Abstract
Pearl millet [Cenchrus americanus (L.) Morrone; also known as Pennisetum glaucum], originated 4900 years ago, is a C4 crop with high photosynthetic efficiency and fulfills the food and fodder needs of resource-poor farmers of sub-Saharan Africa, Southeast Asia, and the Indian subcontinent. Pearl millet is a climate-ready crop and grows well in poor and low-fertility soil. It is profoundly nutritious, fiber-rich, and non-glutinous. Based on transcriptome and bioinformatics studies, it is estimated that 1.79 Gb of the pearl millet genome consists of 38,579 genes. Unfortunately, functional genomics and genotype-phenotype association in pearl millet are poorly explored areas. Pearl millet suffers from low yield for many reasons. There is an urgent need to validate the functions of important genes to improve the crop and better utilize it for future agriculture in a changing climate scenario. In recent years, genome editing, especially CRISPR-Cas, has come under the spotlight for improving crop varieties. In this chapter, we discuss how the available genome editing tools can play a significant role in deciphering the functions of pearl millet genomic regions and crop improvement. We also highlight major bottlenecks to using genome editing in pearl millet and discuss possible ways to overcome those constraints.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
7.1 Introduction
Global shifts in climate and population increase are the two significant issues of concern in the present scenario. Estimates indicate that the human population will reach approximately 10 billion by 2050, which has a counter effect on food security (FAO 2017). Climate change could be a serious threat to global food security. About one-third of the total greenhouse gas emissions is contributed by our food system. We have been focusing heavily on a few cereal crops (wheat, rice, and maize) as sources of macro- and micronutrients, ignoring their potential for global warming. For example, wheat has the highest CO2 emission capacity (4 tons CO2eq/ha), followed by rice and maize (3.4 tons CO2 eq/ha) (Jain et al. 2016). Compared to these cereals, there are orphan crops, like millets and sorghum, with lower carbon emission rates (Prasad and Staggenborg 2009). Therefore, focusing on improving orphan crop varieties could help ensure food security in the changing climate. Among these orphan crops, pearl millet is a diploid (2n = 2x = 14) C4 grass species having high photosynthetic activity and biomass production ability. It can grow on poor sandy soils. It is well suited for growth in unbearable conditions such as high soil pH, high temperature, low moisture, high salinity, and low rainfall compared with sorghum and maize (Vadez et al. 2012; Nambiar et al. 2011).
Millet grains are highly nutritious, low in starch, and rich in fiber (Nambiar et al. 2011; Kumar et al. 2016). Pearl millet has a broad range of phenotypic and genotypic variations because the cultivation varies in different agro-climatic conditions. Despite all the above advantages, only a limited proportion of its germplasm could be utilized for improving the pearl millet crop for different important traits. Pearl millet suffers from a low crop productivity rate with an average grain yield of just 900 kg/ha, the presence of anti-nutritional factors in the grains, and the vulnerability to several foliar diseases like downy mildew, blast, and smut. Although pearl millet grains are gluten free and could be a recommended diet for people with celiac disease, it is not popular due to their rapid rancidity upon storage.
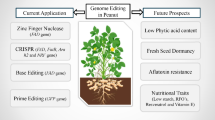
Historically, crop plants have been improved by harnessing genetic diversity. Genetic diversity is an effect of mutations found in various regions of the genome, which can be natural or artificially induced. Natural mutations are extremely slow. To create rapid and useful genetic variations, researchers use four major techniques: conventional breeding through crossing, mutation breeding, transgenesis, and genome editing (Chen et al. 2019). Conventional breeding or cross-breeding is a targeted crossing between two plants via sexual recombination. Major restrictions of this technique are—it is time-consuming, only applicable for sexually compatible plants, the occurrence of linkage drag, and low genetic variability. Later, mutation breeding came to the forefront, relying on chemical and radiation treatment (Holme et al. 2019). This technique generates random mutations, necessitates extensive screening, and demands intensive labor. Subsequently, transgenic breeding technology empowered us to introduce beneficial genes from virtually any organism to crop plants for desired traits. While it has huge potential for crop improvement, developing crop varieties through transgenesis is expensive and faces stringent regulatory hurdles in many countries. For example, golden rice (vitamin A enriched transgenic rice), developed nearly two decades ago to combat malnutrition in the developing world, received approval for open cultivation in the Philippines in July 2021 (https://www.irri.org). Moreover, the integration site of the transgene in the genome is not controllable. Most recently, genome editing technology has emerged as the latest breeding technique, allowing for precise, targeted, and predictable mutations in crop varieties. Genome editing has been rapidly adopted across plant species to generate valuable genetic variations (Gao 2021; Molla et al. 2021). In this chapter, we will discuss the fundamentals of genome editing technology, advanced genome editing tools, a few noteworthy examples of crop improvement through genome editing, and the potential applications of genome editing for enhancing traits in orphan crops like pearl millet.
