Abstract
RNA-binding proteins (RBPs) play very crucial role in various physiological and biochemical functions of cells. It is a well-known fact that proteins are the key component of cell through which various functions of cells are regulated as they are receptors, adaptors, and enzymes. However, their activities are also regulated by different types of RNA. There are different types of noncoding RNA in addition to coding RNAs like mRNA, tRNA, and rRNA. The noncoding RNA bind with protein in a very specific manner identifying specific sites, motifs, or its structure. The interactions of RNAs with proteins are the main cause of various biological functions of cells. In this chapter, various aspects of RNA–protein interactions have been discussed, like structures of RBPs, types of RPI, functions of RBP, and various approaches to understand RNA–protein interaction.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
RNA molecules exhibit a wide range of form and function. RNAs have been categorized based on their coding ability into two major groups: protein coding messenger RNAs (mRNAs) and noncoding RNAs (ncRNA) [1]. To initiate protein synthesis, mRNA molecules act as scaffolds for additional details. ncRNAs are classified depending on the sequences, intracellular localizations, structures, and functions, as follows: rRNAs and tRNAs, that are core elements of the translation system; [2, 3] small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs) are involved in splicing of RNA and its modification [4]. Further, developments in deep sequencing have demonstrated that at least 80% of mammalian genomes produce RNAs, and scores of new ncRNAs have been discovered in living organisms that play undefined roles [5, 6]. However, the underlying mechanisms of these roles have remained elusive. RNA-binding proteins (RBPs) play a significant role in the RNA life cycle like its synthesis, function, and turnover. During all three phases of the RNA life cycle, such roles are always accompanied by involvement with RNA-binding proteins, including synthesis, function, and turnover [7]. RBPs bind directly to RNA sequences and/or structures with its RNA-binding domains in order to make decisions about RNA fate and function.
Interactions between proteins and RNA are the basis of various functions like organization and protein complexes stabilization, mRNA processing and maturation for trafficking and silencing and stabilization of matured mRNA. RBP could recognize single-stranded RNA, double-stranded RNA, structural characteristics of folded RNAs, or may not interact RNA explicitly unlike DNA binding proteins that usually bind double-stranded DNA [8].
RNA–protein interactions (RPI) regulates essential biological processes such as DNA replication, transcription, tolerance to pathogens, viral replication, and gene expression regulation at the posttranscriptional level. Recent high-throughput research has indicated various cellular RNA-binding proteins and are recognizing and characterizing pairs of proteins and RNAs that are involved in RPIs. However, our knowledge regarding RNA-binding proteins is far less in comparison to regulatory DNA-binding proteins, like replication factors and transcription factors. Most computational studies have dealt with the problem of predicting the positions amino acid residues present in a protein that may bind to an RNA.
Till date, there are very limited studies that have focused on the issue of partner prediction, i.e., characterization of specific RNA for an already known RNA-binding protein or protein-binding partner(s) required for nontranslating RNAs. Although many studies like as RIP-Chip, RNA compete, PAR-CLIP, and HITS-CLIP may offer critical information on RNA–protein interaction, they are limited by their high cost and labor-intensive nature. Computational techniques are thus required to correctly predict RPIs and design networks of RNA–protein interaction. It would be particularly helpful to establish sequence-based approaches that can be employed to recognize potential RNA–protein partners without the need for any experimental interactions, because there are only a small number of known RNA protein complexes in the PDB [9].
2 About RNA-Binding Proteins: Structure, Diversity, and Evolution
The majority of RBPs are proteins with a globular RNA-binding domain that binds RNA, which modifies the fate or function of the bound RNA. Some assume that unique and high-affinity RBPs are more likely to possess biological functions. This popular conception of RBPs, though, assumes that they seek to modify the outcome or functionality of RNA. The RBPs are identified as “the mRNA’s clothes.” This makes sure that the 5′ and 3′ UTRs and the coding region are in separate states: one time hidden, the next time exposed, enabling the mRNA to pass through different life stages [7]. Ribonucleoprotein (RNP) complexes that are primarily involved in gene expression consist of a traditional RNA-binding protein (RBP). For RBP function, it utilizes well-defined RNA-binding domains such as the RNA recognition motif (RRM), KH domain, or DEAD-box helicase domain. Additionally, complex protein–RNA interactions can be found in various unconventional RBP types, such as those that employ RNA-binding domains [7]. Four main RNA–protein interactions have been proposed on the basis of fundamental features of RNAs like structure, sequence, modification, and target engagement, as well as the recognition mechanism of RBPs [10].
2.1 RNA–Protein Interactions Based on RNA Motif
RNA motifs are short sequences which regulate the fate of RNA and cellular processes. Interaction of RNA and proteins usually involves modular combination of one or more RBD like RNA recognition motifs (RRMs), hnRNP K-homology (KHs), PUM-homology domains (PUM-HD), and dead box proteins (DDXs). One of the best examples including this principle is the RRM domains of RBFOX2 that bind to a UGCAUG motif [11], while PUM2 take the help of PUM-HD to bind with UGUANAUA [11, 12].
The discovery of an increasing number of RBP-binding motifs has also exposed the intricacies of RNA–protein interaction that depends on RNA motifs. A single RBP possess a variety of binding motifs as in the case of LIN28 where, N-terminal cold shock domain (CSD) and the C-terminal zinc knuckle domain (ZKD) play a part in the binding of two different RNA motifs, namely the ‘GGAG’ motif and the ‘(U) GAU’ motif [13]. In posttranscriptional regulations, LIN28 impedes the biosynthesis of let-7 miRNAs, regulating production and impacting various disease states [13]. Also, the “insulin-like growth factor 2 mRNA-binding protein 1” (IGF2BP1) is one of such RBPs that could bind several motifs. IGF2B P1 is more complex protein than LIN28 as it contains four domains of hnRNPK homology (KH) and two motifs for RNA recognition.
In addition to the number as well as sequence-specific RBP-binding motifs, RNA motif-based RNA–protein interactions occur along with the motif’s flanking sequences. RNA motifs are especially well-suited for RBP-specific interactions, where RNA motif-dependent RNA–protein interactions often allow the use of motif contexts and other RBP-specific interactions.
2.2 RNA–Protein Interactions Based on RNA Structure
Typically, RBPs bind to small sequences of single-stranded RNA, but some RBPs perform their biological activities by interacting on the basis of their common structural characteristics with groups of RNAs, including secondary and tertiary structural characteristics [14]. RNA sequences could fold into various secondary structures, including long stems with bulges or hairpins through base pairing. After complementary base pairing, double-stranded RNA (dsRNA) can fold into various structures such as hairpins and long stems with bulges, known as classic secondary structures. dsRNA is essential in multiple biological functions, including transport of mRNA, editing of RNA, innate immune response, and RNA interference [15]. The detection and operation of RBPs are necessary for all of the process mentioned above. “Double-stranded RBPs” (dsRBPs) are the proteins that bind to dsRNAs and are characterized by the availability of minimum one “double-stranded RBD” (dsRBD).
