Abstract
Brain tumour is a serious disease which can cause severe damage to the brain cells which eventually turns into a life threatening cancer. The tumour stages when identified early can helps to increase the survival rates of the patients. The performance of the automated brain tumour diagnosis depends on the classification accuracy of the model.. In this article, a deep convolutional neural network (DCNN) is developed for brain tumor classification of brain tumors in MRI images. Specifically, the auto-weight dilated convolutional unit utilized multi-scale convolutional feature maps to acquire brain tumor features at different scales and employed a learnable set of parameters to fuse convolutional feature maps in encoding layers. The AD unit is an effective architecture for feature extraction in the encoding stage. We used the advantages of the U-Net network for deep and shallow features, combined with AD units to multimodal image classification. In this model, the four-channel model inputs correspond to the MRI images of four modes, respectively. The main body of the network is composed of auto-weight dilated (AD) unit, Residual (Res) unit, linear upsampling, and the first and last convolution units.. The network that applied Block-R3 had higher segmentation performance than the networks of Block-R1 and Block-R2. In the U-shaped network, feature extraction at the coding stage is the most important component. Designing the network to extract the features of interest efficiently is crucial. The proposed tumour diagnosis with the optimal feature extraction achieved better results with less time consumption.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Gliomas originate from intracranial tumors of glial cells and are highly lethal. It is the occurrence of mutations that are sufficient for carcinogenesis at the level of the cell’s genetic material (DNA) and epigenetic material (EPI) through the interaction of internal genetic predisposing factors with external environmental pathogenic factors. Gliomas are mainly classified into the following categories: Astrocytoma, Oligodendroglioma, Mixed gliomas (such as oligodendro astrocytomas, which contain mixed types of glial cells), and Ependymoma. Patients with low-grade have a survival rate in months or even years, while the history of high-grade gliomas is often in weeks to months. Magnetic Resonance Imaging (MRI) has no ionizing radiation damage to human body and could be imaged without injecting radioisotopes. The soft tissue structure appears clear on MRI images. The multimodal gliomas sequences, including T1, T1c, T2, and T2-Flair, are advantageous for assessing health risks and clinical diagnosis [1, 2].
In recent years, more image segmentation methods are developed based the neural network technology to improve the deficiency of the traditional image segmentation algorithm, these methods are well applied in biomedical image segmentation [3]. The current image segmentation technology is mainly based on the color, gray, texture and other features of the image to extract the main part of the image. The image contains a lot of geometric information, and it is the most fundamental application to segment and extract the geometric features of the image. Its technology is widely used in autonomous driving, medical imaging, biometric recognition and remote sensing images. Traditional image segmentation methods are based on image threshold, edge detection and region based growth method [4]. However, these traditional methods have limitations for analyzing biological images. For example, the segmentation method based on image threshold is not appropriate to analyze images with no obvious difference in gray values or with different target gray value overlap, this method is also susceptible to noise producing false targets, when cell colors are not uniform in the images, using this method can produce holes during segmentation. Although the method based on edge detection can get the target profile, the structure inside the target is still missing. The segmentation method based on region growth is sensitive to noise and produces segmentation cavities or over-segmentation for complex images.
Manual segmentation of multimodal brain tumors is time-consuming and expensive compared to the automated method. It usually takes an expert radiologist about 3 h to the segment at the pixel level. The manual segmentation for the Dice Similarity Coefficient (DSC) score is 74%–85%. Therefore, accurate methods of tumor segmentation are of vital importance in clinical diagnostics and planning for treatment. The automated segmented methods aim to partition these multimodal MRI images into four tissues, including the normal tissues, the whole tumor (WT), the tumor core (TC), and the enhancing tumor (ET). TC describes that most tumors need to be removed. The WT describes the peritumoral edema (ED). And TC represents the necrotic and the non-enhancing parts. Automatic gliomas segmentation accurately is still challenging work. In the 2010s, many methods have been proposed for automated segmentation in brain tumors [5, 6].
