each virus gene product to the each of the steps of the arenavirus life cycle.
References
Barton, L. L., and Mets, M. B. (1999). Lymphocytic choriomeningitis virus: pediatric pathogen and fetal teratogen. Pediatr Infect Dis J 18(6), 540–1.
Barton, L. L., and Mets, M. B. (2001). Congenital lymphocytic choriomeningitis virus infection: decade of rediscovery. Clin Infect Dis 33(3), 370–4.
Barton, L. L., Mets, M. B., and Beauchamp, C. L. (2002). Lymphocytic choriomeningitis virus: emerging fetal teratogen. Am J Obstet Gynecol 187(6), 1715–6.
Bergthaler, A., Gerber, N. U., Merkler, D., Horvath, E., de la Torre, J. C., and Pinschewer, D. D. (2006). Envelope exchange for the generation of live-attenuated arenavirus vaccines. PLoS Pathog 2(6), e51.
Beyer, W. R., Popplau, D., Garten, W., von Laer, D., and Lenz, O. (2003). Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. J Virol 77(5), 2866–72.
Borden, K. L., Campbell Dwyer, E. J., and Salvato, M. S. (1998). An arenavirus RING (zinc-binding) protein binds the oncoprotein promyelocyte leukemia protein (PML) and relocates PML nuclear bodies to the cytoplasm. J Virol 72(1), 758–66.
Borrow, P., and Oldstone, M. B. (1994). Mechanism of lymphocytic choriomeningitis virus entry into cells. Virology 198(1), 1–9.
Bowen, M. D., Rollin, P. E., Ksiazek, T. G., Hustad, H. L., Bausch, D. G., Demby, A. H., Bajani, M. D., Peters, C. J., and Nichol, S. T. (2000). Genetic diversity among Lassa virus strains. J Virol 74(15), 6992–7004.
Buchmeier, M. J., Bowen, M. D., and Peters, C. J. (2001). Arenaviridae: the virus and their replication. 4th ed. In Fields Virology (D. M. Knipe, and P. M. Howley, Eds.), Vol. 2, pp. 1635–1668. Lippincott Williams & Wilkins, Philadelphia.
Buchmeier, M. J., Peters, C. J., and de la Torre, J. C. (2007). Arenaviridae: the viruses and their replication. 5th ed. In Fields Virology (D. M. Knipe, and P. M. Holey, Eds.), Vol. 2, pp. 1792–1827. Lippincott Williams & Wilkins, Philadelphia.
Campbell Dwyer, E. J., Lai, H., MacDonald, R. C., Salvato, M. S., and Borden, K. L. (2000). The lymphocytic choriomeningitis virus RING protein Z associates with eukaryotic initiation factor 4E and selectively represses translation in a RING-dependent manner. J Virol 74(7), 3293–300.
Capul, A. A., Perez, M., Burke, E., Kunz, S., Buchmeier, M. J., de la Torre, J.C (2007). Arenavirus Z–GP association requires Z myristoylation but not functional RING or L domains. J Virol 81(17), 9451–9460.
Conzelmann, K. K. (2004). Reverse genetics of mononegavirales. In Curr. Top. Microbiol. Immunol. (Y. Kawaoka, Ed.), Vol. 283, pp. 1–41.
Cornu, T. I., and de la Torre, J. C. (2001). RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J Virol 75(19), 9415–26.
Cornu, T. I., and de la Torre, J. C. (2002). Characterization of the arenavirus RING finger Z protein regions required for Z-mediated inhibition of viral RNA synthesis. J Virol 76(13), 6678–88.
Cornu, T. I., Feldmann, H., and de la Torre, J. C. (2004). Cells expressing the RING finger Z protein are resistant to arenavirus infection. J Virol 78(6), 2979–83.
Damonte, E. B., Mersich, S. E., and Coto, C. E. (1983). Response of cells persistently infected with arenaviruses to superinfection with homotypic and heterotypic viruses. Virology 129(2), 474–8.
de la Torre, J. C., and Oldstone, M. B. A. (1996). The anatomy of viral persistence: mechanisms of persistence and associated disease. Adv Virus Res 46, 311–43.