7.2 Basics of Genome Editing
Genome editing represents a significant advancement over conventional breeding, as it allows us to modify genetic regions of interest more precisely and rapidly in a targeted fashion. The fundamental of genome editing is to create a targeted double-strand break (DSB) in a desired genomic location. DSBs are created with different sequence-specific nucleases (SSN). For example, ZFNs (zinc finger nucleases) (Kim et al. 1996), TALENs (transcription activator-like effector nucleases) (Christian et al. 2010), and CRISPR-Cas (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein) (Jinek et al. 2012) are well studied programmable SSN systems for recognizing and modifying specific DNA sequences. ZFNs and TALENs systems rely on protein-based DNA recognition, whereas CRISPR-Cas depends on RNA-guided DNA recognition. Therefore, for targeting each new DNA sequence by ZFNs and TALENs, new proteins need to be engineered. However, new DNA sequences could be targeted by CRISPR-Cas by manipulating a small RNA sequence kmown as guide RNA (gRNA). This RNA-dependent DNA targeting platform gained rapid popularity for manipulating the genome of diverse organisms, including plants. The creation of a DSB by SSN triggers cellular pathways to repair the damaged DNA. There are two methods for repairing DSBs: one is more common but error-prone, known as non-homologous end joining (NHEJ), and the other is less frequent but errorless, called homology-directed repair (HDR) (Molla and Yang 2020). Besides canonical NHEJ and HDR, another type of error-prone repair system is called microhomology-mediated end joining (MMEJ). Out of these repair systems, NHEJ and MMEJ lead to the creation of indels (insertions and deletions) at the DSB sites of the target gene, potentially resulting in frameshift mutations and enabling effective gene knockout. In summary, genome editing through CRISPR-Cas methods generates indels in the genome, leading to genetic variation in organisms.
CRISPR-Cas9 genome editing tool is a two-component system: a customizable small guide RNA (gRNA) and a Cas9 nuclease. The 20-nucleotide sequence present at the 5′ end of the guide RNA is crucial for site-specific DNA recognition and subsequent cleavage by Cas9. Therefore, for each new target DNA, only the 20-nucleotide sequence of the guide RNA needs to be changed, while the other components remain the same. Although single guide RNA (gRNA)-based CRISPR is a common tool for targeting a single gene, multiple genes can be edited in a single experiment by including an array of guide RNAs with Cas9. Multiplex editing is a powerful technology for large-scale genome engineering, parallel targeting of gene family members, and metabolic engineering. A schematic diagram of the steps involved in plant genome editing through CRISPR-Cas9 is presented in Fig. 7.1. Available genome editing tools are listed in the Table 7.1.
Steps for performing CRISPR-Cas-based genome editing in plants. (a, b) Target selection and guide RNA design. After the target genes or genetic region is selected, the sequence is retrieved from the public database. The sequence is used in web tools for protospacer design (the 20 bp at the 5′ end of guide RNA). Web tools facilitate predicting suitable guides with high on-target efficiency and low off-target efficiency. (c, d) CRISPR-reagent (plasmid construct or RNPs) are delivered to explant by biolistic, Agrobacterium, or PEG-mediated transfection. For RNP, in-vitro transcribed guide RNA is complexed with Cas9 protein before transforming into plant cells. PEG-mediated transfection is done with isolated protoplast. (e–g) Selection and regeneration of the transformed calli. (h, i) Genomic DNA isolation and amplification of the targeted region from putative edited plants (T0). (j) Screening of the putative edited plants by restriction fragment length polymorphism (RFLP) and/or Sanger sequencing. (k, l) Generation advancement to obtain homozygous lines
7.3 Precise Editing with CRISPR-Cas9
CRISPR-Cas technique efficiently generates random insertions or deletions, which cannot be controlled. Although the mutation outcome can be predicted (Molla and Yang 2020), conventional CRISPR-Cas tools are unsuitable for generating precise sequence modifications. Base editing and prime editing, the two advanced CRISPR-Cas-based approaches, can create precise mutations in DNA without generating double-stranded breaks (DSBs) (Molla et al. 2021). Most of the agronomic characteristics are determined by single nucleotide polymorphisms (SNPs) or combinations of a few SNPs and indels. Introducing specific SNPs from wild to elite varieties through conventional breeding techniques is an extensive and time-consuming procedure; base editing and prime editing could accelerate this process.
7.3.1 Base Editing
Base editors were developed for installing single nucleotide changes in target loci. Recently, three types of DNA base editors have been developed, viz., cytosine base editors (CBEs), adenine base editors (ABEs), and C to G base editors (CGBEs). CBEs create C to T transitions, while ABEs create A to G transitions. CBE, the first among the base editors, contains four different components: a cytosine deaminase (which converts C to U), a modified Cas9 (nCas9 or Cas9 nickase D10A), UGI (uracil DNA glycosylase inhibitor), and a guide RNA (Komor et al. 2016; Nishida et al. 2016; Molla and Yang 2019). It induces the creation of C to T transition (G to A in the complementary strand) in the genome. ABEs are made up of nCas9 (D10A) fused with protein-engineered deoxyadenosine deaminase and can convert targeted A to G (T to C in the complementary strand) (Gaudelli et al. 2017). Similarly, recently developed platforms enable us to execute C to G conversion (G to C in the opposite strand) in the genome (Molla et al. 2020a). Diverse plant-based editing platforms have evolved and been applied for performing precise base alteration in various plant species (Molla et al. 2021).