Since the powerful generalization, the deep learning method has gained a significant advantage compared to other approaches. Deep learning puts forward a way to let computers learn the features automatically based on data-driven to reduce the complexity of artificial design features. The deep learning model with essentially enlarged depth advances segmentation performance, such as CNN, FCN, GNN, RNN, GAN and other procedures of network. In brain tumor segmentation, the deeper neural network model is more and more important to advance the state-of-art performance. In recent years, deep learning methods gained significant interest in the segmentation of brain tumors. The U-shaped model is an efficient and straightforward segmentation network in 3D medical images especially in brain tumors, learning features from deep and shallow neural units. The UNet model consists of four encoder layers and four decoder layers [7, 8]. The manuscript is organized as follows. Section 2 provides the related work. Section 3 illustrates the proposed work, Sect. 4 details the simulation results and Sect. 5 ends with conclusion.
2 Literature Review
The categorization of brain tumors has been the subject of empirical investigation. In this particular division, modern works on the classification of brain tumors are mentioned. Their proposed methodologies are also explained. Researchers have suggested many architectural models for the classification of brain tumors and the most eminent methods have been identified. Majib et al. (2021) [9] have proposed a VGG Net-Based deep learning framework. They considered 90 images, 81 of which have been labeled as YES and the rest labeled as No. They received the best training and also tested performance by the usage of CNN architecture. They achieved 97.8% F1 score using the VGG16 model and got the issue on the trade-off between the algorithmic performance and time complexity.
Noreen et al. (2020) [10] developed a concatenated approach for brain tumor classification, using Inception-V3 and DensNet201 deep learning models and extracted the features. They also concatenated those features using a softmax classifier for classification of the brain tumor cells. They achieved 99.34% and 99.51% accuracy for Inception-V3 and DensNet201 respectively. Kumar et al. (2019) [11] have suggested the combination of Deep Wavelet Autoencoder (DWA) with the Deep Neural Network (DNN). DWA adds the feature reduction property known for the autoencoder with the image decomposition property seen in the wavelet transform. They used all the images in the DICOM format and the python for processing the data. They achieved 96% accuracy by combining DWA with DNN. The entire research had been carried out using tenfold cross-validation. They also saw DNN as combination with other varieties of the encoder to compare these results. Liu et al. (2020) [12] have suggested deep Convolutional Long Short-Term Memory (C-LSTM) for the detection of a tumor in the brain. They also made a comparative study with the other types of deep learning techniques and concluded that the proposed C-LSTM achieved satisfactory performance in the classification of the five classes of brain tumors. They ran the experiment 20 instances in order to get the average and standard deviation of the categorization findings. The main drawback of the model was its proneness to unanticipated noise and prototype errors. They saw computation as exorbitant, with datasets limited and so the training data sets as also limited requiring improvement.
Sultan et al. (2019) [13] have developed a brain tumor classification system or the classification of brain tumors using a Convolutional Neural Network (CNN). They worked on two different datasets. The first dataset was used for the classification of various types of tumors including glioma, meningioma, and pituitary tumor. The second dataset was used for the classification of different glioma grades. They found the proposed model requiring training with the use of a large number of datasets including samples from different age groups. Wang et al. (2019) [14] addressed the issues seen in the Deep Convolutional Neural Network (DCNN) considering the computational complexity as high. They have proposed an assignment process that can be used for correlation of the weights of the Fully Connected Layer (FCL), instead of the weight adjustment process for a reduction in complexity. According to the author’s expertise, there exists some computational complexity in the existing system, and as also issues arising while handling large datasets. Time complexity can be reduced while handling huge datasets when the parameters are small.
3 Proposed Methodology
Deep learning method has gained a significant advantage compared to other approaches. Deep learning puts forward a way to let computers learn the features automatically based on data-driven to reduce the complexity of artificial design features. The deep learning model with essentially enlarged depth advances segmentation performance. Figure 1 depicts the summary of the proposed methodology.
3.1 Pre-processing
We used the randomization strategy as image preprocessing, which could ensure that the deep learning model still maintains strong generalization performance after a large number of repeated training. Multimodal brain images of the same patient use the same processing in one epoch training and different random measures in different epochs. It helps to learn the image features of different modes in the same brain while obtaining generalization. The figure shows the image preprocessing methods: 3D random clipping, 3D random rotation, 3D image intensity random enhancement, 3D image random mirror inversion, and normalization.
