Abstract
Brain tumor segmentation methods using deep neural networks have recently achieved significant performance breakthroughs. However, the existing brain tumor segmentation networks are directly implemented on whole brain images, resulting in possibly reduced segmentation performance due to the disturbance of background regions. To solve this problem, inspired by the Mask R-CNN, a novel brain tumor segmentation model called BrainSeg R-CNN is proposed in this work, which classifies the brain tumor areas and boundaries based on the detected region of interest in an end-to-end manner to achieve segmentation result. Also, an effective feature extraction strategy is presented in BrainSeg R-CNN, which in detail extracts various kinds of information from separate channels for each modality and immediately adopts a cross-connection operator to realize the information transmission among different channels. Moreover, concatenation and add calculation are integrated to improve the fusion efficiency of multi-scale features from brain tumor images. Additionally, a multi-weighted and multi-task loss function which fully considers tumor size and overlap label is introduced, significantly improving the segmentation performance. Experimental results on BraTS 2017 dataset demonstrate that our BrainSeg R-CNN obtains competitive performance with state-of-the-arts.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
With the rapid development of deep learning in the field of medical imaging, brain tumor segmentation task, as a key step in brain function analysis and disease diagnosis, has also made a major breakthrough in recent years [1, 2]. The initial deep segmentation networks take brain tumor segmentation as patch classification problem, mainly employing typical convolutional neural networks (CNN) architectures in visual classification task. Also, sliding window and post processing are adopted to achieve the entire segmentation result. The main disadvantages of these methods lie in redundant calculations and global information loss. Then, fully convolutional networks (FCNs) are introduced to provide a pixel-to-pixel solution for brain tumor segmentation with effective expansion of receptive fields, leading to the superior segmentation accuracy and efficient calculation reduction [3, 4]. Specially, an evolutionary version of FCN called UNet [5, 6], which well integrates high-level and low-level features of medical images and achieves significant performance improvement in a variety of medical segmentation tasks, gradually becomes the mainstream of brain tumor segmentation methods. To further improve its segmentation performance, residual module [7], attention mechanism [8] and multi-scale fusion cascade ideology [9] are injected into the baseline model, which largely promotes the development of brain tumor segmentation methods. Although promising segmentation performance has been achieved, existing brain tumor segmentation networks [10,11,12,13] are directly performed on whole images, resulting in possibly reduced segmentation performance due to the disturbance of background regions.
To resolve this problem, inspired by the recent Mask R-CNN [15], a small and flexible object instance detection network with a segmentation branch for natural images, we propose a novel brain tumor segmentation model named BrainSeg R-CNN in this work. BrainSeg R-CNN classifies brain tumor areas and boundaries based on the detected region of interest (RoI) in an end-to-end manner to achieve segmentation result, providing a new pipeline for brain tumor segmentation. In addition, an effective feature extraction strategy is given in BrainSeg R-CNN, and it in detail extracts various kinds of information from separate channels for each modality with cross-connection operator to realize the information transmission among different channels. Also, concatenation and add calculation are integrated to improve the fusion efficiency of multi-scale features from brain tumor images. Moreover, a multi-weighted and multi-task loss function which fully considers tumor size and overlap label is introduced, and it significantly improves the segmentation performance. The proposed BrainSeg R-CNN is extensively evaluated in the brain tumor segmentation challenge (BraTS) [16], and experiment results illuminate that it gains competitive performance with state-of-the-arts. Specially, it achieves the whole tumor segmentation accuracy of 91.54% in slices with brain tumors. The overall architecture of the proposed BrainSeg R-CNN is illustrated in Fig. 1. The main contributions of this work are three folds: (1) A novel brain tumor segmentation network called BrainSeg R-CNN is proposed, which significantly distinguishes from the existing networks for this task. (2) BrainSeg R-CNN introduces effective feature extraction and fusion strategies as well as an effective loss function for brain tumor segmentation, largely improving the performance of the network. (3) Experimental results on a widely used dataset demonstrate its competitive performance with state-of-the-arts.