7.3.2 Prime Editing
Although base editors opened a new path to perform single nucleotide changes (C to T, A to G, and C to G) in the genome, they are restricted to transition mutations and two types of transversion mutations. The other six types of transversion mutations (C to A, G to T, A to T, T to A, A to C, and T to G) can not be installed using the available base editors at the time of writing this chapter. A recently developed genome editing technique called prime editing enables us to install all 12 types of base conversions, small insertions, deletions, and combinations of those mutations (Anzalone et al. 2019). Prime editors consist of a nickase Cas9 (nCas9-H840A) fused with reverse transcriptase (RT) and an extended guide RNA. The extended guide RNA is called prime editing gRNA (pegRNA), which contains information to rectify the target sequence (Anzalone et al. 2019). The 5′ end of the pegRNA carries the guide region, which binds to the target site of the genome, while the 3′ extension contains the primer binding site and RT template. nCas9-H840A creates a nick in the DNA. The primer binding site binds to the 3′ end of the nicked DNA strand, and the RT template encodes the desired edit that is directly copied to the genomic DNA by the RT enzyme. Prime editing is considered a highly promising tool in the field of genome editing because it can create transversion mutations and precise insertions and deletions. Although its efficiency is currently low in plant systems (Molla et al. 2021), with technical improvements, prime editing holds great promise for plant genome engineering in the near future.
7.4 Some Spotlight Case Studies on Crop Improvement Through Genome Editing
Genome editing technologies alternatively called targeted mutagenesis enable us to modify the gene of interest rapidly and precisely The repurposing of the bacterial CRISPR-Cas9 immune system in 2012 has brought about a revolution in the field of genome engineering. The rapid advancement of CRISPR-Cas technologies, from double strand break (DSB) to base and prime editing, has opened new paths of precise plant and animal genome editing. It would not be an exaggeration to say that we are now in an era where we can modify genes precisely predictably. In 2013, several groups began applying the CRISPR-Cas system to plants (Feng et al. 2013; Shan et al. 2013; Xie and Yang 2013). Since then, we have witnessed numerous succesful examples of utilizing this tool for crop improvement. For example, broad-spectrum bacterial blight-resistant rice (Oliva et al. 2019), low-gluten wheat (Sánchez-León et al. 2018), waxy corn (Gao et al. 2020), male sterile wheat (Li et al. 2020a), clonally propagated hybrid rice (Khanday et al. 2019; Wang et al. 2019), and high yielding tomato (Rodríguez-Leal et al. 2017) have been developed with conventional CRISPR-Cas9. Recently, high oleic acid soybean in the USA and high GABA tomato in Japan have been commercialized and are available for human consumption. This is remarkable that within a few years of its development, genome editing tools have provided us with commercialized improved crop varieties.
In the last few years, base editing evolved as a promising device for performing genome editing with single base resolution. CBEs have been demonstrated to alter C to T in several crops like rice (Shimatani et al. 2017), and maize (Zong et al. 2018). Similarly, A to G conversion was executed by ABE in rice (Li et al. 2018; Molla et al. 2020b) and wheat (Li et al. 2018). Recently, C to G base conversion in rice, tomato, and poplar was achieved (Sretenovic et al. 2021). Single base editing in acetolactate synthase (ALS), acetyl-CoA carboxylase (ACCase), and TubA2 genes have been found to confer herbicide resistance in rice, wheat, Arabidopsis, and maize (Shimatani et al. 2017; Zhang et al. 2019; Liu et al. 2021; Dong et al. 2020; Li et al. 2020b). Herbicide-tolerant germplasm generated through base editing will serve greatly in effective weed management. Base editors were also used to alter the nutritional composition of fruits. For example, the sugar content in strawberries and carotenoid content in tomatoes increased with base editors (Hunziker et al. 2020; Xing et al. 2020). In a recent review, Molla et al. (2021) comprehensively portrayed the current and potential crop improvement applications of base editors and prime editors. Therefore, along with canonical CRISPR-Cas tools, base, and prime editors hold great promise for pearl millet improvement.
7.5 Harnessing Climate-Resilience and Nutritional Value of Pearl Millet by Genome Editing
The C4 crop pearl millet was domesticated approximately 4900 years ago in West Africa, and it plays a central role in ensuring food security in arid and semi-arid regions of Asia and Africa (Burgarella et al. 2018). It is well adapted to abiotic and biotic stresses; for example, some wild pearl millet varieties can thrive in extreme environmental conditions (they can withstand temperatures exceeding 42 °C) (Gupta et al. 2015). Therefore, pearl millet could serve as a fascinating biological model for ensuring future food security in a changing climate. Varshney et al. (2017) reported that the 1.79 GB pearl millet genome has a high GC content (47.9%), which is a characteristic associated with desiccation tolerance in monocots. The pearl millet genome is also enriched with cutin, suberin, and wax biosynthetic genes, which might contribute to its drought and heat tolerance (Varshney et al. 2017). Furthermore, Sharma et al. (2020) reviewed that high genetic differentiation is found between cultivated and wild Pennisetum species, revealing that only 74% of the genetic diversity found in wild species is recognized in cultivated species. Thus, domestication has led to a reduction in genetic diversity. A group of researchers suggests that the wild Pennisetum species could serve as a repository for preserving new allelic variants associated with climate-resilient and disease-resistant traits (Oumar et al. 2008; Mariac et al. 2006). Although pearl millet is a good source of nutrition, including proteins, vitamins, and minerals, major concerns about it include low grain yield, the presence of anti-nutritional factors like polyphenols and phytic acid, and the rapid rancidity of pearl millet flour. Therefore, increasing genetic diversity will pave the way to develop pearl millet varieties that combine improved nutritional values with high yields and disease resistance.