Image normalization is a widely used technique in computer vision, pattern recognition and other fields. The z-score normalization was applied in this work. It is defined as per Eq. (1):
where \(\sigma\) is the standard deviation, and \(\mu\) is the mean value. Then, the 3D random clipping method randomly cuts the MRI image (240, 240, 155) into a matrix (144, 144, 128). The 3D random rotation method rotates the reduced image by the angle \(U(-10,+10)\). The random intensity enhancement method of 3D image sets the image pixel value is defined as per Eq. (2):
where \(U\) is the uniform distribution. The random mirror processing symmetrizes the image according to deep, height and width directions. We applied these image enhancement routines to extend the training data set to improve the performance and generalization ability of the deep neural network.
3.2 Deep Learning Model
Deep learning which is a subset of Artificial intelligence is gaining momentum each day by making different tasks much easier and more efficient. CNN which is a type of deep learning mechanism is an inevitable part of image vision problems. In recent years, deep learning methods gained significant interest in the segmentation of brain tumors. The U-shaped model is an efficient and straightforward segmentation network in 3D medical images especially in brain tumors, learning features from deep and shallow neural units [15,16,17]. The UNet model consists of four encoder layers and four decoder layers. The proposed model is shown in Fig. 2. In this model, the four channel model inputs correspond to the MRI images of four modes, respectively. The main body of the network is composed of auto-weight dilated (AD) unit, Residual (Res) unit, linear upsampling, and the first and last convolution units. In the downsampling stage (feature coding extraction), we use 8 AD units to obtain multi-scale feature maps. In the upsampling stage (feature decoding), we use the AD unit, Res unit and a linear upsampling layer to form a primary decoding layer. Finally, a convolution unit outputs the results of the network model. Moreover, each convolution unit, AD unit and Res unit contains batch normalization and ReLU functions. We used extended convolution to extract fine-grained and multi-scale glioma features, and employed residual structure to obtain long-dependent glioma features.
As for the Res Unit layer, we used two convolution units to reduce and then enlarge the number of convolution kernels so as to realize feature learning and feature map reorganization. From an experimental point of view, this is an efficient coding method. Then, we used two group convolution units with stripe 1 and group 16, and the kernel size is 3 × 3 × 3. Finally, we used a convolution residual element to obtain the characteristic graph of long dependence. As for the AD Unit layer, we used two convolution units firstly (like the Res unit). Then, we used three extended convolution units (the divided parameters are 1 and 2, respectively) and used two learnable parameters to adjust and fused the characteristics of the two group extended convolution units. Finally, a group convolution unit was used to output the result of the AD unit. We also set up residual calculations in the AD unit. The dilated convolution could expand the receptive field of the convolutional kernel without sacrificing computational resources, while normal convolution could provide a more accurate feature map. The fusion of the two types of convolutions could strengthen the ability of the network to extract features.
In the encoder stage, each residual block is a dual-pathway structure. In this stage, we set channel depth to 32, 64, 128, and 256. The residual block is the critical structure of down-sampling. In the decoder stage, we connect a convolutional unit and a de-convolutional unit for upsampling. The stripe of the de-convolutional unit is 2 \(\times\) 2 \(\times\) 2, and the kernel size is 3 \(\times\) 3 \(\times\) 3. Batch Normalization and RReLU activation functions are connected behind the convolutional unit or the de-convolutional unit for all convolutional units and all de-convolutional units in this stage. Similarly, we set the channel depth in the decoder stage to 32 \(\times\) 64 \(\times\) 128 \(\times\) 256. A combined deep neural network with the residual blocks enables the network to obtain more significant gradients in deep layers.
So, the phenomenon of gradient disappearance is relatively rare and gets more practical features of gliomas. The formula of the gradient propagation in the convolutional layer can be defined as per Eq. (3),
where \({\sigma }^{^{\prime}}\) means the first derivative of the loss function, \(w\) describes the weight, \(O\) indicates the output matrix vector, and \(l\) is the layer \(l\). Then, the gradient in Block-R1, Block-R2 and Block-R3 can be defined as per Eq. (4),
where \(f\) means the activation function, \({\delta }_{1}\) and \({\delta }_{2}\) represent the first and second convolution calculations, respectively. It is worth noting that the difference between Eqs. (3) and (4) lies in the order of normalization, which is not reflected in the equation
Multiplication is widely used in the calculation of series convolution, such as \({\delta }_{2}(f({\delta }_{1}))\). The cumulative multiplication between \((-\mathrm{1,1})\) makes it possible for the gradient to appear the result of approximate 0, so that the classical gradient disappears. The residual-connection weakens this problem through weight addition, and enhances the stability of the network. Obviously, it is a very effective way to use residual blocks to build architecture in very deep neural layers, especially in calculating the depth feature map.
