Abstract
The novel coronavirus is a family of animal transferred viruses that can cause illness in humans. This virus took over the world in 2019 and WHO deemed it as an epidemic naming it as COVID-19. A lot of research has gone in the prediction of the trends and classification of cases using various machine learning and deep learning techniques. With the outbreak of this pandemic, efficient detection of the disease at a faster rate has become very crucial. This study proposes a convolutional neural network (CNN)-based deep learning approach for classification of COVID-19 positive cases from normal cases using X-Ray radiology scans of the patients. The model consists of a large custom dataset of images extracted from an open source dataset and is then trained using our proposed model. Different optimizer algorithms are also compared in order to check which one of them gives the most accuracy. The model is further tested using the categorical accuracy metrics and then a graphical analysis of the results is provided. A comparative study is also conducted with an already existing support vector machines (SVM) model. The images were trained according to three classifications: normal, COVID infected, and viral pneumonia infected patients. The main objective of our research is to help further the research in early diagnosis of COVID-19 using modern deep learning techniques.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The novel coronavirus or the COVID-19 was first identified in Wuhan, China. Further, it didn’t take long for it to spread to the other parts of the country, and eventually, the whole world. COVID-19 is an infectious disease which can easily be transmitted through coming in direct contact (coughing or sneezing) with an infected person. Even though the symptoms for COVID-19 are very similar to the common cold, the virus is capable of causing other chronic respiratory illnesses as well. There have been a total of 167,531,200 cases that the world has experienced till May, 24, 2021, of which, 15,505,637 were still active.
Currently, a special test called the reverse transcription polymerase chain reaction (RT-PCR) is conducted to test the presence of the virus, but it generally takes 24 h to get the result. Some studies show that when a person is infected with the virus, there are some significant changes that could be noticed in the infected person’s chest. If these changes can be identified well in time, it might lead to huge clinical benefits. Since, getting X-rays is not only cheap but also is a less time consuming process, this method could aid doctors in identifying and classifying any abnormalities that may be present. It could further be implemented as a screening stage for the RT-PCR test, thus saving time and resources as shown in Fig. 1.
The aim of our study is to create an efficient and best possible system that will help distinguish these changes and predict if a person has been infected by COVID-19 or not. A 23-layer CNN, consisting of six different kinds of layers, is proposed. The main objective is to figure out the most efficient optimizer that would give the best accuracy, to check if increasing the number of classes and the scale of the datasets will make the efficiency and accuracy of the model vary and whether we can use the same model to classify multiple classes.
2 Literature Survey
It has been noticed through medical research that chest X-ray is an imaging technique that can play a vital role in the detection of COVID-19. More and more large-scale datasets are being made available for research in these field and convolutional neural networks have emerged to be great tools for image classification. The research field is new and the time period is less, hence the biggest limitation is the availability of labelled medical images, which make it easier to model for classification [1].
Currently, even though the reverse transcription-polymerase chain reaction (RT-PCR) shows a very low sensitivity and is relatively a time consuming process, it is still the most widely used technique for the detection of COVID-19 [2]. It has been demonstrated how machine learning techniques, which have been used so often for detection of potential risk factors, can also be implemented to predict the number of cases of the ongoing COVID-19 threat. ML modelling was used to analyze the exponential smoothing risk factors. [3] A system which uses blood testing to support the diagnosis of COVID-19 was also implemented where the parameters are obtained from hemogram and the biochemical tests [4]. The healthcare sector is overburdened and the need for fast and efficient detection procedures is the need of the hour. Through recent medical research it has been established that detection of COVID-19 by means of X-ray and CT scans is a possibility [5]. The countries with shortage of proper equipment and testing kits have also been seen relying on CT scan images and x-rays. Research also shows that in order to make the COVID detection process efficient and faster, a combination of RT-PCR along with image featuring models can be used [6].
Medically speaking, there have been a lot of research to find out the key features to look for when detecting COVID chest X-ray scans from a normal scan. The COVID X-rays were found to have an abnormal increase in opacification near the lung region. This was coupled with reticulation interstitial thickening due to a collection of small opacities in a linear pattern. Ground glass is a radiological term that is used to define a hazy opaque lung appearance. These opacities may become dense in some cases that might obscure bronchial walls and vessels. We use feature extraction and modern image processing techniques to enhance this opacity features and train the model with a large number of images for detection of COVID-19 scans in comparison to Normal scans.
The recent large scale data mining and deep learning techniques such as CNNs are applied for rapid detection of the disease. This research saw the progress in development of a CNN model called CoroDet. It serves for accurate detection for multi-class classification. The 22-layer architecture of the CNN model that was proposed could classify with a max accuracy of 99.1% for the two classes [7].