Traditional breeding has the potential to harness allelic variants from wild germplasm to incorporate beneficial traits. However, challenges such as linkage drag and crossing barriers often impede this process. Therefore, to complement traditional breeding efforts, CRISPR-Cas-based genome editing tools can be used to remove undesired characterstics by knocking out genes and to mimic beneficial alleles through precise editing within the genetic background of cultivated species (Table 7.2).
7.5.1 Pearl Millet Traits Improvement for Nutrition
Tackling the hunger of the world population amid changing climate conditions is the biggest challenge facing humanity. Thus, improvements in cereal crops like pearl millet, which not only serves as a major source of carbohydrates, proteins, fats, minerals, and vitamins but also thrives in harsh climatic conditions, could play a crucial role in addressing these challenges. Although pearl millet is a nutritionally rich crop, it has remained underutilized and is not widely popular among the general people and food processing sectors. Here, present some futuristic ideas on how CRISPR-Cas-derived genome editing tools could accelerate the genetic improvement of pearl millet.
Pearl millet is inherently nutritious and contains an appropriate amounts of proteins, dietary fiber, macro-, and micronutrients. Alarmingly, in India, 54% of children and 74% of women suffer from iron (Fe) deficiency anemia, while 54% of children below 5 years are stunted due to zinc (Zn) deficiency (WHO 2020). Pearl millet is a rich source of the principal micronutrients iron and zinc compared to other cereals like rice and wheat (Tako et al. 2015). However, the availability of these micronutrients is limited due to the presence of antinutritional factors like phytic acid, which chelate them into multivalent cations (Fe2+, Zn2+, etc.). CRISPR-Cas9-mediated disruption of the TaIPK1 gene has been shown to reduce phytic acid and enhance Iron and Zinc content in wheat grains (Ibrahim et al. 2021). Similarly, the knockout of the BnITPK gene has been found to reduce phytic acid in oilseed rape (Sashidhar et al. 2020). A similar strategy could be employed to reduce phytic acid content in pearl millet grain, thereby increasing the availability of those key nutrients.
Govindaraj et al. (2020) developed a core collection of 39 accessions of pearl millet with multiple nutrients. However, the genetic regions or genes associated with higher nutrients are largely unknown. Genetic studies aimed at identifying these genes and functional genomics studies would greatly facilitate the efficient use of this germplasm. Similarly, Mahendrakar et al. (2020) identified several putative candidate genes for grain iron and zinc content. In this regard, CRISPR-Cas9 would play a significant role in functional genomics to elucidate their exact function. Interestingly, a recent study identified single nucleotide polymorphisms (SNPs) that co-segregate with Fe and Zn (Pujar et al. 2020). Once the responsible and functional SNPs are pinpointed, these variations could be introduced into cultivars using precise editing tools like base and prime editors.
The starch composition of pearl millet can be altered using genome editing tools. For example, waxy grains of millet have nearly 0% amylose content, are recommended for infants because they are easily digestible. The waxy phenotype is produced by the recessive allele wx, which is a loss of function allele of the dominant Wx allele (Nakayama et al. 1998; Fukunaga et al. 2002). CRISPR-Cas-mediated knockout of this dominant Wx allele can create waxy grains in pearl millet.
One of the key reasons for pearl millet’s limited popularity among consumers and the food processing industry is its rapid rancidity during storage. Pearl millet flour contains a high level of lipids, which are prone to oxidation by lipoxygenase (LOX) to produce lipid hydroperoxides (Vinutha et al. 2022). Subsequent nonenzymatic oxidation of lipid hydroperoxides leads to rancidity of flours during storage. Genome editing can be used to knock out suitable candidate LOX gene(s) to increase the shelf life of pearl millet flour. However, care should be taken to avoid generating any concomitant phenotypic anomalies.
A recent study on green millet (Setaria viridis) showed the use of CRISPR-Cas9 to knock out the Less Shattering1 (SvLes1) gene, which controls seed shattering (Mamidi et al. 2020). The strategy and methodology with appropriate modifications could be beneficial for the successful application of CRISPR-Cas9 in pearl millet genotypes.