We defined the convolution block (BN, RL, Conv) in AD unit as per Eq. (5)
where \({c}_{i}\) is the convolution layer \(i\), and \(k\) describes the kernel size, \(f\) is the activation function. The \(w\) and \(b\) represent the convolution weight and bias, \({I}_{i}\) is the input data. Then, the AD block can be define as per Eq. (6)
where \(\ell _{{d_{0} }}\) is the dilated convolution. In the gradient back-propagation process, \(a\) and \(b\) can automatically adjust the weight ratio of the convolution integral branch in the main path. In addition, the channel parameters of the AD-Net were set to 32, 64, 128, 256, and the skip connection adopted the 3D matrix concatenate method. The residual structure was a necessary element. The residual calculation ensured the stability of gradient in deep feature calculation.
4 Experimental Results and Discussion
4.1 Dataset
The MICCAI BraTS datasets consist of many pre-operative multimodal MR images from multi- institution [5, 15]. MICCAI is a comprehensive academic conference held by the international medical image computing and Computer Assisted Intervention society. It is a top-level conference in the field of medical image computing (MIC) and computer assisted intervention (CAI). The BraTS datasets already applied various preprocessing steps by the organizers. BraTS challenge has always focused on evaluating advanced methods for the multimodal brain tumor segmentation. The results of the validation set and test set required to be evaluated and returned by the online platform. In this study, two datasets (BraTS19 and BraTS20) were used. The BraTS19 training set consists of 335 multimodal MRI images, including high-grade gliomas (HGG) and low-grade gliomas (LGG), 259 HGGscans and 76 LGG scans. In addition, the validation set includes 125 MRI scans. The BraTS20 training set consists of 369 multimodal MRI images, including HGG and LGG and paying no attention to divide into HGG and LGG. In addition, the validation set contains 125 MRI scans and the testing set includes 166 MRI scans. The test set is for participants. Each scans contains five MRI images (T1, T1c, T2, Flair and segmentation label).
4.2 Classification Results
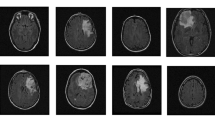
The proposed brain MRI images segmentation method is simulated using MATLAB 2020b driven by a single Intel(R) Core(TM) i5-3570 64bit CPU with 8 GB RAM and 3.40 GHz clock frequency. Additional memory was not required during the entire process. In this work, three different publicly available Dataset is used for training and testing the classifier. The different types of brain tumor images are given in Fig. 3.
The following evaluation metrics are used for validating the classifier performance. It is given in Eq. (7)
where accuracy explains how well the architecture can classify the images. It is given as the ratio of total correct prediction made to the total number of predictions made. Precision is given by the ratio of total number of correctly classified positive cases to the sum of total positive cases predicted. High value for precision is required in order to minimize the number of false positive classifications. Recall is defined as the ratio of total positive cases correctly classified to all correctly classified observations and F1 score gives the harmonic mean of precision and recall. It can be also defined as the weighted average of precision and recall [18,19,20]. The performance comparison of proposed model and other models are given in Table 1. The confusion matrix of the proposed system with different Dataset is given in Fig. 4 and the classification rate of various models using different statistics method is given in Fig. 5.
5 Conclusion
In this paper, a new multi-scale approach to segment the brain tumor in MRI image was described and evaluated in several publicly available databases. This paper also presents an assessment of the most appropriate scales for the brain tumor segmentation, complementing previous work that defines these scales empirically. Furthermore, it was also demonstrated that a multi-scale analysis can improve the brain tumor segmentation. Although recent research has been focusing on deep learning methods, rule-based methods can also be important for the definition of features, that can significantly improve the outcome of these methods. The achieved results show that the proposed approach is very competitive when compared with the current state of the art methods, particularly in high-resolution images. Our method still needs further improvement in the enhancing tumor region segmentation. It were practical tools in 3D brain tumor segmentation.