The shortcoming of most researches in this field was that the dataset was used was small and only as far as the knowledge goes. Future work includes working on larger and more comprehensive data as we move forward into more research in this field. This can be a big step in providing a supportive alternative to tackle the problem of testing kit scarcity for detection of COVID-19 [8].
Research has also shown that it is also possible to differentiate whether the problems caused is because of COVID infection or any other pulmonary disease. Supervised machine learning techniques can be used for the same and they consists of mainly two phases: training and testing. The training phase discriminates between the X-ray images of COVID-19 images and other pathologies, whereas the testing phase then evaluates the performance of the model built in the training phase [9]. Data pre-processing is a major aspect here since data-imbalances can sway the predictions. Hyperparameter optimization using grid search algorithm provided more accurate predictions as compared to traditional random forest model. These models can integrated into real mobile healthcare applications. Rapid detection and screening for COVID-19 infections can be made possible through such semi-autonomous diagnostic systems [10].
Studies also showed that multiple feature extraction methods using support vector machines (SVM) can be used in order to classify the extracted features of different classes [11]. The SVM model can also be used to produce real-time forecasting. Data attributes which included confirmed, deceased, recovered COVID-19, and location wise data were collected. And then to explore the impact on recovery, deceased and identification, SVM was used. [12] A major limitation for such models is the availability of labeled medical images. A solution to this is the concept of transfer learning. It is an effective mechanism and shows promise in future results. The main solution is to transfer knowledge from conventional object detection tasks to specific domains. The research explores a kind of deep CNN called DeTraC which stands for decompose, transfer, and compose. An advantage of using such a model is that it can easily deal with irregularities in the image datasets. It uses a class decomposition mechanism to analyze class boundaries [13].
Further, in order to enable the identification of false prediction or unknown cases, Monte-Carlo Drop weights Bayesian convolutional neural network was used on publically available chest X-ray dataset. A pre-trained ResNet50V2 model was used for transfer learning [14].
Even though COVID-19 detection techniques based on CNN and various other models can become well established, they are still in the testing phases. There is a great room for improvement in this domain. These deep learning techniques are becoming more and more popular and can be easily associated with well-established medical techniques [15].
3 Proposed Methodology
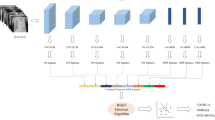
Figure 2 explains the various steps that we underwent for the experiment. The first step was to extract the dataset from open-source platforms. We found clear images which were classified into COVID, Normal and pneumonia chest X-ray scans. Only PA (Posterior Anterior) view scans were extracted since they are most relevant for our feature extraction. The second step consisted of data pre-processing and labeling. The dataset was first given proper labels and was sorted into training and testing datasets of 80-20% and further into directories of the three categories. Next, the generator functions of Keras were used to take the inputs in real-time batches where we adjusted the size, zoom range, and other parameters in order to get the most optimal training dataset. The third step is the most important where we tested a number of permutations and combinations of different layers and optimizers in order to find the most accurate CNN model for classification. The fourth step included the training phase using our final proposed model. We used categorical parameters to get results on the training data. Lastly, the learning curves for model accuracy and loss were plotted along with other parameters. The proposed model was compared with previously tested SVM models in order to find the relevance of our model.
3.1 Dataset
Various open-source datasets were scanned for X-Ray images of the three categories: COVID-19 infected, viral pneumonia infected and normal cases.
A large open-source COVID-19 radiography database on Kaggle was identified which consisted of 3616 COVID-19 positive X-rays, 1345 viral pneumonia x-rays, and 10,192 normal x-ray scans. All the images in the dataset were of the posterior anterior (PA) view, the best suited for our model [16,17,18].
3.2 Data Labeling and Structuring
Since the original dataset was unevenly distributed, we extracted our own customized dataset from the same. These images were then stored in categorical folders of testing (20%) and training data (80%). The directory structure for the 3-class dataset included training (total: 2400 images) and testing (total: 600 images) directories which were then further divided into three categorical directories of COVID, normal and pneumonia, each containing equal number of images. The 2-class dataset consisted of a similar structure but only contained the categorical directories of COVID and normal (Table 1).
3.3 Data Pre-processing
We used the Image Data Generator function to augment the images in real time. This helps in memory utilization since the process runs while the model is training and no excess memory is required to store the processed dataset. It also performs fewer transformations before going through the model.
Further, the images were rescaled by dividing them by a factor of 255 so that their spatial values ranged between 0 and 1. The zoom and shear range was set to 0.2 and a horizontal flip augmentation of the images was performed for the training dataset.