7.5.2 Pearl Millet Traits Improvement Related to Different Abiotic Stress
The orphan crop pearl millet thrives in adverse conditions such as water scarcity and high temperatures. Its high adaptability to various abiotic stresses makes it a useful crop for functional genomics studies to understand the molecular basis of stress adaptation and tolerance (Shivhare and Lata 2017). However, it is unfortunate that only a limited number of stress-related genes have been identified and validated in pearl millet to date. By combining transcriptome data and bioinformatics analysis, Varshney et al. 2017 predicted that a strong advancement of the cutin, suberin, and wax biosynthesis genes has occurred in pearl millet compared to other crop varieties (Debieu et al. 2017). The knockout approach using CRISPR-Cas9 will be useful in rapidly analyzing stress-responsive genes. Drought adversely affects crop yields by at least 25% (Choudhary et al. 2015). In such environmental conditions, improved drought-tolerant varieties of pearl millet could be a favorable choice for future generations (Habiyaremye et al. 2016). Sehgal et al. (2015) reported that an SNP in the acetyl CoA carboxylase gene is associated with grain yield and panicle number under drought stress. A single nucleotide change from C to T in acetyl CoA carboxylase alleles has shown a significant difference in grain yield; with the T-containing allele producing more grains than the C allele. In the future, using cytosine base editors (CBEs), researchers can target the C nucleotide in the acetyl CoA carboxylase gene to improve grain yield in pearl millet under drought.
Apart from drought stress, temperature stresses can affect the germination, photosynthesis, and groundwater uptake of pearl millet varieties (Dwivedi et al. 2012; Garcia-Huidobro et al. 1982). Although pearl millet has the inherent capacity to withstand abiotic stresses, some varieties are sensitive to these stresses. For example, H 77/833-2, a popular inbred line in northwestern India, is sensitive to drought stress but tolerates high-temperature stress (Yadav et al. 2014). Therefore, the C to T SNP mentioned earlier could be introduced by CBE into this genotype to transform it into a drought-tolerant variety.
Unlike other crops, pearl millet is also susceptible to biotic stresses such as fungal, bacterial, and viral infections. Fungal diseases like downy mildew, smut, rust, blast, and ergot are considered important pathogens of pearl millet. Downy mildew (DM), caused by Sclerospora graminicola, is a major concern for pearl millet farmers worldwide (Kumar and Manga 2011). It has been reported that the loss of the Mildew Resistance Locus O (MLO) results in resistance to powdery mildew in barley (Jacott et al. 2021). The MLO gene is a susceptibility gene for powdery mildew fungus and has been targeted by researchers using genome editing to develop resistance in wheat (Wang et al. 2014; Li et al. 2022) and grapevine (Wan et al. 2020). However, there have been no reports regarding the susceptibility genes of the downy mildew fungus in millets. MLO gene families could be identified through genome-wide analysis, and a similar strategy to knockout candidate MLO genes could be used for developing resistance. Numerous strategies are available to develop disease resistance in plants using the power of genome editing tools (Karmakar et al. 2022), which could also be harnessed for resistance breeding in pearl millet.
7.5.3 Pearl Millets for Biofuel
Like other cereals, pearl millet holds excellent potential for biofuel production using its lignocellulosic biomass. However, lignin is a major barrier to converting cellulose into biofuel. It would be interesting to mutate key lignin biosynthesis genes like cinnamyl alcohol dehydrogenase (CAD) and caffeic acid O-methyltransferase (COMT) to reduce lignin content and develop low-lignin pearl millet. A report by Liu et al. (2019) has already demonstrated CRISPR-Cas-mediated knock out of CAD gene in Sorghum.
7.6 Bottlenecks to Use Genome Editing in Pearl Millet Improvement
Successful genome editing in plants requires efficient genetic transformation and a regeneration system. To unlock the potential of genome editing in pearl millet, the availability of reliable methods for delivering genome editing reagents and regenerating transformed cells is crucial. Like other monocots, pearl millet faces challenges in tissue culture, transformation, and regeneration. Reports of successful transgenic pearl millet development are scarce in the literature (Latha et al. 2006; Ramineni et al. 2014). The recalcitrance bottleneck must be broken to apply genome editing tools for basic research and improving pearl millet.
In an earlier study, ectopic expression of developmental regulators like Wuschel (Wus) and Baby boom (Bbm) has been shown to enhance organogenesis efficiency and reduce the time required for regeneration in maize (Lowe et al. 2016). Similarly, a GRE-GRF chimeric protein enhanced the efficiency of regeneration in wheat (Debernardi et al. 2020). Co-delivery of these developmental regulators with gene editing reagents may be beneficial in reducing the recalcitrance of pearl millet to tissue culture and transformation. Along with the conventional biolistic and Agrobacterium-mediated gene transfer methods, nanoparticle-based and RNA virus-based delivery methods could also be explored.
7.7 Conclusion and Future Prospective
Pearl millet is a nutritious, climate-resilient crop that can thrive in harsh climatic conditions where other cereals may fail. In recent decades, we have seen tremendous growth in various genetic and genomic studies. Whole-genome sequencing and resequencing efforts of the pearl millet genome have provided insights into the genes and their associations with yield, abiotic and biotic stresses. Despite all these efforts, there is an urgent need to functionally validate genes important for their nutrient contents, stress resistance, and other beneficial traits. Rapid functional validation using genome editing tools would play a significant role in this direction. From knockout to base editing and prime editing, CRISPR-Cas-derived tools and techniques are valuable resources for the rapid characterization of genomic regions and their utilization in pearl millet improvement programs. However, to realize the full potential of genome editing, efficient genetic transformation and regeneration protocol needs to be established for different pearl millet genotypes. The small genetic changes made by editing are impossible to differentiate from those occuring naturally. Therefore, classifying genome-edited crops as GMOs seems irrational, and they should be regulated no differently from crop varieties derived from mutation or conventional breeding. A positive regulatory environment is crucial for the rapid adoption of these powerful new breeding tools. Recently, the Government of India exempted genome-edited crops (free from foreign-DNA ) from stringent GMO regulations. There is no doubt that the availability of gene transformation protocols and rapid adoption of genome editing tools will open new avenues for pearl millet improvement, facilitating nutrient fortification and climate-smart agriculture.