The ADAR family, which includes dsRBPs of various sizes, all of which possess conserved modular domain organization carrying a catalytic domain at C-terminal, possesses various dsRBD [16, 17]. Though they usually focus sequences with fewer interruptions and under certain sequence constraints, ADAR proteins search out and process dsRNAs with any given sequence [18]. ADAR1/ADAR 2 bind to mRNA and miRNA precursors to promote adenosine to inosine conversion [19, 20]. Mostly conversion from adenosine to inosine occurs in noncoding sequences of mRNAs, like 5′ and 3′ UTRs and retrotransposon elements of introns, such as long interspersed elements (LINEs) and Alu elements. It is also important to point out that multiple biological changes can be caused by A-to-I editing, which can include the possibility to edit pre-mRNA splicing patterns, and thus create new isoforms [21] Many editing sites are present in miRNAs where some of the sites influence synthesis and function of miRNA [22].
dsRNAs and dsRBPs also mediate translation, mRNA, splicing, stability, and degradation of mRNA. STAU1 is a dsRBP that is localized to the rough endoplasmic reticulum. In order to analyze STAU1-bound RNA structures in human cells, researchers used hiCLIP technology to look for structures formed by STAU1 within these samples and found STAU1 to bind mainly to intramolecular RNA duplexes. An RNA duplex that spans 858 nucleotides in the X-box-binding protein 1 (XBP1) mRNA was discovered, which controls splicing and stability in cytoplasm [23]. Depending on their particular three-dimensional tertiary structures, multiple RNAs have important regulatory roles in diverse biological processes.
A helix internal loop helix motif is formed in the double-stranded region by the Kink-turn (K-turn) RNA structure which consists of a three-base loop surrounded by a noncanonical stem (NC-stem) and a canonical stem (C-stem) that starts with a tandem base pair of GA/AG [24, 25]. There are various RNA structures that include the K-turn motif, including box C/D snRNAs, snoRNAs, mRNAs, and rRNAs. Some K-turn motifs are different, but they have the same three-dimensional distinctive shape.
In addition to organized RBDs, there are amino acid sequences in proteins that are not self-structural and need an external molecule to attain secondary structure. These are termed intrinsically disordered regions (IDRs). IDRs may promote RNA–protein interactions [26]. While certain structural characteristics support specific interactions, the RGG/RG motifs of IDR bind RNA through weak multivalent interaction. The fragile X mental retardation protein (FMRP) binds with the secondary structure of the G4 RNA by utilizing the RGG/RG motifs present in an IDR [27]. The interplay of the G4 and FMRP IDR is important for the attachment of several mRNAs and regulates translation control and alternative splicing [28, 29]. Disordered sequences are observed in one-third of the RBPs, with many of these have missing canonical RBDs [30] demonstrating the major role of IDRs in the ability to bind RNA. The recent advancements of RNA structurome and RBDs with respect to variety, dynamics, and expansion have indicated that various facets of regulation in gene expression may be discovered from protein interactions with RNA structure-dependent RNA.
2.3 RNA–Protein Interactions Based on RNA Modification
There are approximately 160 RNA variations that have been discovered to date [31]. A new layer of RNA stability and functional control is provided by the use of nucleotide-base chemical modifications in RNA [32]. Researchers also discovered several RNA mutations associated with human disease, such as cancer and neurological disorders [33]. RNA and protein interaction also occurs by posttranscriptional modifications such as 5-methylcytosine (m5C) and N6-methyladenosine (m6A). There have been further m6A and m5C studies suggesting that these modifications are indispensable in various biological processes. M6A is the most prevalent and reversible RNA modification, which is involved in a number of RNA functions including mRNA polyadenylation, splicing, transport, translation, and degradation. M6A modification is a complex process, and after cellular stress, m6A levels go through a wide-ranging redistribution of the transcriptome. RNA–protein interaction is mediated by m6A methylation of RNA. The newly altered RNA following methylation of m6A, acts as a reactants for m6A-specific interactors, including m6A readers and erasers. YTHDC1–2 and YTHDF1–3 are well known m6A readers that include YT521-B homology (YTH) domain-containing proteins. All of these m6A readers recognize m6A via a non-motif-specific process. Typically, YTHDC2 and YTHDF1–3 are found in the cytoplasm. YTHDF1 is a cytoplasmic protein containing two domains, a C-terminal YTH-binding domain and an N-terminal domain that promotes recruitment of complex 3 (eIF3) translation initiation factor, all of which enables cap-independent translation. The terminal YTH-binding domain of YTHDF2 interacts with m6A mRNA and the CCR4-NOT deadenylase complex is recruited by its N-terminal domain enhances the deadenylation and degradation of mRNA modified at m6A.
Recent research indicates that m6A-modified mRNAs decay and translations are facilitated by YTHDF3. The nuclear reader YTHDC1 recruits and suppresses a pre-mRNA splicing factor called SRSF3. The nuclear reader YTHDC1 recruits the pre-mRNA splicing factor SRSF3, which inhibits accessibility of SRSF10 to m6A-altered mRNAs. This subsequently enables inclusion of exon in specific mRNAs and governs slicing of mRNA. YTHDC1 also communicates with SR SF3, CPSF6, and SRSF7 in the oocyte nucleus to regulate pre-mRNAs and affect fetal growth. M6A modification created on chromatin-associated RNAs are mediated by METTL3 and recognized by YTHDC1, facilitating degradation of these m6A-modified RNAs [10].
2.4 RNA Guide-Bas Ed RNA–Protein Interactions
There are various kinds of small noncoding RNAs, including snRNAs, piRNAs miRNAs, snoRNAs, crRNAs, and other ncRNAs, that help facilitate protein–RNA interactions. In addition to regulating diverse life processes, this mode of RNA–protein interaction helps to control disease growth. However, despite there being some continuity in these interactions, there is notable variation in the structures and roles that various ncRNAs use.
2.4.1 Role of miRNA in RNA–Protein Interaction
miRNAs are one of the small noncoding RNA molecules found in plants, animals, and viruses [34, 35]. Drosha, DGC R8, Dicer, and TRBP are some of the dsRBPs used in the biogenesis of miRNAs [34]. miRNA is inserted into the RISC and binds to the core sequences of the target mRNAs, thus inducing translational repression [36]. siRNAs are made in a way close to that of miRNAs [37]. Dicer cut the DsRNAs or hairpin RNAs into small fragments [38]. At this stage, the guide strand is anchored to AGO2 and other proteins, and RISC is synthesized, which takes mRNA substrates that have a complementary sequence and starts to degrade them [39].
Genes are regulated by miRNAs by base-pairing with mRNAs while preserving complementarity to the seed region of miRNA (2–8 nucleotides) [40, 41]. Two kinds of miRNA–mRNA interactions can be found: canonical and atypical.
miRNAs make base pairs fully with target mRNA during both atypical and canonical matching even if the seed region is located at the 5′ end of miRNA. The mRNA repression is different for these matching process. In one case, endonucleolytic cleavage is activated by key constituents of RISC-AGO2 when miRNAs have a significant complementary matching with the coding sequence or UTR of mRNA targets. While in other cases, proteins directed by miRNA can cause translation inhibition or deadenylation of mRNA, if mismatches between miRNAs and their targets are observed [42, 43].