Overview of BrainSeg R-CNN. It is mainly comprised by feature learning, contextual fusion and network head. It employs multi-channel and cross-modality connection to extract more discriminate features, followed by an improved feature pyramid structure for contextual fusion. An extra Dice loss is introduced on the top of network in parallel with other losses.
2 Method
The BrainSeg R-CNN is mainly inspired by the Mask R-CNN to provide a novel pipeline for brain tumor segmentation task. It adopts the similar two-stage procedure as Mask R-CNN. Differently, as shown in Fig. 1, our BrainSeg R-CNN consists of three different parts, i.e., feature learning, contextual fusion and network head, aiming at gaining superior performance for this task.
2.1 Mask R-CNN
Here, we briefly review the Mask R-CNN [15] that is highly related to our work. Mask R-CNN takes advantage of the principle of Faster R-CNN [17] while introducing the extra mask branch so that it can predict object mask on RoI generated by region proposal network (RPN) for fast instance segmentation. Besides, Mask R-CNN improves the coarse spatial quantization of RoIPool in Faster R-CNN and alternatively proposes the quantization-free layer RoIAlign for avoiding misalignment. Mask R-CNN has provided strong baselines for multiple vision tasks such as human poses estimation and instance segmentation. As such, we follow the similar principle to deal with brain tumor segmentation task. Unfortunately, compared to natural image tasks, medical image tasks face almost very different situations, such as multi-modality images, fewer labeled samples as well as various instance shapes. Therefore, Mask R-CNN cannot be directly transferred to the brain tumor segmentation task, and we have to redesign the architecture to fit for this task.
2.2 BrainSeg R-CNN
Multi-path and Cross-Modality Feature Learning. Although four modalities (T1, T1c, T2 and Flair) contain spatially and semantically similar information, they describe brain tumor from different views and provides complementary information to each other. Effective feature learning will provide better representation of brain tumor image for following segmentation of RoI. Meanwhile, in the family of mainstream CNN models, different convolutional layers capture different visual features and varying scales information. The backbone models encode the entire input or larger feature maps spatially in lower layers, thereby harvesting finer spatial information for pixel-wise segmentation. However, due to the local convolution with small receptive fields, lower layers have poor semantic capturing capability. In higher layers, the stacked multiple convolutional layers progressively sense the entire input with larger receptive view and possess strong semantic information, but the outputs of higher layers are spatially coarse after the downsampling. Overall, the lower layers provide more accurate spatial characteristics while the high ones predict more accurate semantic labels. To this end, we design the effective features learning strategy from multi-path and cross modality, combining the inherent merits of varying convolutional layers and complementary information of four modalities.
To achieve that goal, the four modalities are separately fed into four CNN models, shown in Fig. 1(a), from left to right are T1, T1c, T2 and Flair, respectively. Motivated by the shortcut in ResNet, the features in the i-th level from T1 are combined with features in j-th (\(j=i+1\)) level from T2 though element-wise addition. Note that the two feature maps always have different spatial size. We conduct extra convolution with downsampling on the larger one, making them have same size. The resulting features then pass though the next convolutional layer. For other modalities, we repeat the similar operation. In this way, each modality integrates features of every level from one or more adjacent modalities except the first T1. The network not only learns features from individual CNN model and modality, but also gets multi-scale and cross-modality features, fully considering the interaction among modalities to obtain discriminative features of brain tumor. Besides, all features of the i-th level of every modality are concatenated along the channel dimension to form a new feature map to characterize brain tumor at i-th level, fed into next contextual fusion part.
Feature Pyramid Structure Based Contextual Fusion. To get better global contextual information, we present an improved feature pyramid structure to fuse features gained from feature learning period under different pyramid resolutions, depicted in Fig. 1(b). After feature learning, we get concatenated feature maps of each layer. Here the number of channels and spatial size per concatenated feature maps are different. The feature maps at deeper layers get more small spatial size with more channel number. We first perform bottleneck block on them to give them the same dimension. The UAC block is then carried out to fuse features, which primarily involves Upsampling, Add and Concatenation operations (UAC) as shown in Fig. 2.