Abbreviations
- ABEs:
-
Adenine base editors
- CBEs:
-
Cytosine base editors
- CGBEs:
-
C to G base editors
- CRISPR-Cas:
-
Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein
- DSBs:
-
Double-strand breaks
- HDR:
-
Homology-directed repair
- MMEJ:
-
Microhomology mediated end joining
- NHEJ:
-
Non-homologous end joining
- pegRNA:
-
Prime editing gRNA
- RFLP:
-
Restriction fragment length polymorphism
- SNPs:
-
Single nucleotide polymorphisms
- SSN:
-
Sequence-specific nuclease
- TALENs:
-
Transcription activator like effector nucleases
- ZFNs:
-
Zinc finger nucleases
References
Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM et al (2019) Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576(7785):149–157
Burgarella C et al (2018) A Western Sahara Centre of domestication inferred from pearl millet genomes. Nat Ecol Evol 2:1377–1380
Chen K, Wang Y, Zhang R, Zhang H, Gao C (2019) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol 70:667–697
Choudhary M, Jayanand, Padaria JC (2015) Transcriptional profiling in pearl millet (Pennisetum glaucum L.R. Br.) for identification of differentially expressed drought responsive genes. Physiol Mol Biol Plants 21:187–196
Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F et al (2010) Targeting DNA double-strand breaks with TAL effector nucleases. Genet 186:757–761
Debernardi JM, Tricoli DM, Ercoli MF, Hayta S, Ronald P, Palatnik JF et al (2020) A GRF–GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat Biotechnol 38:1274–1279
Debieu M, Kanfany G, Laplaze L (2017) Pearl millet genome: lessons from a tough crop. Trends Plant Sci 22(11):911–913
Diack O, Kanfany G, Gueye MC, Sy O, Fofana A, Tall H et al (2020) GWAS unveils features between early-and late-flowering pearl millets. BMC Genomics 21(1):1–11
Dong H et al (2020) Generation of imidazolinone herbicide resistant trait in Arabidopsis. PLoS One 15:e0233503
Dwivedi S, Upadhyaya H, Senthilvel S, Hash C (2012) Millets: genetic, and genomic resources. In: Janick J (ed) Plant breeding reviews. Wiley, Hoboken, pp 247–375
FAO (2017) The future of food and agriculture: trends and challenges. FAO
Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P et al (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23(10):1229–1232
Fukunaga K, Kawase M, Kato K (2002) Structural variation in the Waxy gene and differentiation in foxtail millet [Setaria italica (L.) P. Beauv.]: implications for multiple origins of the waxy phenotype. Mol Genet Genomics 268(2):214–222
Gao C (2021) Genome engineering for crop improvement and future agriculture. Cell 184(6):1621–1635
Gao H, Gadlage MJ, Lafitte HR, Lenderts B, Yang M, Schroder M et al (2020) Superior field performance of waxy corn engineered using CRISPR–Cas9. Nat Biotechnol 38(5):579–581
Garcia-Huidobro T, Monteith T, Squire GR (1982) Time temperature and germination of pearl millet (Pennisetum typhoides S. & H.): I. Constant temperature. J Exp Bot 33:288–296
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR (2017) Programmable base editing of A• T to G• C in genomic DNA without DNA cleavage. Nature 551(7681):464–471
Govindaraj M, Rai KN, Kanatti A, Upadhyaya HD, Shivade H, Rao AS (2020) Exploring the genetic variability and diversity of pearl millet core collection germplasm for grain nutritional traits improvement. Sci Rep 10(1):1–13
Gupta SK et al (2015) Seed set variability under high temperatures during flowering period in pearl millet (Pennisetum glaucum L. (R.) Br.). Field Crops Res 171:41–53
Habiyaremye C et al (2016) Proso millet (Panicum miliaceum L.) and its potential for cultivation in the Pacific northwest, US: a review. Front Plant Sci 7:1961
Holme IB, Gregersen PL, Brinch-Pedersen H (2019) Induced genetic variation in crop plants by random or targeted mutagenesis: convergence and differences. Front Plant Sci 10:1468
Hunziker J et al (2020) Multiple gene substitution by target-AID base-editing technology in tomato. Sci Rep 10:20471
Ibrahim S, Saleem B, Rehman N, Zafar SA, Naeem MK, Khan MR (2021) CRISPR/Cas9 mediated disruption of inositol Pentakisphosphate 2-kinase 1 (TaIPK1) reduces phytic acid and improves iron and zinc accumulation in wheat grains, vol 37. J Adv Res, p 33
Jacott CN, Ridout CJ, Murray JD (2021) Unmasking mildew resistance locus O. Trends Plant Sci 26(10):1006–1013
Jain N, Arora P, Tomer R, Mishra SV, Bhatia A, Pathak H, Chakraborty D, Kumar V, Dubey D, Harit R et al (2016) Greenhouse gases emission from soils under major crops in Northwest India. Sci Total Environ 542:551–561
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096):816–821