2.4.2 RNA–Protein Interactions Guided by piRNA
A new category of small noncoding RNAs known as piRNAs has been discovered in the male gametes of animals [44]. piRNAs are 30 nucleotides long (26–31 nucleotides). Murine PIWI (MIWI), which includes Aub, AGO3, and piwi [45], are also linked with PiRNAs of the PIWI subfamily, and piRNA guides the PIWI proteins to play a critical role in the silencement of transcriptional and posttranscriptional transposons and to defend themselves against the regeneration of viral stem cells [45]. Almost every species relies on this mechanism to prevent transposons from being expressed in their genome of gametes. Additionally, piRNA-directed nuclear PIWI proteins associate with nascent transposon transcripts to produce heterochromatin by DNA or histone methylation, ultimately leading to transcriptional silencing [46, 47]. Mosquitoes mount an antiviral response based on piRNA whenever they are infected with positive sense ssRNA virus. Piwi5 and Ago3 are precursors of piRNAs, and the heterotypical ping pong system synthesizes piRNAs. Thus, as the number of piRNAs increases, RNA virus replication is suppressed, achieving the antiviral response target [48]. The entire process is supervised by piRNA. Additionally, piRNAs participate in the metabolic activities of PIWI and facilitate its degradation [49]. Recently published research indicates that piRNAs obtained from transposons and pseudogenes can degrade specific mRNAs as well as lncRNAs through interaction with PIWIL1L [50]. In addition, degradome sequencing [50] also provides a systematic method of analyzing RNA degradation patterns mediated by piRNA and has significantly expanded insight into the interaction of universal piRNA-guided RNA–protein.
2.4.3 RNA–Protein Interactions Based on SnoRNA Guide
SnoRNAs are a group of highly expressed ncRNAs present in archaeans and eukaryotes, mainly located inside nucleolus. They are derived from pre-mRNA introns having a size of 60–300 nt. SnoRNAs can be classified as box H/ACA or box C/D snoRNAs based on their conserved sequence. The motifs of box C (RUGAUGA) and D (CUGA) are combined with less conserved box C and box D motifs to form a stem-internal loop-stem structure. The folding of the box H/ACA snoRNAs results in a distinctive hairpin-hinge-hairpin-tail arrangement, with box H (ANANNA) situated amongst the two hairpins and ACA motifs near the 3′ end. A subclass of snoRNA named Cajal body-specific RNAs (scaRNAs) have been found extensively in Cajal bodies where both C/D box and a H/ACA box domain are present. snoRNA performs various types of functions which include guidance of chemical modification in rRNAs and snRNAs in sequence-specific manner. Box C/D snoRNAs mediate 20-O-methylation inside SNORD-ribonucleoprotein (RNP) complexes, 20-O-methylation in ribose present in snRNA, and rRNA is capable of affecting its production and function, which could have an effect in cellular processes and diseases [10].
SNORA-RNP complexes are created by a combination of box H/ACA snoRNA with DKC1, NHP2, GAR1, and NOP10 that catalyse the conversion of uridine to pseudouridine located at 15 nt upstream of boxH/ACA. Box H/ACA snoRNAs instruct rSNORA-RNPs to modify uridine residues on snRNAs necessary for RNA splicing as well as uridine residues on rRNAs. Apart from directing RNA modification, SnoRNA has been used to facilitate pre-rRNA and pre-mRNA alternative splicing processing [10].
2.4.4 Spliceosome Assembly and Function Using snRNA Guides
The spliceosome is constructed stepwise from components such as pre-mRNAs, proteins, and snRNAs. Specifically, snRNAs act as guides, leading each snRNP to its final destination. There are five distinct types of RNA–protein interactions relying on snRNA guide, as per the type of snRNA involved in RNA splicing: U1 snRNP:: 50-splicing site (50SS) interacting ions, U2 snRNP:: branch point sequence (BPS) interactions, At the 5′ and 3′ splice sites, U6 snRNP:: 50SS interactions, U6 snRNP:: U2 snRNP interactions, and U5 snRNP:: exonsequence interactions [10]. U1 snRNP, the first snRNP to bind to precursors of splicing, identifies mRNA precursors with high specificity through base pairing between 50SS and U1 snRNA bases 3–10. In eukaryotes, the interaction of pre-mRNAs and snRNPs led by U1 is extremely conserved and necessary for splicing. Recent studies, however, have identified U1 as a unique mutated gene in chronic lymphocytic leukemia, hepatocellular carcinoma (HCC), and hedgehog medulloblastoma. The first base of the U1 50SS recognition sequence contains significant mutations A > G and A > C, implying the splicing patterns of different cancer pathways [10]. After recognition by U1 snRNP, U2 snRNP binds to the BPS of a pre-mRNA through a base-pairing interaction between the U2 snRNA and BPS. The tri-snRNP U4/U 6.U5 then participates in spliceosome assembly and the substitution of U1 snRNPs. Finally, the U6 snRNP interacts via base pairing with the 5′ end of the intron and the U6 snRNA. Additionally, the U5 snRNA binds to the exon sequence at the 5′ and 3′ splice sites and is involved in trans-esterification reactions [10].
2.4.5 RNA Targeting by the Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)-Cas System Based on RNA
CRISPRs are bacteria and archaea-specific repetitive sequences that play a crucial role in prokaryotes’ RNA-based adaptive immune systems. They were first used in research on DNA and genome editing. System of CRISPR/Cas9 and novel CRISPR/Cas have been developed to achieve accurate RNA targeting, restriction, monitoring, and editing in mammalian cells. As in case of CRISPR/SpyCas9 (Streptococcus pyogenes Cas9), specially engineered PAMmers can be used to direct Cas9 to selectively bind or cut RNA targets while avoiding matching sequences of DNA. Additionally, the integration of PAMmers and deactivated Cas9 (dCas9) allows monitoring of RNA in living cells without the use of genetically programmed tags, avoiding the use of microsatellite repeat RNA expansion sequences. Besides SpyCas9, some Cas9 homologs derived from other bacterial organisms, such as SauCas9, NmeCas9, and CjCas9, are capable of attaching and breaking intracellular RNA in a PAM-independent manner. Cas3a, Cas13b, and CasRx, all Class 2 type VI CRISPR-Cas effectors, are customizable singular RNA-targeting RNases directed by RNA.