3.4 CNN Model
We used Keras and Tensorflow framework for creating our proposed CNN model. This consisted of a 23-layer architecture which comprised of six basic components, namely, convolutional layer, batch normalization layer, pooling layer, dropout layer, flatten layer, and dense layer.
The purpose of the convolutional layer is to help the model learn. With the help of certain parameters, filters and kernels, this layer performs most of the computations in terms of the convolutional process, which is the most important for a CNN model. In this layer, image segmentation procedures are performed to distinguish differences and similarities between the images. Our model uses this layer to extract high level features like edges of the rib cage and distinguishes it from the lung region, where the infection could be detected. We used eight conv2D layers in our model where the kernel size for the first four layers was taken as three and the last four as five. The number of output filters exponentially increased by a factor of 2 from 16 till 128. In the model, the conv2D layers were used in pairs. The input shape taken was (244, 244, 3) corresponding to the size of the images in our dataset. The activation function used was ‘relu’.
We used a batch normalization layer after each pair of the convolutional layer to maintain the mean output near 0 and the standard deviation near 1. The role of this layer is to help every other layer of the model to learn more independently.
The purpose of the pooling layer is to prevent overfitting by helping in reduction of the image size when the volume is too large. We used the maxpool2D layer with the stride set to (2, 2). This takes the largest value from the feature map received from the previous layer. The stride allows us to specify the movement of the filter over the image.
The dropout layer also helps us to prevent overfitting of the data. We set the rate to 0.25 which means that this fraction of input units were dropped after this layer for the maximum optimization.
Before passing the data to the next layer, the flatten layer helps us in reducing the dimensionality of the pool feature map into a single column.
We used three consecutive dense layers with the activation function ‘softmax’. These are fully connected layers that flatten the input from the previous matrix into a vector. This is the layer which is able to decide which features best match a particular class. This is where the classification takes place. For the 2-class structure, the last layer will have two units and similarly the 3-class structure will have three.
Figure 3 shows the model summary of our CNN architecture. The total parameters in the model are 7,735,891 out of which 7,735,411 are trainable and 480 are non-trainable.
4 Results and Discussion
4.1 Training the Model
We performed two sets of experiments, one for the 3-class and one for the 2-class image classification. The same model is used to classify both the datasets. The three classes were taken as COVID, normal, and Pneumonia.
The training was carried out on Google Colab for its GPU capabilities and the model was implemented using the Tensorflow and Keras frameworks.
We first chose three Keras optimizers for our experiment, namely, RMS, Adam, and Adamax. We ran simulations for all the three individually, under the same conditions and compared the results to choose which optimizer is best suited for our model. The experiment was run for 100 epochs with a batch size set to 16. The steps per epoch and validation steps were tested for multiple values, but it was noticed that optimal results were obtained when both were kept at 16 each.
The RMS optimizer was best suited for our model. The RMS prop algorithm was used to define the optimizer where the learning rate was kept at 0.0001 and the rho at 0.95. The discounted average of the gradient squares is maintained through this algorithm.
The loss function set was ‘catergorical_crossentropy’ and the metrics as ‘categorical_accuracy’. The graphs of accuracy and loss for training and testing were plotted to observe the learning curve.
5 Results and Discussion
In this section, we have mentioned the training and validation accuracy metrics in Tables 2 and 3 for the 3-class and 2-class models, respectively. We observed that in both the models, the RMS and the Adamax optimizers performed better than Adam optimizer, RMS giving the best results of 0.9082 and 0.9875 validation accuracy in the 3-class and 2-class models, respectively.
It was also observed that increasing the number of classes led to a decrease in the accuracy of the models.
Figure 4 shows the graph plotted for the training and testing accuracy against the number of epochs for the RMS optimizer. We observed that the training accuracy remained fairly constant throughout, whereas the testing accuracy increased and then stabilized around the range of 40 epochs.
Figure 5 shows the graph plotted for the training and testing loss against the number of epochs for the RMS optimizer. We observed that the training loss, again, remained fairly constant throughout, whereas the testing loss decreased and then stabilized around the range of 40 epochs.
In Table 4, we have shown the results of the comparison of all the optimizers used for the 2-class model with the already existing SVM model. We observed that the SVM model gave a 2-class accuracy of 0.96 whereas our proposed 2-class RMS model gave the best accuracy of 0.9875.
6 Conclusion
Our study focusses on research of deep learning application in the field of medical science related to COVID-19 and helps by providing a technique which can be used, if not as an alternative, but as an additional method for identifying and classifying any abnormalities that might be present while diagnosing any pulmonary diseases. We proposed a 23-layer CNN architecture which was tested for 3-class (COVID, normal, and pneumonia) and 2-class (COVID, Normal) models. Multiple optimizers (RMS, Adam, Adamax) were used and the RMS optimizer provided us with the best accuracy. This model was further compared with the already existing machine learning based SVM model and, even here, our proposed RMS model showcased better accuracy. It was noticed that when the model was tested on larger datasets, the 2-class accuracy was approximately the same, whereas the 3-class accuracy decreased to 85–90%. Further research can be done to optimize the model in order for it to give constant performance results with larger datasets for both 2-class and 3-class classification.