Di Simone, C., and Buchmeier, M. J. (1995). Kinetics and pH dependence of acid-induced structural changes in the lymphocytic choriomeningitis virus glycoprotein complex. Virology 209(1), 3–9.
Di Simone, C., Zandonatti, M. A., and Buchmeier, M. J. (1994). Acidic pH triggers LCMV membrane fusion activity and conformational change in the glycoprotein spike. Virology 198(2), 455–65.
Eichler, R., Lenz, O., Strecker, T., Eickmann, M., Klenk, H. D., and Garten, W. (2003a). Identification of Lassa virus glycoprotein signal peptide as a trans-acting maturation factor. EMBO Rep 4(11), 1084–8.
Eichler, R., Lenz, O., Strecker, T., Eickmann, M., Klenk, H. D., and Garten, W. (2004). Lassa virus glycoprotein signal peptide displays a novel topology with an extended endoplasmic reticulum luminal region. J Biol Chem 279(13), 12293–9.
Eichler, R., Lenz, O., Strecker, T., and Garten, W. (2003b). Signal peptide of Lassa virus glycoprotein GP-C exhibits an unusual length. FEBS Lett 538(1–3), 203–6.
Fearns, R., Peeples, M. E., and Collins, P. L. (1997). Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology 236(1), 188–201.
Fischer, S. A., Graham, M. B., Kuehnert, M. J., Kotton, C. N., Srinivasan, A., Marty, F. M., Comer, J. A., Guarner, J., Paddock, C. D., DeMeo, D. L., Shieh, W. J., Erickson, B. R., Bandy, U., DeMaria, A., Jr., Davis, J. P., Delmonico, F. L., Pavlin, B., Likos, A., Vincent, M. J., Sealy, T. K., Goldsmith, C. S., Jernigan, D. B., Rollin, P. E., Packard, M. M., Patel, M., Rowland, C., Helfand, R. F., Nichol, S. T., Fishman, J. A., Ksiazek, T., and Zaki, S. R. (2006). Transmission of lymphocytic choriomeningitis virus by organ transplantation. N Engl J Med 354(21), 2235–49.
Flatz, L., Bergthaler, A., de la Torre, J. C., and Pinschewer, D. D. (2006). Recovery of an arenavirus entirely from RNA polymerase I/II-driven cDNA. Proc Natl Acad Sci USA 103(12), 4663–8.
Freed, E. O. (2002). Viral late domains. J Virol 76(10), 4679–87.
Freedman, D. O., and Woodall, J. (1999). Emerging infectious diseases and risk to the traveler. Med Clin North Am 83(4), 865–83.
Froeschke, M., Basler, M., Groettrup, M., and Dobberstein, B. (2003). Long-lived signal peptide of lymphocytic choriomeningitis virus glycoprotein pGP-C. J Biol Chem 278(43), 41914–20.
Fulhorst, C. F., Bowen, M. D., Salas, R. A., Duno, G., Utrera, A., Ksiazek, T. G., De Manzione, N. M., De Miller, E., Vasquez, C., Peters, C. J., and Tesh, R. B. (1999). Natural rodent host associations of Guanarito and Pirital viruses (Family Arenaviridae) in central Venezuela. Am J Trop Med Hyg 61(2), 325–30.
Fulhorst, C. F., Charrel, R. N., Weaver, S. C., Ksiazek, T. G., Bradley, R. D., Milazzo, M. L., Tesh, R. B., and Bowen, M. D. (2001). Geographic distribution and genetic diversity of Whitewater Arroyo virus in the southwestern United States. Emerg Infect Dis 7(3), 403–7.
Gallaher, W. R., DiSimone, C., and Buchmeier, M. J. (2001). The viral transmembrane superfamily: possible divergence of arenavirus and filovirus glycoproteins from a common RNA virus ancestor. BMC Microbiol 1(1), 1.