Cas13a has been engineered to target and monitor endogenous RNAs in plant and mammalian cells. When compared to RNA interference, the CasRx ribonuclease effector derived from Ruminococcus flavefaciens XPD3002 exhibits high specificity and efficiency against a wide variety of endogenous transcripts. Its inactive type (d CasRx) can be used to modulate alternative splicing and relieve dysregulated tau isoform ratios in a neuronal model of frontotemporal dementia. REPAIR and RESCUE, both based on Cas13b, were also developed and used to modify RNA from A to I and C to U. CRISPR/Cas inspired RNA targeting system (CIRTS) is a new RNA engineering toolkit that was recently developed by researchers. It is composed of a tri-domain protein with a single-strand RNA-binding domain, a hairpin RNA-binding domain, and an effector domain, as well as a designed gRNA with a hairpin and a single strand. The discovery of the CRISPR-gRNA system provided new insights into ncR NA-mediated RNA–protein interactions. Along with protein engineering, the CRISPR-gRNA system has enormous potential for research and gene editing, especially for gene therapy [10].
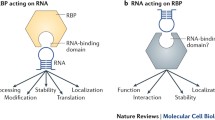
3 Functional Roles of RBPs
3.1 mRNA Localization
Genes can be regulated by localization of mRNA to various subcellular locations [51, 52]. The efficiency and temporal resolution of protein synthesis is enhanced by mRNA trafficking, triggered by cellular signals. Additionally, it facilitates the synthesis of protein complexes by increasing the localized concentration of particular mRNA.
Localization of mRNAs involves three different mechanisms [53, 54]: (1) mRNA-directed transport, (2) local selective stabilization, and (3) local trapping. Different RBPs are required to recognize separate localized signals in the mRNAs. Signals of localization for active and direct transport usually seem as synergistic clustered secondary structure repeats [55,56,57], whereas some similar signals seem to be available in the primary sequence [58, 59]. Various localizing RBPs interact with the UTRs of localized mRNA separately with low specificity and affinity [60]. Multiple RBPs interacting cooperatively is considered important [61]. The effect of RBP-mediated defense on a single cellular position results in selective stabilization.
The well-studied example is Hsp83, whose deadenylation and degradation in Drosophila is controlled by the 3′ untranslated regions (3′ UTRs)-bound Smaug RBP with the exception of the posterior pole, where it localizes embryos. Diffusion and local trapping are used by the third mechanism. However, due to its moderate efficiency to limit mRNAs spatially, selective stabilization normally occurs, similar to the localization of Nanos mRNA at the posterior pole in Drosophila embryos [62].
3.2 Translation of mRNA
Regulation of translation can take place by changes to the translational machinery, or it can specifically target specific mRNAs. RBP-based modulation, an intriguing regulatory mechanism, enables mRNA-specific control of the basic translational machinery [63]. For example, mRNA-specific RBPs can obstruct the interaction between the mRNA and the ribosome 43S complex by physical blockage in a cap-dependent pathway [64] or arrest by 43S scanning in a cap-independent pathway, as observed in Drosophila msl-2 mRNA by SXL [65,66,67]. On the other hand, specific mRNAs are suppressed by global eIF4E structural adaptors, as seen in the case of Bruno and Smaug RBPs, which promote the blockage of Cup and Maskin eIF4E adaptors on nanos, oskar, and poly (A)-tailed mRNAs [68,69,70]. RBPs can also regulate translation at a later phase of initiation steps, by prohibiting the linking of ribosomal subunits [71], or after initiation stages, as demonstrated by the hnRNP E1 RBP, which inhibits ribosomal subunits [71] Dab2 and ILEI at the extension phase by attaching to the 3′ UTR [72, 73].
A group of RBPs recognize aberrant mRNAs as opposed to normal mRNAs in the translation-dependent quality control process, which is coupled with a degradation mechanism to turn on the translation machinery. Cytoplasmic polyadenylation [74] is another effective mechanism for regulating translation. RBPs are thought to serve as “place-markers” in the assembly of catalytic complexes on the poly (A) dynamic combinatorial code in several models.
3.2.1 Degradation of mRNA
In addition to RNA maturation, several different degradation mechanisms, RNA maturation, and regulated mRNA turnover are all involved in quality surveillance. RBP protects nuclear RNA quality by exporting and degrading abnormal RNA in the cytoplasm or adenylation through nuclear TRAMP and exosome-mediated 3′-5′ decay [75, 76].
Surveillance of cytoplasm is either achieved by “nonsense-mediated decay” (NMD) when aberrant stop codons are found or by “ribosome extension-mediated decay” (REMD) when translation extends beyond the stop codons. NMD, for example, includes the “exon-junction complex” (EJC), “poly-A binding protein 1” (PABPC1), and HRP1 to identify regulatory sites in mRNA decay substrates. RBPs may also function as adaptors, as evidenced by Upf1, which is involved in the development of the SURF complex and subsequent association with EJC [77]. Additional RBPs, such as Pub1 [78], the “APOBEC1–ACF editing complex” [79], and several 3′ UTR helicases or chaperones [80] provide selective control of decay performance. REMD decay recognizes the role of 3′ UTRs by designating the correct space between the terminating codon and the polyadenylation region [80]. Some key factors in quality surveillance mechanisms are frequently used in conditionally regulated degradation pathways that depend on mRNA-specific RBPs such as Staufen1 [81] and SLBP [82].
3.2.2 Editing of mRNA
RNA editing that occurs posttranscriptionally involves covalently altering RNA sequences by inserting adenosines or cytidines into uridines or inosines, respectively (C-to-U editing). Adenosines that readily localize to the double-stranded portion of viral RNAs, cellular pre-mRNAs, and noncoding RNAs are affected by adenosine-to-inosine (A-to-I) editing. Adenosine deaminase enzymes acting on the RNA (ADAR) family catalyze A-to-I editing. dsRNA-binding motifs (dsRBMs) are located in amino(N)-terminal ADAR regions, while ADAR portions of carboxy-terminal have a conserved domain with catalytic activity. ADARs can act on any double-stranded RNA sequence, but they prefer nucleotides that are close together. The 5′ nearest neighbor is the most powerful to bring about editing of adenosine in both ADAR1 and ADAR2. Since the catalytic domain is primarily responsible for nearest neighbor preferences, dsRBM helps human hADAR2 discern adenosines with a 3′ G. Also, the nucleotides outside of the nearest neighbor have an effect on ADAR preferences. Various factors like length of the dsRNA and presence of loops, bulges, and mismatches determine the number of adenosines to be edited [8].
Adenosine-to-inosine conversion has been suggested to take part in a number of processes, including regulation of neuronal signaling, formation of higher brain function, RNAi activity shaping, and regulation of microRNA synthetic pathway. Cytidine editing to uridine is carried out by the enzyme family of AID–APOBEC. Following the discovery of cytidine editing to uridine in mRNA of apoB, detailed investigations into the possible target sites of APOBEC1 revealed that such editings are mostly restricted to 3′ UTRs. The proof of localization for editing sites at 3′ UTRs is a presence of cytidine surrounded on either side by uridine or adenosine and accompanied by a properly separated sequence motif (WCWN2-4WRAUYANUAU). Nonetheless, the consensus sequences of these motifs were not a target site when available in translating sequences, except ApoB. Editing of 3′ UTRs which is-mediated by ‘APOBEC1’ can affect posttranscriptional processes such as stability of transcripts, polyadenylation, subcellular localization, and translational output. The passing on of information of nucleotide sequence from DNA to RNA is a crucial operation, as shown by adenosine-to-inosine and cytidine-to-uridine editing. Along these lines, one study paper reported an unusual degree of changes in bases from DNA to RNA that cannot be explained by classical editing, and the reason behind the mechanisms are unknown [8].