Through this study, we were able to successfully implement a deep learning-based CNN model, fully capable of multi-class image classification of chest X-ray scans. Future research can revolve around the implementation of transfer learning in the same field. Further study could also be done on how we can maintain the model accuracy even while scaling the dataset.
References
Aljondi R, Alghamdi S (2020) Diagnostic value of imaging modalities for COVID-19: scoping review. J Med Internet Res 22(8):e19673
Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J (2020) Chest CT for typical coronavirus disease 2019 (COVID-19) pneumonia: relationship to negative RT-PCR testing. Radiology 296(2):E41–E45
Mojjada RK, Yadav A, Prabhu AV, Natarajan Y (2020) Machine learning models for covid-19 future forecasting. Mater Today Proc
de Freitas Barbosa VA, Gomes JC, de Santana MA, de A. Jeniffer E, de Souza RG, de Souza RE, dos Santos WP (2021) Heg. IA: an intelligent system to support diagnosis of Covid-19 based on blood tests. Res Biomed Eng 1–18
Lee EYP, Ng MY, Khong PL (2020) COVID-19 pneumonia: what has CT taught us? Lancet Infect Dis 20(4):384–385
Kong W, Agarwal PP (2020) Chest imaging appearance of COVID-19 infection. Radiol Cardiothoracic Imaging 2(1):e200028
Hussain E, Hasan M, Rahman MA, Lee I, Tamanna T, Parvez MZ (2021) CoroDet: a deep learning based classification for COVID-19 detection using chest X-ray images. Chaos, Solitons Fractals 142:110495
Maguolo G, Nanni L (2021) A critic evaluation of methods for covid-19 automatic detection from x-ray images. Inform Fusion
Brunese L, Martinelli F, Mercaldo F, Santone A (2020) Machine learning for coronavirus covid-19 detection from chest x-rays. Proc Comp Sci 176:2212–2221
Kaliappan J, Srinivasan K, Mian Qaisar S, Sundararajan K, Chang CY (2021) Performance Evaluation of Regression Models for the Prediction of the COVID-19 Reproduction Rate. Front. Public Health, 9
Barstugan M, Ozkaya U, Ozturk S (2020) Coronavirus (covid-19) classification using CT images by machine learning methods. arXiv preprint arXiv:2003.09424
Singh V, Poonia RC, Kumar S, Dass P, Agarwal P, Bhatnagar V, Raja L (2020) Prediction of COVID-19 corona virus pandemic based on time series data using support vector machine. J Discrete Math Sci Cryptogr 1–15
Abbas A, Abdelsamea MM, Gaber MM (2021) Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl Intell 51(2):854–864
Jayakumar K, Revathi T, Karpagam S (2015) Fusion of Heterogeneous Intrusion Detection Systems for Network Attack Detection. Sci World J
Apostolopoulos ID, Mpesiana TA (2020) Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med 43(2):635–640
Rahman T, Khandakar A, Qiblawey Y, Tahir A, Kiranyaz S, Kashem SBA, Islam MT, Maadeed SA, Zughaier SM, Khan MS, Chowdhury ME (2020) Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images.
Chowdhury MEH, Rahman T, Khandakar A, Mazhar R, Kadir MA, Mahbub ZB, Islam KR, Khan MS, Iqbal A, Al-Emadi N, Reaz MBI, Islam MT (2020) Can AI help in screening viral and COVID-19 pneumonia? IEEE Access 8:132665–132676
Tawsifur R, Chowdhury M, Khandakar A (2021) https://www.kaggle.com/tawsifurrahman/covid19-radiography-database (version 4) [online]
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Rathod, L., Jain, H., Kaliappan, J., Suganthan, C. (2022). Multiclass Image Classification of COVID-19 Chest X-ray Scans Using Deep Learning. In: Singh, P.K., Wierzchoń, S.T., Chhabra, J.K., Tanwar, S. (eds) Futuristic Trends in Networks and Computing Technologies . Lecture Notes in Electrical Engineering, vol 936. Springer, Singapore. https://doi.org/10.1007/978-981-19-5037-7_38
Download citation
DOI: https://doi.org/10.1007/978-981-19-5037-7_38
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-19-5036-0
Online ISBN: 978-981-19-5037-7
eBook Packages: Computer ScienceComputer Science (R0)