Garcia, J. B., Morzunov, S. P., Levis, S., Rowe, J., Calderon, G., Enria, D., Sabattini, M., Buchmeier, M. J., Bowen, M. D., and St Jeor, S. C. (2000). Genetic diversity of the Junin virus in Argentina: geographic and temporal patterns. Virology 272(1), 127–36.
Garcin, D., and Kolakofsky, D. (1990). A novel mechanism for the initiation of Tacaribe arenavirus genome replication. J Virol 64(12), 6196–203.
Garcin, D., and Kolakofsky, D. (1992). Tacaribe arenavirus RNA synthesis in vitro is primer dependent and suggests an unusual model for the initiation of genome replication. J Virol 66(3), 1370–6.
Geisbert, T. W., and Jahrling, P. B. (2004). Exotic emerging viral diseases: progress and challenges. Nat Med 10(12 Suppl), S110–21.
Geisbert, T. W., Jones, S., Fritz, E. A., Shurtleff, A. C., Geisbert, J. B., Liebscher, R., Grolla, A., Stroher, U., Fernando, L., Daddario, K. M., Guttieri, M. C., Mothe, B. R., Larsen, T., Hensley, L. E., Jahrling, P. B., and Feldmann, H. (2005). Development of a new vaccine for the prevention of Lassa fever. PLoS Med 2(6), e183.
Hass, M., Golnitz, U., Muller, S., Becker-Ziaja, B., and Gunther, S. (2004). Replicon system for Lassa virus. J Virol 78(24), 13793–803.
Hass, M., Westerkofsky, M., Muller, S., Becker-Ziaja, B., Busch, C., and Gunther, S. (2006). Mutational analysis of the lassa virus promoter. J Virol 80(24), 12414–9.
Holmes, G. P., McCormick, J. B., Trock, S. C., Chase, R. A., Lewis, S. M., Mason, C. A., Hall, P. A., Brammer, L. S., Perez-Oronoz, G. I., McDonnell, M. K., et al. (1990). Lassa fever in the United States. Investigation of a case and new guidelines for management. N Engl J Med 323(16), 1120–3.
Isaacson, M. (2001). Viral hemorrhagic fever hazards for travelers in Africa. Clin Infect Dis 33(10), 1707–12.
Jacamo, R., Lopez, N., Wilda, M., Franze-Fernandez, M. T. (2003). Tacaribe virus Z protein interacts with the L polymerase protein to inhibit viral RNA synthesis. J Virol 77(19), 10383–93.
Jahrling, P. B., and Peters, C. J. (1992). Lymphocytic choriomeningitis virus. A neglected pathogen of man. Arch Pathol Lab Med 116(5), 486–8.
Kawaoka, Y. (2004). Biology of negative strand RNA viruses. 1st ed. In Current Topics in Microbiology and Immunology, Vol. 283. Springer-Verlag, Berlin, Heidelberg.
Kentsis, A., Dwyer, E. C., Perez, J. M., Sharma, M., Chen, A., Pan, Z. Q., and Borden, K. L. (2001). The RING domains of the promyelocytic leukemia protein PML and the arenaviral protein Z repress translation by directly inhibiting translation initiation factor eIF4E. J Mol Biol 312(4), 609–23.
Kunz, S., Borrow, P., and Oldstone, M. B. (2002). Receptor structure, binding, and cell entry of arenaviruses. Curr Top Microbiol Immunol 262, 111–37.
Kunz, S., Edelmann, K. H., de la Torre, J.-C., Gorney, R., and Oldstone, M. B. A. (2003). Mechanisms for lymphocytic choriomeningitis virus glycoprotein cleavage, transport, and incorporation into virions. Virology 314(1), 168–78.
Kunz, S., Sevilla, N., Rojek, J. M., and Oldstone, M. B. (2004). Use of alternative receptors different than alpha-dystroglycan by selected isolates of lymphocytic choriomeningitis virus. Virology 325(2), 432–45.
Lee, K. J., Novella, I. S., Teng, M. N., Oldstone, M. B., and de La Torre, J. C. (2000). NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J Virol 74(8), 3470–7.