3.2.3 Stability of a Specific mRNA Species
RBPs that interact with “adenine/uridine-rich elements” (AREs) are preferentially located within 3′ UTRs of mRNA, including TTP, AUF1, and Hu family members. The stability of a particular mRNA is determined by the interaction of many RBPs that both stabilize and destabilize it. The effect of RBP binding to be cooperative or antagonistic is affected by the spatial interaction and variance in affinity within the UTR between their regulatory sites. The effect of RBP binding is also influenced by the comparative quantity of such RBPs in the cellular condition and its confinement where the binding takes place. Furthermore, microRNAs and RBPs can also join together and their structural stability can be affected by RBPs and microRNAs [8].
3.2.4 Role in Diseases
Due to the fact that RBPs are engaged in almost all aspect of RNA metabolism, any mutation or disturbance of RBP function can result in a number of diseases. In cancer, overexpression of RBP or genetic variation can lead to inaccurate or extensive RNA binding at different phases of RNA metabolism, which can have a significant impact on cancer cells. During the development of the nervous system, gene expression is subject to strict dynamical regulation. RBPs involved in normal neuron growth and functioning were identified by Deschenes-Furry and colleagues. Lukon et al. have identified that many illnesses are caused by inhibition of function or overactivity of RBP. CGG triplet expansion on FMR1’s 5′ UTR is linked to Fragile X syndrome, resulting in FMR1 function loss required for normal neuronal development. In autoimmune disorders like paraneoplastic neurologic syndromes (PNSs), RBPs like Nova proteins and Hu family are targeted by autoantibodies causing loss of function in RBP. The neuronal-specific Nova protein family mediates alternative splicing of their target pre-mRNAs present in the regions of CNS like the hindbrain and ventral spinal cord.
Numerous trinucleotide disorders are caused by defective RBPs. “Myotonic dystrophy type 1” (DM1) has several repetitions in the 3′ UTR region of the DMPK gene, whereas myotonic dystrophy type 2 (DM2) has significantly longer repetitions of the tetra-nucleotide CCTG, resulting in toxic mutant RNAs. A GCG repeat extension in the PABPN1 exon gene results in the development of a PABPN1 variant in oculo-pharyngeal muscular dystrophy (OPMD), a degenerative disease which starts during adulthood. After that, the mutant gene induces the continuation of its poly (A) tails to the size of a nascent mRNA. Transcripts with a lengthy poly (A) tail accumulate in the nuclei of skeletal muscle, resulting in the development of muscular dystrophy. ASF/SF2 and eIF4E are two additional cancer-related RBPs that have been studied. EIF4E is a particularly overexpressed oncogene in breast cancer that is correlated with a poor prognosis. Various cancers also overexpress ASF/SF2. ASF/SF2 overexpression has the potential to alter the splicing of important cell cycle regulators and tumor suppressor genes, making it an attractive target for cancer therapy. Mutations in the consumer regions of RNA operators, the master regulators of co-expressed genes, may result in the loss of one or more mRNA targets. Two SNPs in the FGF20 gene’s 3′ UTR region have been linked to Parkinson’s disease. Similarly, RBP function may be lost as a result of SNPs on mRNAs in miRNA genes or their target sites [8].
4 Investigative Methods for Interactions of RBP–RNA
This section describes the conceptual structure for experiments designed to classify RNA species bound by RBPs or, alternatively, subsets of RBPs bound to particular RNAs. This section is divided into four. In the first chapter, in vitro methods for studying protein–RNA interactions are discussed, as well as the basic concepts of these experimental protocols. In addition, newly developed techniques that complement in vivo approaches will be considered. The second section shows how to examine large in vivo transcriptomes, and the third section offers a few examples of structural approaches for studying protein–RNA interactions.
4.1 In Vitro Identification of RNA–Protein Interactions
In vitro methodologies usually use one of the two approaches to understanding interactions between RNA and RBP. An established RBP can be used as a starting point for identifying RNAs that interact with it. Traditional “electrophoretic mobility shift assays” (EMSA) or supershift assays are frequently used to illustrate that protein incubation in the presence or absence of an antibody specific for RBP disrupts RNA movement in PAGE. The second strategy entails finding any RBPs that are bound to the target RNA. To attach an antisense oligonucleotide to a matrix, affinity chromatography can be used. The oligonucleotide attaches to any RBPs or related proteins after the cell lysate flows through the matrix. One of in vitro methodologies’ flaws is their inability to differentiate between physiologically important and nonphysiologically important interactions. Interactions between RNA and RBP must be measured in vivo in order to understand their biological significance [8].
4.1.1 Systematic Evolution of Ligands by Exponential Enrichment (SELEX)
SELEX had aided in our knowledge of the molecular mechanism by which proteins interact with RNA. To execute in vitro selection, a DNA pool containing a random and mutant sequence segment surrounded on both ends by a conserved sequence and maybe a promoter of T7 RNA polymerase is being chemically synthesized. Following many PCR cycles, the DNA is amplified and then in vitro transcribed to generate the RNA pool. According to their capacity to bind to a protein, RNAs are classified as binders or nonbinders. The RNAs are obtained, reverse transcribed, amplified by PCR, and transcribed again. With each round of filtering, the ratio of high- to low-affinity sequences increases until the pool is populated by the RNA species with highest-affinity. It is possible to detect sequences with a wide range of affinities when the sequence pool is at an intermediate stage of selection. Each sequence’s relative concentration is proportional to its affinity, with a lower concentration suggesting a greater affinity [83].
4.1.2 RNA Compete
The RNA compete technique is used to determine the binding specificity of RBPs. This approach is based on an RNA library that contains all potential 8-base sequences identified a minimum of 12 times in unorganized RNAs, as well as all possible 6- and 7-nucleotide loop sequences (and about 60% of 8-base loops) within RNA hairpins of RNA with special 10-base pair stems. These sequences are utilized to generate ssDNA using a microarray, which is subsequently converted to dsDNA and amplified by polymerase chain reaction. Ultimately, an in vitro transcription step is used to create the ssRNA library from dsDNA. Thus, after the generation of the RNA library, a single drive of RNA target sequences employing a tagged RBP of interest is conducted. Then, RNA sequences selected by RBP are tagged and hybridized to a microarray of the same form as the RNA library. The richness of the specified RNAs from the start library is determined using computational analysis.
RNA compete provides a detailed estimation of RBP-binding tendencies to small RNAs spanning the entire k-mer range in both structured and nonstructured conformation. RNA can be employed to validate and assess in vivo approaches to understand protein–RNA interactions. Furthermore, positional weight matrices (PWMs) and consensus motifs are supported. In a broad sense, RNA compete includes the following three steps:: (1) the construction of an RNA pool from a collection of RNA sequences and structures; (2) a single pull-down of RNAs associated with a labeled RBP of interest; and (3) hybridization of the microarray and computational analysis of the proportional enrichment of the bound percentage with respect to the initial pool of RNAs [84].