In UAC block, given two inputted feature maps from adjacent i-th and j-th levels, denoted respectively as \(\mathbf{A} \) and \(\mathbf{B} \), the low resolution feature map \(\mathbf{B} \) is \(2\times \) bilinear upsampled, producing feature map \(\mathbf{B}* \), to match the spatial size with high resolution \(\mathbf{A} \) followed by \(1\times 1\) convolutional layer. The resulting \(\mathbf{B}* \) and \(\mathbf{A} \) are added in element-wise manner, obtaining feature map \(\mathbf{C} =\mathbf{B}* +\mathbf{A} \). The added feature map \(\mathbf{C} \) then are concatenated with feature map \(\mathbf{A} \), getting new map \(\mathbf{D} =\left[ \begin{array}{cc} \mathbf{A} ,&\mathbf{C} \end{array} \right] \), which contains global and local information with stronger semantic and finer spatial resolution, particularly helpful for segmentation. Subsequently, the fused feature maps \(\mathbf{D} \) are connected to one bottleneck block for feature adaption. From the deepest layer to the shallowest layer, we keep repeating above operation progressively. The outputs of all UAC blocks hold the same dimension but have different resolutions. We upsample all of them up to the same resolution as the largest with different times ratio except the shallowest one. After that, we combine them with concatenation along the channel direction. The final fused features go though vanilla RPN to generate RoI of brain tumor, and produced each RoI is fed into the network head for bounding-box recognition and mask prediction.
The basic structure of the given UAC block. The UAC block is designed to fuse multi-channel features, which primarily involves Upsampling, Add and Concatena-tion operations. The outputs of UAC block holds the same dimension but has different resolutions. Additionally, it contains global and local information with stronger semantic and finer spatial resolution, which will be helpful for brain tumor segmentation.
Network Head. Our network head is similar in structure to Mask R-CNN, focusing on the guidance of training by loss function. However, due to the high similarity between tumors and tissues, their various shapes and small size, the loss function employed in Mask R-CNN actually pays too less attention on desired tumor regions, possibly resulting in poor segmentation performance and unsuitable for brain tumor segmentation task. Therefore, following [14], BrainSeg R-CNN adds a multi-weighted loss function in conjunction with ones of Mask R-CNN in parallel fashion for brain tumor segmentation (Fig. 1(c)). The total loss is defined as following:
where \(L_{rpn}\), \(L_{cls}\) and \(L_{box}\) are identical as Mask R-CNN, which are used to train the branch of detection. \(L_{mask}\) means the average binary cross-entropy loss, and \(L_{dice}\) is the added Dice loss to optimize segmentation branch. \(\lambda _{i}\) (\(1 \le i \le 4\)) is the hyper-parameter that controls the importance of each loss.
3 Experiments
3.1 Dataset and Settings
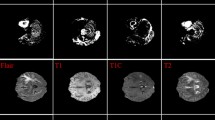
We evaluate the proposed BrainSeg R-CNN on the commonly used BraTS 2017 dataset. For each MRI image, there are four modalities: FLAIR, T1-weighted (T1), T1 with gadolinium enhancing contrast (T1c), and T2-weighted (T2). The dimensions of all images are \(240 \times 240 \times 155\) voxels. The BraTS 2017 training set is composed of 210 cases of high-grade gliomas (HGG) and 75 cases of low-grade gliomas (LGG). Each ground-truth for brain tumors is given by experts [18, 19]. Here, we divided the original training set into three subsets for model training, validation and testing, respectively. Figure 3 demonstrates two typical multi-mode brain tumor image samples in BraTS 2017 dataset.
Our experiments mainly consist of two parts: (1) Compared experiments using slices with tumors; (2) Compared experiments using all slices (whole brain image). As the BrainSeg R-CNN is based on the detection model, which will result in a higher level of false positive for slices without brain tumors. Therefore, the first part of our experiments is carried out on slices which definitely contain brain tumors to verify the effectiveness of BrainSeg R-CNN, specially to evaluate the three designed parts, i.e., feature learning, contextual fusion and network head. In the second experiment, we compare BrainSeg R-CNN with several state-of-the-art methods by using whole brain image with the same protocol as [20]. Moreover, Dice score is adopted in all of the experiments.