Lee, K. J., Perez, M., Pinschewer, D. D., and de la Torre, J. C. (2002). Identification of the lymphocytic choriomeningitis virus (LCMV) proteins required to rescue LCMV RNA analogs into LCMV-like particles. J Virol 76(12), 6393–7.
Licata, J. M., Simpson-Holley, M., Wright, N. T., Han, Z., Paragas, J., and Harty, R. N. (2003). Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J Virol 77(3), 1812–9.
Lopez, N., and Franze-Fernandez, M. T. (2007). A single stem-loop structure in Tacaribe arenavirus intergenic region is essential for transcription termination but is not required for a correct initiation of transcription and replication. Virus Res 124(1–2), 237–44.
Lopez, N., Jacamo, R., and Franze-Fernandez, M. T. (2001). Transcription and RNA replication of tacaribe virus genome and antigenome analogs require N and L proteins: z protein is an inhibitor of these processes. J Virol 75(24), 12241–51.
Lukashevich, I. S., Patterson, J., Carrion, R., Moshkoff, D., Ticer, A., Zapata, J., Brasky, K., Geiger, R., Hubbard, G. B., Bryant, J., and Salvato, M. S. (2005). A live attenuated vaccine for Lassa fever made by reassortment of Lassa and Mopeia viruses. J Virol 79(22), 13934–42.
Martinez-Sobrido, L., Zuniga, E. I., Rosario, D., Garcia-Sastre, A., and de la Torre, J. C. (2006). Inhibition of the type I interferon response by the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol 80(18), 9192–9.
McCormick, J. B., and Fisher-Hoch, S. P. (2002). Lassa fever. In Arenaviruses I (M. B. Oldstone, Ed.), Vol. 262, pp. 75–110. Springer-Verlag, Berlin, Heidelberg, New York.
Merkler, D., Horvath, E., Bruck, W., Zinkernagel, R. M., Del la Torre, J. C., and Pinschewer, D. D. (2006). “Viral deja vu" elicits organ-specific immune disease independent of reactivity to self. J Clin Invest 116(5), 1254–63.
Mets, M. B., Barton, L. L., Khan, A. S., and Ksiazek, T. G. (2000). Lymphocytic choriomeningitis virus: an underdiagnosed cause of congenital chorioretinitis. Am J Ophthalmol 130(2), 209–15.
Meyer, B. J., de La Torre, J. C., and Southern, P. J. (2002). Arenaviruses: genomic RNAs, transcription, and replication. In Arenaviruses I (M. B. Oldstone, Ed.), Vol. 262, pp. 139–149. Springer-Verlag, Berlin Heidelberg.
Meyer, B. J., and Southern, P. J. (1994). Sequence heterogeneity in the termini of lymphocytic choriomeningitis virus genomic and antigenomic RNAs. J Virol 68(11), 7659–64.
Muller, G., Bruns, M., Martinez Peralta, L., and Lehmann-Grube, F. (1983). Lymphocytic choriomeningitis virus. IV. Electron microscopic investigation of the virion. Arch Virol 75(4), 229–42.
Neuman, B. W., Adair, B. D., Burns, J. W., Milligan, R. A., Buchmeier, M. J., and Yeager, M. (2005). Complementarity in the supramolecular design of arenaviruses and retroviruses revealed by electron cryomicroscopy and image analysis. J Virol 79(6), 3822–30.
Neumann, G., Whitt, M. A., and Kawaoka, Y. (2002). A decade after the generation of a negative-sense RNA virus from cloned cDNA – what have we learned? J Gen Virol 83(Pt 11), 2635–62.
Oldstone, M. B. (2002). Biology and pathogenesis of lymphocytic choriomeningitis virus infection. In Arenaviruses (M. B. Oldstone, Ed.), Vol. 263, pp. 83–118.
Perez, M., Craven, R. C., and de la Torre, J. C. (2003). The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc Natl Acad Sci USA 100(22), 12978–83.
Perez, M., and de la Torre, J. C. (2002). Characterization of the genomic promoter of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV). J Virol 77(2), 1184–94.
Perez, M., Greenwald, D. L., and de la Torre, J. C. (2004). Myristoylation of the RING finger Z protein is essential for arenavirus budding. J Virol 78(20), 11443–8.