4.2 In Vivo Identification of Protein–RNA Interactions
In vivo protein–RNA interaction methods may be used to characterize either the RBPs that bind to specific RNAs or the RNAs that bind to specific RBPs to and complement each other with a previously identified RBP. In the following segment, we will go through these two distinct but complementary approaches.
4.2.1 RIP-Chip
In this technique, immunoprecipitation is used to assay RNA–protein binding in vivo. The RIP-Chip employs antibodies to bind unique RBPs and enrich RNA fragments bound to these RBPs. When hybridized to a microarray, the associated RNA fragments are classified, allowing for genome-wide analysis of RNA–protein interactions. The RIP Chip has some drawbacks, including the likelihood of co-immunoprecipitation of additional RBPs alongside the RBP of interest. Furthermore, RBP–RNA associations sometimes fail to accurately reflect in vivo associations due to RBP and RNA re-association after cell lysis. Furthermore, RBP binding sites could not be identified within the specified RNA fragments with this technique. Hence motif analysis is also required to ascertain RNA binding preferences [85].
4.2.2 Cross-Linking and Immunoprecipitation (CLIP) and HITS-CLIP
Ultraviolet (UV) radiation CLIP enables the stringent in vivo purification of both RBPs and small RNA fragments that could be used for amplification and sequencing. UV-induced crosslinking of RBPs and RNAs is performed in vivo prior to protein purification in order to boost the performance of conventional immunoprecipitation methods. For example, photocrosslinking inhibits in vitro RNA–protein reassociation and co-immunoprecipitation. UV cross-linking helps in easy purification of protein complexes ensuring more stringent purification schemes to be employed. This results in high pure protein–RNA complexes and binding sites are identified by incomplete proteinase K digestion. In some cases, the reverse transcriptase (RT) that is used to prepare samples was shown to effectively transcribe via cross-linked regions. Cross-linked sites with reverse transcription errors may be used to precisely localize protein–RNA interface (such as by the iCLIP method).
“High-throughput Sequencing CLIP” (HITS-CLIP) is a technique that blends regular CLIP with HITS-CLIP. CLIP-based quantification of high-throughput sequencing of DNA (HTS/NGS) enhances the sensitivity, and RBP binding sites have a spatial resolution. CLIP suffers from HTS technique limitations, including high error rates in sequencing, uneven CLIP tag alignments, and also the description of acceptable context CLIP tag distributions for evaluating the statistical significance of RBP binding sites. Additionally, variations in CLIP analysis procedures might affect the RBP’s assumed specificity. Some RNAases are employed to degrade unbound RNA, unattached RNA exhibit sequence selectivity, which may have an effect on CLIP-tagged RBP-binding sites [86]. Additionally, although the CLIP cross-linking protocol is more sensitive, it may have a lower specificity [87].
4.2.3 Photo-Activatable Ribonucleoside-Enhanced Cross-Linking and Immunoprecipitation (PAR-CLIP)
The PAR-CLIP method is a variation of the cross-linking and immune precipitation technique in which photo-activated nucleosides are applied to the medium, followed by cell absorption and protein–RNA crosslinking. This improvement has a number of advantages over conventional CLIP. To begin, PAR-CLIP recovers 100–1000 times more cross-linked RNA when intensities of radiation are equivalent. The second advantage is that UV radiation induces T-to-C mutations, which are typical in cross-linked nucleoside analog-containing sites. PAR-CLIP leverages mutation analysis to enhance the detection of RBP attachment site locations or footprints [8].
4.2.4 Individual-Nucleotide Resolution Ultraviolet Cross-Linking and Immunoprecipitation (iCLIP)
Although all other CLIP techniques operate in the same way, iCLIP is a version that focuses on the RNA–protein interaction detection during sample preparation and the formation of crosslinking sites. iCLIP accomplishes this by taking advantage of reverse transcription’s natural tendency to terminate before cross-bound nucleotides owing to the remaining amino acids. After circularization and linearization, the circularized and linearized cDNAs are PCR-amplified and then HTS-analyzed. The location may be used in place of the adaptor sequence utilized in the circularized PCR amplification to identify the RBP-binding site [8].
4.2.5 Finding the Proteins Bound to RNAs
Although studying protein components of RNA protein complexes in vivo can be challenging, some strategies have been developed. This problem is addressed by integrating and improving magnetic bead-based assays and crosslinking of protein-nucleic acid induced by UV radiation, as well as improving the PNA-assisted RBP identification method. The use of PNA oligonucleotides linked to peptides and peptide-PNA-linked oligonucleotides that can bind RNAs with greater specificity and selectivity than complementary RNA or DNA, as well as targeting of oligonucleotides to living cells efficiently, are among the method’s unique features. PNAs hybridize with their RNA cognates once within the cell, and UV light is used to crosslink the targeted RNAs. After magnetic beads have been used to separate the RBP–PNA complexes, they are combined with an antisense PNA oligo and characterized using mass spectrometry techniques. Many protein–RNA complexes discovered by protein capture methods are severely misidentified, according to researchers. In contrast, quantitative mass spectrometry [88] aids in the differentiation of proteins particularly bound to the RNA of choice from other compounds with similar binding affinity.
4.2.6 Protein–RNA Interactions: Structural Analysis
CLAMP (crosslinking and mapping the protein domain) allows the mapping of RNA-binding domains that are cross-linked to unique nucleotides in the RNA within RBPs. This method is particularly useful when dealing with RNA-binding domains and protein–RNA interactions. The chromophore must be inserted into the site, photochemical protein–RNA crosslinking must be added, and a site-specific chemical protein cleavage is required for CLAMP to function.
4.2.7 Online Resources for Experimental Protein–RNA Interactions
Only a few resources were utilized to record the data provided by the given technologies about protein–RNA interactions. The RNA-binding protein database can be found at http://rbpdb.com, while the CLIPZ database can be found at http://www.clipz.unibas.ch [89]. RBPDB might be a good place to start if someone wants to learn more about manually collected RNA-binding interactions and/or regions for a particular RNA-binding protein The RBPDB contains experimental associations identified in vitro (e.g., RNA compete) or in vivo (e.g., RIP-Chip, CLIP) (human, mouse, fly and worm). RBPDB extends the capabilities of searching for motifs in an input RNA sequence by adding the ability to retrieve probable binding sites annotated by PWM ratings CLIPZ, in comparison to RBPDB, seems to be a more structured database of RNA-binding sites developed by the HITS–CLIP approach that enables display and study of the data collected using this approach. Using motif enrichment review, RBP binding sequence motifs can be predicted. The statistical significance of putative binding site motifs is also restored. Other methods are also capable of assessing spatial relationships between RBPs.
http://pridb.gdcb.astate.edu/index.php is a database including interactions between proteins and RNA. The Protein Data Bank (PDB) has a database of complex-derived protein–RNA interactions. It makes it easier to find and visualization of covalently linked amino acids and ribonucleotides in the primary sequences of the proteins and RNA chains involved. PRIDB uses both a distance-based criterion and the ENTANGLE algorithm to characterize interfaces [90]. Additionally, PRIDB searches for ProSite [91] and FR3D [92] motifs, respectively.
