Abstract
Percutaneous coronary intervention (PCI) is a vital treatment method for coronary artery disease, but the unstructured nature of its clinical data makes it challenging to utilize directly. The data for this study was obtained from the Cardiovascular Treatment Center of the People’s Hospital of Liaoning Province, China. A representative dataset of 5.8% of PCI patients’ surgical records was selected for labeling, and a language model-based PCI surgical information entity recognition model was developed. First, Encoder Representations from Transformers (BERT) was employed to express the semantic relationship between characters accurately. Then, BiLSTM was used as a feature extractor to extract contextual relations, and finally, conditional random field (CRF) was applied to optimize the prediction results. Experimental results demonstrated that the F1 score in the PCI surgical information entity recognition model reached 85.49%, which is 25.66% higher than the traditional HMM and 0.94% higher than BiLSTM in deep learning.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Percutaneous Coronary Intervention (PCI) is a cardiology procedure that involves balloon dilation or stent implantation to alleviate symptoms of coronary artery stenosis or occlusion. Although PCI operation information records the complete PCI process, clinical texts related to PCI are unstructured, which makes it difficult for clinicians to effectively utilize the information. Therefore, the use of artificial intelligence to process PCI information in a structured manner is necessary to uncover the full potential value of PCI operation information.
In 1996, the term Named Entity Recognition (NER) was introduced as a fundamental task of Natural Language Processing (NLP) at MUC-6 [1]. NER has demonstrated excellent performance in medical data mining. Early approaches relied on rule-based and dictionary-based methods [2] that used templates based on contextual semantic structures. However, these methods could not effectively summarize difficult-to-extract information, and were relatively expensive. To address these limitations, scholars have applied machine learning methods [3,4,5] such as Hidden Markov Models (HMM), which outperformed traditional methods but could not effectively use contextual semantics for named entity recognition. Currently, deep learning-based NER methods [6, 7] have become more prevalent than the previous two methods. The most popular method is the Bidirectional Long Short-Term Memory (BiLSTM) method. However, BiLSTM cannot constrain the relationship between predicted labels. The Conditional Random Field (CRF) can better constrain the relationship between labels through the emission probability matrix and transition probability matrix. For example, Li et al. [8] proposed an LSTM-CRF-based named entity recognition method.
The intersection of computer science and medicine has led to the emergence of Clinical Named Entity Recognition (CNER) as an important research field. CNER has been successfully utilized to identify body parts, diseases, drugs, and more in various medical fields [9]. However, its specific application in PCI clinical texts remains largely unexplored. The structure of PCI clinical information is complex, involving a combination of Chinese, English, and symbols. Clinical records in this field are relatively incomplete and disorganized, with a high degree of grammatical errors and context ambiguity. These factors pose significant challenges for the identification of named entities related to PCI clinical and surgical information within China’s electronic medical records.
2 Methods
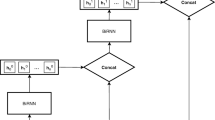
The BERT-BiLSTM-CRF model’s structure diagram is presented in Fig. 1. This model consists of an input layer, an LSTM layer, a Linear layer, and a CRF layer. The input layer is responsible for converting the input corpus into character vector embedding matrices to facilitate subsequent global feature extraction using Bi-LSTM. The LSTM layer is tasked with extracting the global features and contextual semantic relationships of the time series. The Linear layer functions as a classifier to assign each entity a probability matrix based on the number of entities. The CRF layer then employs the probability matrix to constrain the relationship between the labels and determine the most probable label sequence.
2.1 BERT
In 2018, Devlin et al. [10] introduced BERT, a pre-trained language model based on the Transformer architecture. Unlike traditional language models, BERT employs a 12-layer Transformer Encoder for learning, with each Encoder consisting of a multi-head attention mechanism and a feedforward network. The multi-head attention mechanism calculates the relationship between words using Query, Key, and Value, and adjusts the weight to extract essential features from the text. Compared to previous pre-training models, BERT captures contextual information more accurately and learns the relationship between consecutive text fragments.
Traditional embedding methods, such as Word2vec [11], Glove [12], and FastText [13], represent all possible word-level meanings in vector form. However, the resulting embeddings are often limited in their ability to express the semantic and distance relationships between words. Additionally, in PCI operation information, there is no clear boundary between Chinese, English, and symbols, which can lead to poor recognition performance. To address these issues, this paper proposes a model that utilizes BERT as an embedding method through transfer learning. Unlike traditional methods, BERT is capable of capturing contextual information and understanding relationships between consecutive text fragments, which can improve performance in recognizing PCI operation information.