3.2 Compared Experiments Using Slices with Tumors
Comparison with Mask R-CNN. Here, we take Mask R-CNN architecture without multi-path and cross-modality feature (MCF), multi-scale fusion (MF) and multi-weighted dice (MD) loss as our naive baseline. Based on the different combinations of adding MCF, MF and MD, we conduct a series of comparative experiments on BraTS 2017 dataset whose results are reported in Table 1. As shown in Table 1, by introducing MCF and MF as well as the MD loss, our BrainSeg R-CNN achieves the optimal segmentation performance of 91.54%, 86.22% and 81.05% on whole, core and enhance tumors, which outperforms that of Mask R-CNN over 5.58%, 6.10% and 2.85%, respectively. In addition, following conclusions can be drawn from Table 1. All of the MCF, MF and MD gain performance improvement over the baseline. Among them, MF is superior to the others while MCF achieves the smallest effect. Further performance improvements can be achieved through the combination of MCF, MF and MD.
Comparison with U-Net Models. Here, we mainly compare BrainSeg R-CNN with several typical 2D U-Net models including basic U-Net, Res-UNet and Res-UNet with weighted-Dice (Res-UNet+WD) on BraTS 2017 dataset to give a further evaluation, and the compared results are shown in Table 2. Table 2 illuminates that BrainSeg R-CNN achieves promising performance improvement over basic U-Net and Res-UNet. Compared with Res-UNet+WD, BrainSeg RCNN respectively gains 3.03% and 0.88% performance improvement on whole and enhance tumor segmentation results. Meanwhile, it is inferior to Res-UNet+WD on core tumor segmentation. However, the overall experimental results demonstrate the effectiveness of our BrainSeg R-CNN method for brain tumor segmentation.
3.3 Compared Experiments Using Whole Brain Image
To further test BrainSeg R-CNN, we compare it with several state-of-the-art methods on all slices (whole brain image) with the same setting as [20], and experiment results on BraTS 2017 dataset are given in Table 3. Among them, dense FCN (DFCN) employs typical 2D FCN model and introduces dense connection to improve the segmentation accuracy [20]. In contrast, FCN+CRF adopts 2D FCN model followed by conditional random field (CRF) as post processing [12, 13]. As BrainSeg R-CNN is based on detection model, it will result in a high level of false positive for slices without brain tumors. However, this problem can be resolved by adding a pre-classifier before feature learning. Here, we take U-Net as the pre-classifier and denote this method as BrainSeg R-CNN+Classifier.
Table 3 illustrates that BrainSeg R-CNN overall outperforms DFCNN, FCN+ CRF and U-Net methods. Due to the high false positive on slices without tumors, it is inferior to the Res-UNet+WD method. However, with a simple pre-classifier as supplement, our BrainSeg R-CNN+Classifier obtains the optimal performance for both of whole and enhance tumor segmentation. Specially, it gains 91.22% Dice score for whole tumor segmentation, which is significantly higher than the others.
4 Conclusion
In this paper, inspired by Mask R-CNN, we propose a novel brain segmentation method called BrainSeg R-CNN, which classifies brain tumor areas and boundaries based on the detected RoI to finish segmentation, avoiding invalid segmentation calculation in the background area as well as providing a new pipeline for this task. Additionally, three improvements are presented in BrainSeg R-CNN to achieve better segmentation performance. Extensive experiment results on widely used brain tumor segmentation dataset demonstrate the effectiveness of our proposed BrainSeg R-CNN method. In the future, the more powerful pre-classifier will be integrated into current BrainSeg R-CNN model to further improve its performance on the entire brain image. In addition, we will extend the proposed BrainSeg R-CNN method into 3D model, and this could further avoid the wrong detection existing in 2D method.