The Atlas of UTR Regulatory Behavior (AURA) is a manually compiled Catalog of Human UTRs and UTR Regulatory Annotations that can be found at http://aura.science.unit.it (AURA). A simple, interactive online interface gives complete access to a vast amount of data on UTRs, including information on phylogenetic preservation, RNA sequence and structure data, single nucleotide variation, gene expression, and functional descriptions of genes. It has also taken into account interactions between RBPs and miRs that have been experimentally determined to be nonredundant, as well as their effects on human UTRs [93].
5 Computational Inference of RBP-Binding Sites
There are a variety of analytical methods for identifying RNA sequence elements that operate as RBP binding sites. These techniques will be explored briefly in this section.
5.1 Binding Site Search
PWMs are often used to summarize the statistical features of observed binding sites. PWMs denote the odds of each nucleic acid occurring at each position. PWMs are used to scan RNA sequences for potential RBP binding sites. This search can be carried out using regulatory sequence analysis methods like RSAT (http://rsat.ulb.ac.be/rsat/). The accuracy of this RNA-binding specificity representation is on the basis of a large fraction of experimental data.
5.2 Models of Binding Sites
When introducing the most up-to-date techniques for modeling RBP attachment sites, models of transcription factor attachment site provide valuable guidance for the solution of pattern prediction and discovery. New techniques or modifications of existing techniques are used to model the binding elements of RBPs. Due to their distinctiveness and commonalities, several strategies for identifying RBP binding sites are described here in comparison to discovery of DNA attachment site. As with transcription factors, RBP attachment sites are modeled using both unsupervised and supervised (regression) methods. There may be two models of RBP-binding sites: one that ignores RNA structure and another that does not, because RNA structure may affect binding. RBP attachment sites are distinct from binding sites of transcription factor in that they allow for the binding of RNA structure. Therefore, as a result, models can be classified into those that neglect the structure of the RNA and those that do give importance to the RNA structure. The methods that consider RNA structure can be divided into two groups: First model predict the structure of RNA and second model is about the structure of RNA in its structural context.
The unsupervised methods take collection of RNA sequences as inputs that are optimized for a given RBP’s attachment sites (obtained, for example, via a SELEX procedure) and a standard model of usual composition of RNA sequence. Techniques of transcription factor techniques could be used directly with minimal adjustments (i.e., replacing Us with Ts) in case the impact of RNA structure is overlooked. For example, Multiple Expectation Maximization for Motif Elicitation (MEME) [94] maximizes the probability of the observed sequence set fitting a position-specific scoring matrix (PSSM) motif model using the expectation-maximization (EM) algorithm. Centered on the assumption of nucleotide independence, the PSSM model describes a product multinomial distribution over bound k-mers. It is interesting to note that MEME does not allow gaps in sequence pattern, which may present a problem when RNA-binding domains such as RRMs bind to randomly separated and very short RNA sequence. Another well-known example of a structure-naive approach used for RBP binding site modeling is the assignment of a conservation index to all possible k-mers (RNA words of length k) in order to perform an independent genome-wide search for k-mers retained in 3′ UTRs [94]. These k-mers can serve as regulators.
MEMERIS is an upgraded version of the MEME algorithm that incorporates RNAfold-derived probabilities for base-pairing when fitting PSSM motifs [94]. The probabilities of base-pairing constrain the search space for an RBP-binding site’s initial position. MEMERIS looks for a motif that is important to a specific sense of the RNA structure (i.e., unpaired regions); this technique is distinct from those that focus on sequence-specific structural elements (e.g., stem-loops).
Three types of motif-finding algorithms can be used to model RNA structure. The first group employs co-variation to arrive at a consensus structure for all aligned sequences following RNA sequence alignments. The efficacy of such approaches is largely dependent on the alignment accuracy, which requires a high level of homology between the input RNA sequences, which is an uncommon occurrence. The chances of such event become more common when searching for shared local patterns by multiple mRNAs attached by the very same RBP within long 3′ UTRs. Alternatively, methods such as RNAProfile [95] estimate the minimum free energy folds for every sequence before looking for particular folds. The primary issues here are accurately predicting folds and representing an entire set of folds using a single fold having the minimum free energy fold. The third method uses dynamic programming to match and fold two RNA sequences simultaneously, with the usual secondary structure anticipated utilizing energy-based factors, culminating in a structure-based alignment [96]. This pair-wise alignment is then extended using a variety of heuristics to multiple alignments. Since the secondary structure of an RNA sequence is frequently defined by algorithmic assumptions, the analysis of noisy inputs is essential. Probabilistic covariance models, such as CMfinder [97], are more effective at capturing observable difference in the sequence and structure of RNA patterns. RNApromo [98] was recently used to model co-regulated RBP sequence preferences across a range of RNA sequences.
RBP binding models are used in supervised approaches as part of regression models designed to forecast quantitative estimates of RBP binding, as well as RNA binding. Due to the difficulty of obtaining the required input data in the past, these approaches have been limited to RBP binding data. Earlier efforts in this field were either structure-naïve [99] or relied on simplistic stem-loop models [100]. Examples from more recent years include ATS [101] and RNAcontext [101]. In vitro assays, RNAcompete, and RNAcontext provide information on RBP binding affinity, and that information is used to learn the RNAcompete, for example, by setting a physical model to information of RBP attachment affinity and sequence of RBP. In vitro assays, RNAcompete, and RNAcontext provide information on RBP binding affinity, and that information is used to learn the RNA-protein interaction. RNAcontext is fascinating for two reasons: it is capable of modeling RBP preferences for sequences based on their structural contexts, and it makes extensive use of high-throughput quantitative data to evaluate different parameters of model. RNAcontext operates in three steps, beginning with the input of a series of sequences and their corresponding affinity measurements. The first step calculates the probability that a word of length k contains an RBP binding site using the product of two terms. The first term denotes the inferred RBP sequence’s priorities (in the form of a positional weight matrix), while the second term denotes the relative structural priorities of RBPs in different structural contexts. The second step is to estimate a sequence affinity based on the affinities assigned to each phrase by the previous motif model. The third step is to determine which array of parameters reduces the amount of squared differences between measured and expected input affinities when the sequence score function is modelled as a linear function. ATS is comparable to RNAcontext, except that it employs a selfish search strategy and considers only one structural background at a time while attempting to locate a degenerate consensus sequence motif. ATS, on the other hand, is a better fit for in vivo binding assays than RNAcontext, as the former’s sequence scoring function is optimal for the longer RNA sequences associated with these assays.