2.2 LSTM
The LSTM layer’s individual unit receives the output from the Input layer and extracts the sequence information of the text to learn the contextual features of the corpus. It can consider the semantic relationship between each character before and after the sentence simultaneously and combine them to create a more comprehensive representation. The LSTM layer takes the WordEmbedding of the corpus as an input sequence and produces another output sequence vector \(\text {h} = (\text {h}_1, \text {h}_2, \ldots , \text {h}_{\text {n}})\) that represents the sequence at each time step in the input sequence. LSTM addresses the challenges of long sentence dependencies and gradient explosion in RNNs by incorporating three gate control units that regulate the retention and forgetfulness of specific information. The computation formula for the LSTM hidden layer output representation at a given input time \(\text {X}_{\text {t}}\) is expressed as Formula (1)–(5).
In the equation, W represents the weight matrix and b is the offset vector. C represents the state of the memory unit, \(\sigma \) is the sigmoid activation function, and tanh is another activation function. The input gate, forget gate, and output gate are represented by \(\text {i}_{\text {t}}\), \(\text {f}_{\text {t}}\), and \(\text {o}_{\text {t}}\), respectively. The hidden state output, \(\text {H}_{\text {t}}\), includes the text information in the clinical information. The gate’s frequency threshold mechanism is capable of effectively filtering out irrelevant information and retaining important information that needs to be preserved.
However, unidirectional LSTM can only capture information in a forward direction. As sentences become longer, local features at the beginning of the sentence make up a smaller proportion. Therefore, it is necessary to use a Bidirectional LSTM to capture information both forward and backward, and concatenate the outputs. This enhances the information captured from both the beginning and end of the sentence and improves the ability to capture semantic dependencies within the context.
2.3 CRF
While BiLSTM is effective at extracting semantic features from the entire text, it does not impose constraints on the label relationships. During the entity output prediction stage, the softmax function is typically used as a classifier to address multi-classification problems, resulting in incorrect outcomes and affecting model performance. To address this issue, this paper employs the CRF model in decoding.
The Conditional Random Field (CRF) is a type of undirected probabilistic graphical model that can constrain the relationship between labels and improve the accuracy of entity predictions. By defining a starting probability matrix and a transition probability matrix, the constraints between tags and the prediction of entities can be enforced. Specifically, because the entity labels starting with “B” must be followed by labels of the same type starting with “I”, the launch probability matrix and transition probability matrix effectively model these constraints and reduce errors. Given an input sequence \(\text {H}=\text {h}_1,\text {h}_2,\text {h}_3,\ldots ,\text {h}_{\text {n}}\) and the corresponding output from the LSTM layer, the CRF model calculates the conditional probability distribution of the output sequence \(\text {Y} =\text {y}_1,\text {y}_2,\text {y}_3,\ldots ,\text {y}_{\text {n}}\) and assigns a score to each label. The label with the highest score is then selected as the final prediction label using the calculation formula (6):
When decoding, the Viterbi algorithm [14] is used to obtain the optimal output sequence \(\text {y}_{\text {R}}\). This algorithm computes the score of each label in the sequence based on the transition matrix T and the score vector P. The optimal path is determined by selecting the label with the highest score for each position in the sequence, taking into account the constraints between labels. The calculation formula for the Viterbi algorithm is given by Eq. (7):
3 Experiments and Results
3.1 Dataset
The clinical data utilized in this research was sourced from the People’s Hospital of Liaoning Province (Shenyang, China), a prominent and comprehensive third-class hospital in China. The clinical data relates to PCI (percutaneous coronary intervention) procedures involving coronary angiography, balloon dilatation, and stent implantation. Specifically, the clinical texts pertaining to three distinct categories of fine-grained PCI surgical information were extracted. By conducting a comprehensive analysis of various clinical and surgical records, this study chose to use data from 1340 inpatients who were admitted to the Cardiology Department as the corpus for analysis. This study was finally approved by the Ethics Committee of the People’s Hospital of Liaoning Province (Ethics number: (2023)K021).
For the experimental study, a corpus of 1340 cases of PCI surgery information was utilized. From this, 930 cases were selected as the training set, while 180 cases were assigned as the validation set. Following model training, the remaining 230 cases were utilized as the test set. Table 1 displays the distribution of the eight types of entities in the training, validation, and test sets.
3.2 Result Analysis
Table 2 shows that traditional HMM achieved excellent results due to the simpler format of anticoagulants than the normative structure, but it performed poorly in identifying entities that require contextual semantics. Bi-LSTM showed the best performance in Chinese and English entity recognition, but it did not perform well in identifying complex entities such as contrast results. However, after adding CRF, the relationship between labels was effectively constrained, resulting in significant improvements in angiography results and entities with complex stent signal structures. This greatly reduced false recognition. Additionally, with the addition of BERT, sensitivity to entities such as the combination of numbers and symbols, such as the bracket model, was increased, and the accuracy of other entities was improved.