Peters, C. J. (2002). Human infection with arenaviruses in the Americas. In Arenaviruses I (M. B. Oldstone, Ed.), Vol. 262, pp. 65–74. Springer-Verlag, Berlin Heidelberg.
Peters, C. J. (2006). Lymphocytic choriomeningitis virus – an old enemy up to new tricks. N Engl J Med 354(21), 2208–11.
Pinschewer, D. D., Perez, M., and de la Torre, J. C. (2003a). Role of the virus nucleoprotein in the regulation of lymphocytic choriomeningitis virus transcription and RNA replication. J Virol 77(6), 3882–7.
Pinschewer, D. D., Perez, M., and de la Torre, J. C. (2005). Dual role of the lymphocytic choriomeningitis virus intergenic region in transcription termination and virus propagation. J Virol 79(7), 4519–26.
Pinschewer, D. D., Perez, M., Jeetendra, E., Bachi, T., Horvath, E., Hengartner, H., Whitt, M. A., de la Torre, J. C., and Zinkernagel, R. M. (2004). Kinetics of protective antibodies are determined by the viral surface antigen. J Clin Invest 114(7), 988–93.
Pinschewer, D. D., Perez, M., Sanchez, A. B., and de la Torre, J. C. (2003b). Recombinant lymphocytic choriomeningitis virus expressing vesicular stomatitis virus glycoprotein. Proc Natl Acad Sci USA 100(13), 7895–900.
Radoshitzky, S. R., Abraham, J., Spiropoulou, C. F., Kuhn, J. H., Nguyen, D., Li, W., Nagel, J., Schmidt, P. J., Nunberg, J. H., Andrews, N. C., Farzan, M., and Choe, H. (2007). Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 446(7131), 92–6.
Raju, R., Raju, L., Hacker, D., Garcin, D., Compans, R., and Kolakofsky, D. (1990). Nontemplated bases at the 5′ ends of Tacaribe virus mRNAs. Virology 174(1), 53–9.
Salvato, M. S. (1993a). The Arenaviridae. 1st ed. In The Viruses (F.-C. H. a. W. R. R., Ed.) Plenum Press, New York.
Salvato, M. S. (1993b). Molecular biology of the prototype arenavirus, lymphocytic choriomeningitis virus. In The Arenaviridae (M. S. Salvato, Ed.), Vol. 1, pp. 133–56. Plenum, New York.
Salvato, M. S., Schweighofer, K. J., Burns, J., and Shimomaye, E. M. (1992). Biochemical and immunological evidence that the 11 kDa zinc-binding protein of lymphocytic choriomeningitis virus is a structural component of the virus. Virus Res 22(3), 185–98.
Salvato, M., Shimomaye, E., and Oldstone, M. B. (1989). The primary structure of the lymphocytic choriomeningitis virus L gene encodes a putative RNA polymerase. Virology 169(2), 377–84.
Sanchez, A. B., and de la Torre, J. C. (2005). Genetic and biochemical evidence for an oligomeric structure of the functional L polymerase of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol 79(11), 7262–8.
Sanchez, A. B., and de la Torre, J. C. (2006). Rescue of the prototypic Arenavirus LCMV entirely from plasmid. Virology 350(2), 370–80.
Sevilla, N., Domingo, E., and de la Torre, J. C. (2002). Contribution of LCMV towards deciphering biology of quasispecies in vivo. Curr Top Microbiol Immunol 263, 197–220.
Smallwood, S., Cevik, B., and Moyer, S. A. (2002). Intragenic complementation and oligomerization of the L subunit of the sendai virus RNA polymerase. Virology 304(2), 235–45.
Smallwood, S., and Moyer, S. A. (2004). The L polymerase protein of parainfluenza virus 3 forms an oligomer and can interact with the heterologous Sendai virus L, P and C proteins. Virology 318(1), 439–50.
Southern, P. J. (1996). Arenaviridae: the viruses and their replication. 3rd ed. Trans. n/a. In Fields Virology (D. M. K. Bernard N. Fields, and Peter M. Howley, Eds.), Vol. 2, pp. 1505–51. Lippincott-Raven Publishers, Philadelphia.