References
Pashaei A, Sajedi H, Jazayeri N (2018) Brain tumor classification via convolutional neural network and extreme learning machines. In 2018 8th International conference on computer and knowledge engineering (ICCKE). IEEE, pp 314–319
Irmak E (2021) Multi-classification of brain tumor MRI images using deep convolutional neural network with fully optimized framework. Iran J Sci Technol Trans Electric Eng 1–22
Rajasekaran KA, Gounder CC (2018) Advanced brain tumour segmentation from MRI images. Basic physical principles and clinical applications, high-resolution neuroimaging, pp 83–108
Hossain T, Shishir FS, Ashraf M, Al Nasim MDA, Shah FM (2019) Brain tumor detection using convolutional neural network. In 2019 1st International conference on advances in science, engineering and robotics technology (ICASERT). IEEE, pp 1–6
Jayachandran A, Dhanasekaran R (2013) Brain tumor detection using fuzzy support vector machine classification based on a Texton co-occurrence matrix. J Imag Sci Technol 7(1):10507-1-10507–7
Aurna NF, Anika FS, Rubel MDTM, Habibul Kabir K, Shamim Kaiser M (2021) Predicting periodic energy saving pattern of continuous IOT based transmission data using machine learning model. In 2021 International conference on information and communication technology for sustainable development (ICICT4SD). IEEE, pp 428–433
Jayachandran A, Kharmega Sundararaj G (2016) Abnormality segmentation and classification of multi model brain tumor in MR images using fuzzy based hybrid kernel SVM. Int J Fuzzy Syst 17(3):434–443
Mahiba C, Jayachandran A (2019) Severity analysis of diabetic retinopathy in retinal images using hybrid structure descriptor and modified CNNs. Measurements 135:762–767
Sajjad M, Khan S, Muhammad K, Wu W, Ullah A, Baik SW (2019) Multi-grade brain tumor classification using deep CNN with extensive data augmentation. J Comput Sci 30:174–182
Noreen N, Palaniappan S, Qayyum A, Ahmad I, Imran M, Shoaib M (2020) A deep learning model based on concatenation approach for the diagnosis of brain tumor. IEEE Access 8:55135–55144
Kumar Mallick P, Ryu SH, Satapathy SK, Mishra S, Nguyen GN, Tiwari P (2019) Brain MRI image classification for cancer detection using deep wavelet autoencoder-based deepneural network. IEEE Access 7:46278–46287
Liu Y et al (2020) Deep C-LSTM neural network for epileptic seizure and tumor detection using high-dimension EEG signals. IEEE Access 8:37495–37504
Sultan HH, Salem NM, Al-Atabany W (2019) Multi-classification of brain tumor images using deep neural network. IEEE Access 7:69215–69225
Balasooriya NM, Nawarathna RD (2017) A sophisticated convolutional neural network model for brain tumor classification. In: 2017 IEEE international conference on industrial and information systems (ICIIS). IEEE, pp 1–5
Wang W, Bu F, Lin Z, Zhai S (2020) Learning methods of convolutional neural network combined with image feature extraction in brain tumor detection. IEEE Access 8:152659–152668
Afshar P, Plataniotis KN, Mohammadi A (2019) Capsule networks for brain tumor classification based on MRI images and coarse tumor boundaries. In: ICASSP 2019–2019 IEEE International conference on acoustics, speech and signal processing (ICASSP). IEEE, pp 1368–1372
Prabhu AJ, Jayachandran A (2018) Mixture model segmentation system for parasagittal meningioma brain tumor classification based on hybrid feature vector. J Med Syst 42(12)
Namboodiri S, Jayachandran A (2020) Multi-class skin lesions classification system using probability map based region growing and DCNN. Int J Comput Intell Syst 13(1):77–84
Vijayakumar T (2019) Classification of brain cancer type using machine learning. J Artif Intell 1(2):105–113
Karuppusamy DP (2020) Hybrid manta ray foraging optimization for novel brain tumor detection. J Soft Comput Paradigm 2(3):175–185
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Jayachandran, A., Sreema, M.A., Anandaraj, S.P., Sudarson Rama Perumal, T. (2023). Deep Convolutional Neural Network for Multi-class Brain Tumor Classification System in MRI Images. In: Joby, P.P., Balas, V.E., Palanisamy, R. (eds) IoT Based Control Networks and Intelligent Systems. Lecture Notes in Networks and Systems, vol 528. Springer, Singapore. https://doi.org/10.1007/978-981-19-5845-8_39
Download citation
DOI: https://doi.org/10.1007/978-981-19-5845-8_39
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-19-5844-1
Online ISBN: 978-981-19-5845-8
eBook Packages: EngineeringEngineering (R0)