References
Bakas, S., Reyes, M., Jakab, A., et al.: Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS Challenge. arXiv preprint arXiv:1811.02629 (2018)
Tiwari, A., Srivastava, S., Pant, M.: Brain tumor segmentation and classification from magnetic resonance images: review of selected methods from 2014 to 2019. Pattern Recogn. Lett. 131, 244–260 (2020)
Shen, H., Wang, R., Zhang, J., et al.: Multi-task fully convolutional network for brain tumour segmentation. In: Annual Conference on Medical Image Understanding and Analysis (MIUA), pp. 239–248 (2017)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE conference on Computer Vision and Pattern Recognition (CVPR), pp. 3431–3440 (2015)
Dong, H., Yang, G., Liu, F., et al.: Automatic brain tumor detection and segmentation using U-Net based fully convolutional networks. In: Annual Conference on Medical Image Understanding and Analysis (MIUA), 506–517 (2017)
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), pp. 234–241 (2015)
Kermi, A., Mahmoudi, I., Khadir, M.T.: Deep convolutional neural networks using U-Net for automatic brain tumor segmentation in multimodal MRI volumes. In: International MICCAI Brainlesion Workshop (BrainLes), pp. 37–48 (2018)
Zhang, J.X., Jiang, Z.K., Dong, J., et al.: Attention gate ResU-Net for automatic MRI brain tumor segmentation. IEEE Access 8, 58533–58545 (2020)
Zhou, C., Ding, C., Lu, Z., et al.: One-pass multi-task convolutional neural networks for efficient brain tumor segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), pp. 637–645 (2018)
Mlynarski, P., Delingette, H., Criminisi, A., et al.: 3D convolutional neural networks for tumor segmentation using long-range 2d context. Comput. Med. Imaging Graph. 73, 60–72 (2019)
Tseng, K.L., Lin, Y.L., Hsu, W., et al.: Joint sequence learning and cross-modality convolution for 3d biomedical segmentation. In: IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 6393–6400 (2017)
Kamnitsas, K., Ledig, C., Newcombe, V.F.J., et al.: Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 36, 61–78 (2017)
Zhao, X., Wu, Y., Song, G., et al.: A deep learning model integrating FCNNs and CRFs for brain tumor segmentation. Med. Image Anal. 43, 98–111 (2018)
Sudre, C.H., Li, W., Vercauteren, T., et al.: Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In: International Workshop on Deep Learning in Medical Image Analysis & International Workshop on Multimodal Learning for Clinical Decision Support (DLMIA & ML-CDS), pp. 240–248 (2017)
He, K.M., Gkioxari, G., Dollár, P., et al.: Mask R-CNN. In: IEEE International Conference on Computer Vision (ICCV), pp. 2961–2969 (2017)
Menze, B.H., Jakab, A., Bauer, S., et al.: The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 34(10), 1993–2024 (2015)
Ren, S.Q., He, K.M., Girshick, R., et al.: Faster R-CNN: towards real-time object detection with region proposal networks. In: Advances in Neural Information Processing Systems (NIPS), pp. 91–99 (2015)
Bakas, S., Akbari, H., Sotiras, A., et al.: Advancing the cancer genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Scientific Data 4(1), 1–13 (2017)
Bakas, S. Akbari, H., Sotiras, A., et al.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-GBM collection. Cancer Imaging Arch. 286 (2017). https://doi.org/10.7937/K9/TCIA.2017.KLXWJ
Shaikh, M., Anand, G., Acharya, G., et al.: Brain tumor segmentation using dense fully convolutional neural network. In: International MICCAI Brainlesion Workshop (BrainLes), pp. 309–319 (2017)
Acknowledgements
This work was partially supported by the National Natural Science Foundation of China (61972062), the National Key R&D Program of China (2018YFC0910506), the Key R&D Program of Liaoning Province (2019 JH2/10100030), the Young and Middle-aged Talents Program of the National Civil Affairs Commission, the Liaoning BaiQianWan Talents Program, and the University-Industry Collaborative Education Program (201902029013).
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Zhang, J., Cheng, X., He, T., Liu, D. (2021). BrainSeg R-CNN for Brain Tumor Segmentation. In: Wang, Y., Song, W. (eds) Image and Graphics Technologies and Applications. IGTA 2021. Communications in Computer and Information Science, vol 1480. Springer, Singapore. https://doi.org/10.1007/978-981-16-7189-0_17
Download citation
DOI: https://doi.org/10.1007/978-981-16-7189-0_17
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-7188-3
Online ISBN: 978-981-16-7189-0
eBook Packages: Computer ScienceComputer Science (R0)