The overall performance of a model can be evaluated by its ability to recognize all types of entities correctly. The F1 score for each model is calculated based on the proportion of each entity in the PCI operation information. The results are presented in Table 3.
To ensure the rigor of the experiment, the overall performance of each model is evaluated under both strict and loose standards. As shown in the table, the traditional HMM performed poorly, with an F1 score of only 59.83 under the strict standard, due to its limited ability to extract features and combine contextual semantics, resulting in a large number of recognition errors and limitations in identifying entities. Although BiLSTM achieved higher performance than HMM, its performance on complex imaging entities was not satisfactory, with incorrect label order leading to a reduced F1 score. The addition of CRF effectively constrained label relationships and improved accuracy for some complex entities, such as those with mixed numbers and symbols, resulting in an overall F1 score improvement of 0.35. In comparison, BERT-BiLSTM-CRF performed better, as the WordEmbedding of the former two was randomly initialized and could not accurately reflect the relative distance of the word vector space in character information representation. This improved accuracy in the recognition of most entities.
4 Discussion and Conclusion
This study focuses on the extraction of clinical information from PCI surgery information based on Chinese EMRs. Determine and identify eight entities including angiography result, catheter type, guidewire brand, stent model, stent brand, balloon model, contrast agent, and anticoagulant from three clinical surgical records. The performance of the BERT-BiLSTM-CRF model for extracting clinical PCI operation information has been further improved, and the accuracy rate basically meets the needs of clinical applications. The results demonstrate that deep learning methods can be used to automatically extract PCI surgical information from EMRs for clinical named entity recognition.
References
Grishman R, Sundheim BM (1996) Message understanding conference-6: a brief history. In: COLING 1996: the 16th international conference on computational linguistics, vol 1
Song M, Yu H, Han WS (2015) Developing a hybrid dictionary-based bio-entity recognition technique. BMC Med Inf Decis Making 15(1):1–8
McCallum A, Li W (2003) Early results for named entity recognition with conditional random fields, feature induction and web-enhanced lexicons
Chieu HL, Ng HT (2002) Named entity recognition: a maximum entropy approach using global information. In: COLING 2002: the 19th international conference on computational linguistics
Bender O, Och FJ, Ney H (2003) Maximum entropy models for named entity recognition. In: Proceedings of the seventh conference on natural language learning at HLT-NAACL 2003, pp 148–151
Wang X, Zhang Y, Ren X, Zhang Y, Zitnik M, Shang J, Langlotz C, Han J (2019) Cross-type biomedical named entity recognition with deep multi-task learning. Bioinformatics 35(10):1745–1752
Cai X, Dong S, Hu J (2019) A deep learning model incorporating part of speech and self-matching attention for named entity recognition of chinese electronic medical records. BMC Med Inf Decis Making 19(2):101–109
Li L, Jiang Y (2018) Integrating language model and reading control gate in BLSTM-CRF for biomedical named entity recognition. IEEE/ACM Trans Comput Biol Bioinform 17(3):841–846
Chen X, Ouyang C, Liu Y, Bu Y (2020) Improving the named entity recognition of Chinese electronic medical records by combining domain dictionary and rules. Int J Environ Res Publ Health 17(8):2687
Devlin J, Chang MW, Lee K, Toutanova K (2018) BERT: pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805
Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J (2013) Distributed representations of words and phrases and their compositionality. Adv Neural Inf Process Syst 26
Pennington J, Socher R, Manning CD (2014) Glove: global vectors for word representation. In: Proceedings of the 2014 conference on empirical methods in natural language processing (EMNLP), pp 1532–1543
Bojanowski P, Grave E, Joulin A, Mikolov T (2017) Enriching word vectors with subword information. Trans Assoc Comput Linguist 5:135–146
Viterbi AJ (2006) A personal history of the Viterbi algorithm. IEEE Sig Process Mag 23(4):120–142
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Zheng, Y., Wang, L., Li, F., Xu, H., Ge, J. (2024). Named Entity Recognition of PCI Surgery Information Based on BERT+BiLSTM+CRF. In: Wang, W., Liu, X., Na, Z., Zhang, B. (eds) Communications, Signal Processing, and Systems. CSPS 2023. Lecture Notes in Electrical Engineering, vol 1032. Springer, Singapore. https://doi.org/10.1007/978-981-99-7505-1_11
Download citation
DOI: https://doi.org/10.1007/978-981-99-7505-1_11
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-99-7539-6
Online ISBN: 978-981-99-7505-1
eBook Packages: EngineeringEngineering (R0)