Karmakar S, Das P, Panda D, Xie K, Baig MJ, Molla KA (2022) A detailed landscape of CRISPR-Cas-mediated plant disease and pest management. Plant Sci 323:111376
Khanday I, Skinner D, Yang B, Mercier R, Sundaresan V (2019) A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 565(7737):91–95
Kim YG, Cha J, Chandrasegaran S (1996) Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A 93:1156–1160
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533:420–424
Kumar A, Manga VK (2011) Downy mildew of pearl millet. Biores Bull 4:182–200
Kumar S, Hash CT, Thirunavukkarasu N, Singh G, Rajaram V, Rathore A et al (2016) Mapping quantitative trait loci controlling high iron and zinc in self and open pollinated grains of pearl millet [Pennisetum glaucum (L) R. Br.]. Front Plant Sci 7:16–36
Latha AM, Rao KV, Reddy TP, Reddy VD (2006) Development of transgenic pearl millet (Pennisetum glaucum (L.) R. Br.) plants resistant to downy mildew. Plant Cell Rep 25(9):927–935
Li C, Zong Y, Wang Y, Jin S, Zhang D, Song Q et al (2018) Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol 19:1–9
Li J, Wang Z, He G, Ma L, Deng XW (2020a) CRISPR/Cas9-mediated disruption of TaNP1 genes results in complete male sterility in bread wheat. J Genet Genomics 47(5):263–272
Li Y et al (2020b) Precise base editing of non-allelic acetolactate synthase genes confers sulfonylurea herbicide resistance in maize. Crop J 8:449–456
Li S, Lin D, Zhang Y, Deng M, Chen Y, Lv B et al (2022) Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 602(7897):455–460
Liu G, Li J, Godwin ID (2019) Genome editing by CRISPR/Cas9 in sorghum through biolistic bombardment. In: Sorghum. Humana Press, New York, pp 169–183
Liu L et al (2021) Developing a novel artificial rice germplasm for dinitroaniline herbicide resistance by base editing of OsTubA2. Plant Biotechnol J 19:5–7
Lowe K, Wu E, Wang N, Hoerster G, Hastings C, Cho MJ et al (2016) Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28(9):1998–2015
Mahendrakar MD, Parveda M, Kishor PK, Srivastava RK (2020) Discovery and validation of candidate genes for grain iron and zinc metabolism in pearl millet [Pennisetum glaucum (L.) R. Br.]. Sci Rep 10(1):1–16
Mamidi S, Healey A, Huang P, Grimwood J, Jenkins J, Barry K et al (2020) The Setaria viridis genome and diversity panel enables discovery of a novel domestication gene. bioRxiv 744557
Mariac C, Luong V, Kapran I, Mamadou A, Sagnard F, Deu M et al (2006) Diversity of wild and cultivated pearl millet accessions (Pennisetum glaucum [L.] R. Br.) in Niger assessed by microsatellite markers. Theor Appl Genet 11:49–58
Molla KA, Yang Y (2019) CRISPR/Cas-mediated base editing: technical considerations and practical applications. Trends Biotechnol 37:1121–1142
Molla KA, Yang Y (2020) Predicting CRISPR/Cas9-induced mutations for precise genome editing. Trends Biotechnol 38(2):136–141
Molla KA, Qi Y, Karmakar S, Baig MJ (2020a) Base editing landscape extends to perform Transversion mutation. Trends Genet 36:899–901
Molla KA, Shih J, Yang Y (2020b) Single-nucleotide editing for zebra3 and wsl5 phenotypes in rice using CRISPR/Cas9-mediated adenine base editors. aBIOTECH 1:106–118
Molla KA, Sretenovic S, Bansal KC, Qi Y (2021) Precise plant genome editing using base editors and prime editors. Nat Plants 7(9):1166–1187
Nakayama H, Afzal M, Okuno K (1998) Intraspecific differentiation and geographical distribution of Wx alleles for low amylose content in endosperm of foxtail millet, Setaria italica (L.) Beauv. Euphytica 102(3):289–293
Nambiar VS, Dhaduk JJ, Sareen N, Shahu T, Desai R (2011) Potential functional implications of pearl millet (Pennisetum glaucum) in health and disease. J Appl Pharm Sci 1:62–67
Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M et al (2016) Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353(6305):aaf8729
Oliva R, Ji C, Atienza-Grande G, Huguet-Tapia JC, Perez-Quintero A, Li T et al (2019) Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat Biotechnol 37(11):1344–1350
Oumar I et al (2008) Phylogeny and origin of pearl millet (Pennisetum glaucum [L.] R. Br) as revealed by microsat-ellite loci. Theor Appl Genet 117:489–497
Parvathaneni RK, Spiekerman JJ, Zhou H, Wu X, Devos KM (2019) Structural characterization of ABCB1, the gene underlying the d2 dwarf phenotype in pearl millet, Cenchrus americanus (L.) Morrone. G3: Genes, Genomes. Genetics 9(8):2497–2509
Prasad PV, Staggenborg SA (2009) Growth and production of sorghum and millets. In: Soils, plant growth and crop production, vol 2. EOLSS Publishers, Oxford
Pujar M, Gangaprasad S, Govindaraj M, Gangurde SS, Kanatti A, Kudapa H (2020) Genome-wide association study uncovers genomic regions associated with grain iron, zinc and protein content in pearl millet. Sci Rep 10(1):1–15