Spiropoulou, C. F., Kunz, S., Rollin, P. E., Campbell, K. P., and Oldstone, M. B. (2002). New World arenavirus clade C, but not clade A and B viruses, utilizes alpha-dystroglycan as its major receptor. J Virol 76(10), 5140–6.
Strecker, T., Eichler, R., Meulen, J., Weissenhorn, W., Dieter Klenk, H., Garten, W., and Lenz, O. (2003). Lassa virus Z protein is a matrix protein and sufficient for the release of virus-like particles [corrected]. J Virol 77(19), 10700–5.
Strecker, T., Maisa, A., Daffis, S., Eichler, R., Lenz, O., and Garten, W. (2006). The role of myristoylation in the membrane association of the Lassa virus matrix protein Z. Virol J 3, 93.
Tordo, N., De Haan, P., Goldbach, R., and Poch, O. (1992). Evolution of negative-stranded RNA genomes. Semin Virol 3, 341–57.
Tortorici, M. A., Albarino, C. G., Posik, D. M., Ghiringhelli, P. D., Lozano, M. E., Rivera Pomar, R., and Romanowski, V. (2001). Arenavirus nucleocapsid protein displays a transcriptional antitermination activity in vivo. Virus Res 73(1), 41–55.
Urata, S., Noda, T., Kawaoka, Y., Yokosawa, H., and Yasuda, J. (2006). Cellular factors required for Lassa virus budding. J Virol 80(8), 4191–5.
Vieth, S., Torda, A. E., Asper, M., Schmitz, H., and Gunther, S. (2004). Sequence analysis of L RNA of Lassa virus. Virology 318(1), 153–68.
Weaver, S. C., Salas, R. A., de Manzione, N., Fulhorst, C. F., Duno, G., Utrera, A., Mills, J. N., Ksiazek, T. G., Tovar, D., and Tesh, R. B. (2000). Guanarito virus (Arenaviridae) isolates from endemic and outlying localities in Venezuela: sequence comparisons among and within strains isolated from Venezuelan hemorrhagic fever patients and rodents. Virology 266(1), 189–95.
Weaver, S. C., Salas, R. A., de Manzione, N., Fulhorst, C. F., Travasos da Rosa, A. P., Duno, G., Utrera, A., Mills, J. N., Ksiazek, T. G., Tovar, D., Guzman, H., Kang, W., and Tesh, R. B. (2001). Extreme genetic diversity among Pirital virus (Arenaviridae) isolates from western Venezuela. Virology 285(1), 110–8.
Welsh, R. M., and Pfau, C. J. (1972). Determinants of lymphocytic choriomeningitis interference. J Gen Virol 14(2), 177–87.
Yarnell, W. S., and Roberts, J. W. (1999). Mechanism of intrinsic transcription termination and antitermination. Science 284(5414), 611–5.
York, J., Romanowski, V., Lu, M., and Nunberg, J. H. (2004). The signal peptide of the Junin arenavirus envelope glycoprotein is myristoylated and forms an essential subunit of the mature G1–G2 complex. J Virol 78(19), 10783–92.
Young, P. R., and Howard, C. R. (1983). Fine structure analysis of Pichinde virus nucleocapsids. J Gen Virol 64(Pt 4), 833–42.
Zinkernagel, R. M. (2002). Lymphocytic choriomeningitis virus and immunology. Curr Top Microbiol Immunol 263, 1–5.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2009 Springer Science+Business Media, LLC
About this chapter
Cite this chapter
de la Torre, J.C. (2009). Arenaviruses: Genome Replication Strategies. In: Raney, K., Gotte, M., Cameron, C. (eds) Viral Genome Replication. Springer, Boston, MA. https://doi.org/10.1007/b135974_10
Download citation
DOI: https://doi.org/10.1007/b135974_10
Published:
Publisher Name: Springer, Boston, MA
Print ISBN: 978-0-387-89425-6
Online ISBN: 978-0-387-89456-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)