Ramineni R, Sadumpati V, Khareedu VR, Vudem DR (2014) Transgenic pearl millet male fertility restorer line (ICMP451) and hybrid (ICMH451) expressing Brassica juncea nonexpressor of pathogenesis related genes 1 (BjNPR1) exhibit resistance to downy mildew disease. PLoS One 9(3):e90839
Rodríguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB (2017) Engineering quantitative trait variation for crop improvement by genome editing. Cell 171(2):470–480
Saïdou AA, Mariac C, Luong V, Pham JL, Bezancon G et al (2009) Association studies identify natural variation at PHYC linked to flowering time and morphological variation in pearl millet. Genetics 182:899–910
Sánchez-León S, Gil-Humanes J, Ozuna CV, Giménez MJ, Sousa C, Voytas DF, Barro F (2018) Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol 16(4):902–910
Sashidhar N, Harloff HJ, Potgieter L, Jung C (2020) Gene editing of three BnITPK genes in tetraploid oilseed rape leads to significant reduction of phytic acid in seeds. Plant Biotechnol 18(11):2241–2250
Sehgal D, Skot L, Singh R, Srivastava RK, Das SP, Taunk J et al (2015) Exploring potential of pearl millet germplasm association panel for association mapping of drought tolerance traits. PLoS One 10(5):e0122165
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z et al (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31(8):686–688
Sharma B, Chugh LK (2017) Two isoforms of lipoxygenase from mature grains of pearl millet [Pennisetum glaucum (L.) R. Br.]: purification and physico-chemico-kinetic characterization. J Food Sci Technol 54(6):1577–1584
Sharma S, Sharma R, Govindaraj M, Mahala RS, Satyavathi CT, Srivastava RK et al (2020) Harnessing wild relatives of pearl millet for germplasm enhancement: challenges and opportunities. Crop Sci 61(1):177–200
Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H et al (2017) Targeted base editing in rice and tomato using a CRISPRCas9 cytidine deaminase fusion. Nat Biotechnol 35:441–443
Shivhare R, Lata C (2017) Selection of suitable reference genes for assessing gene expression in pearl millet under different abiotic stresses and their combinations. Sci Rep 6(1):1–12
Sretenovic S, Liu S, Li G, Cheng Y, Fan T, Xu Y et al (2021) Exploring C-To-G base editing in rice, tomato, and poplar. Front Genome Ed 3:756766
Tako E et al (2015) Higher iron pearl millet (Pennisetum glaucum L.) provides more absorbable iron that is limited by increased polyphenolic content. Nutr J 14(1):1–9
Vadez V, Hash T, Bidinger FR, Kholova J (2012) II 1.5 phenotyping pearl millet for adaptation to drought. Front Physiol 3:386
Varshney RK, Shi C, Thudi M, Mariac C, Wallace J, Qi P et al (2017) Pearl millet genome sequence provides a resource to improve agronomic traits in arid environments. Nat Biotechnol 35:969
Vinutha T, Kumar D, Bansal N, Krishnan V, Goswami S, Kumar RR et al (2022) Thermal treatments reduce rancidity and modulate structural and digestive properties of starch in pearl millet flour. Int J Biol Macromol 195:207–216
Wan DY, Guo Y, Cheng Y, Hu Y, Xiao S, Wang Y, Wen YQ (2020) CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera). Hortic Res 7(1):1–14
Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32(9):947–951
Wang C, Liu Q, Shen Y, Hua Y, Wang J, Lin J et al (2019) Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat Biotechnol 37(3):283–286
WHO (2020) Anaemia. https://www.who.int/health-topics/anaemia#tab=tab_1
Xie K, Yang Y (2013) RNA-guided genome editing in plants using a CRISPR–Cas system. Mol Plant 6(6):1975–1983
Xing S et al (2020) Fine-tuning sugar content in strawberry. Genome Biol 21:230
Yadav AK, Arya RK, Narwal MS (2014) Screening of pearl millet F1hybrids for heat tolerance at early seedling stage. Adv Agric, Article ID. 231301. https://doi.org/10.1155/2014/231301
Zhang R, Liu J, Chai Z, Chen S, Bai Y, Zong Y et al (2019) Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat plants 5(5):480–485
Zong Y, Song Q, Li C, Jin S, Zhang D, Wang Y et al (2018) Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat Biotechnol 36:950–953
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Panda, D., Baig, M.J., Molla, K.A. (2024). Genome Editing and Opportunities for Trait Improvement in Pearl Millet. In: Tonapi, V.A., Thirunavukkarasu, N., Gupta, S., Gangashetty, P.I., Yadav, O. (eds) Pearl Millet in the 21st Century. Springer, Singapore. https://doi.org/10.1007/978-981-99-5890-0_7
Download citation
DOI: https://doi.org/10.1007/978-981-99-5890-0_7
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-99-5889-4
Online ISBN: 978-981-99-5890-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)