6 Conclusion and Future Perspectives
With the introduction of efficient high-throughput technique capable of analyzing whole transcriptome and proteome, it is estimated that the number of RBP and types of its interaction with RNA is more than expected. The integration of structural data, defining site of molecular contacts, and high-throughput sequencing method that unfold RNA sequence specificity could allow for the determination of predictive model for specific RBPs. The growing experimental data of transcriptome should facilitate development in computational methods for prediction of RNA–protein interaction and for modeling regulatory pathway of RPI. According to genome-wide study, SNPs found in the RBP-binding region were associated with diseases. Disease susceptibility is influenced by genetic variation in RPI and interference with normal function. Further investigation on association of genetic variation and investigation will give better understanding of RPI.
Abbreviations
- CSD:
-
Cold shock domain
- DDX:
-
Dead box proteins
- dsRNA:
-
Double-stranded RNA
- FMRP:
-
Fragile X mental retardation protein
- KH:
-
K-homology
- LINE:
-
Long interspersed element
- mRNA:
-
Messenger RNA
- ncRNA:
-
Noncoding RNA
- PUM-HD:
-
PUM-homology domains
- RBP:
-
RNA-binding proteins
- RNP:
-
Ribonucleoprotein
- RPI:
-
RNA–protein interaction
- RRM:
-
RNA recognition motif
- snoRNA:
-
Small nucleolar RNA
- snRNA:
-
Small nuclear RNA
- XBP1:
-
X-box-binding protein 1
- YTH:
-
YT521-B homology
- ZKD:
-
Zinc knuckle domain
References
Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423–37.
Khade P, Joseph S. Functional interactions by transfer RNAs in the ribosome. FEBS Lett. 2010;584:420–6.
Sauert M, Temmel H, Moll I. Heterogeneity of the translational machinery: variations on a common theme. Biochimie. 2015;114:39–47.
Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74.
Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8.
Uszczynska-Ratajczak B, Lagarde J, Frankish A, Guigó R, Johnson R. Towards a complete map of the human long non-coding RNA transcriptome. Nat Rev Genet. 2018;19:535–48.
Hentze MW, Castello A, Schwarzl T, Preiss T. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol. 2018;19:327–41.
Re A, Joshi T, Kulberkyte E, Morris Q, Workman CT. RNA–protein interactions: an overview. In: Gorodkin J, Ruzzo WL, editors. RNA sequence, structure, and function: computational and bioinformatic methods [Internet]. Totowa, NJ: Humana Press; 2014 [cited 2021 May 2]. p. 491–521. Available from: https://doi.org/10.1007/978-1-62703-709-9_23.
Muppirala UK, Honavar VG, Dobbs D. Predicting RNA-protein interactions using only sequence information. BMC Bioinform. 2011;12:489.
Liu S, Li B, Liang Q, Liu A, Qu L, Yang J. Classification and function of RNA–protein interactions. WIREs RNA. 2020;11:e1601.
Wang X, McLachlan J, Zamore PD, Hall TMT. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–12.
Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH. ZBP1 recognition of β-actin zipcode induces RNA looping. Genes Dev. 2010;24:148–58.
Ustianenko D, Chiu H-S, Treiber T, Weyn-Vanhentenryck SM, Treiber N, Meister G, et al. LIN28 selectively modulates a subclass of Let-7 microRNAs. Mol Cell. 2018;71:271–283.e5.
Zhang M, Perelson AS, Tung C-S. RNA structural motifs. eLS [Internet]. American Cancer Society; 2011 [cited 2021 May 2]. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/9780470015902.a0003132.pub2.
Poynter SJ, DeWitte-Orr SJ. Understanding viral dsRNA-mediated innate immune responses at the cellular level using a rainbow trout model. Front Immunol [Internet]. Frontiers; 2018 [cited 2021 May 2]; 9. Available from: https://www.frontiersin.org/articles/10.3389/fimmu.2018.00829/full.
Eisenberg E, Levanon EY. A-to-I RNA editing—immune protector and transcriptome diversifier. Nat Rev Genet. 2018;19:473–90.
Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–49.
Eggington JM, Greene T, Bass BL. Predicting sites of ADAR editing in double-stranded RNA. Nat Commun. 2011;2:319.
Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–7.
Palladino MJ, Keegan LP, O’Connell MA, Reenan RA. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–49.
Lev-Maor G, Sorek R, Levanon EY, Paz N, Eisenberg E, Ast G. RNA-editing-mediated exon evolution. Genome Biol. 2007;8:R29.
Pinto Y, Buchumenski I, Levanon EY, Eisenberg E. Human cancer tissues exhibit reduced A-to-I editing of miRNAs coupled with elevated editing of their targets. Nucleic Acids Res. 2018;46:71–82.
Sugimoto Y, Vigilante A, Darbo E, Zirra A, Militti C, D’Ambrogio A, et al. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature. 2015;519:491–4.
Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–21.
Wang J, Daldrop P, Huang L, Lilley DMJ. The k-junction motif in RNA structure. Nucleic Acids Res. 2014;42:5322–31.
Calabretta S, Richard S. Emerging roles of disordered sequences in RNA-binding proteins. Trends Biochem Sci. 2015;40:662–72.
Phan AT, Kuryavyi V, Darnell JC, Serganov A, Majumdar A, Ilin S, et al. Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat Struct Mol Biol. 2011;18:796–804.
Didiot M-C, Tian Z, Schaeffer C, Subramanian M, Mandel J-L, Moine H. The G-quartet containing FMRP binding site in FMR1 mRNA is a potent exonic splicing enhancer. Nucleic Acids Res. 2008;36:4902–12.
Schaeffer C, Bardoni B, Mandel J-L, Ehresmann B, Ehresmann C, Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–13.
Neelamraju Y, Hashemikhabir S, Janga SC. The human RBPome: from genes and proteins to human disease. J Proteome. 2015;127:61–70.
Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–7.
Song J, Yi C. Chemical modifications to RNA: a new layer of gene expression regulation. ACS Chem Biol. 2017;12:316–25.
Kadumuri RV, Janga SC. Epitranscriptomic code and its alterations in human disease. Trends Mol Med. 2018;24:886–903.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610.
Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–65.
Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–55.
Ramaswamy G, Slack FJ. siRNA: a guide for RNA silencing. Chem Biol. 2002;9:1053–5.
Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–6.
Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20.
Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98.
Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14.
Oliveto S, Mancino M, Manfrini N, Biffo S. Role of microRNAs in translation regulation and cancer. World J Biol Chem. 2017;8:45–56.
Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202.
Ku H-Y, Lin H. PIWI proteins and their interactors in piRNA biogenesis, germline development and gene expression. Natl Sci Rev. 2014;1:205–18.
Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Fejes Toth K, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–99.
Vodovar N, Bronkhorst AW, van Cleef KWR, Miesen P, Blanc H, van Rij RP, et al. Arbovirus-derived piRNAs exhibit a ping-pong signature in mosquito cells. PLoS One. 2012;7:e30861.
Miesen P, Girardi E, van Rij RP. Distinct sets of PIWI proteins produce arbovirus and transposon-derived piRNAs in Aedes aegypti mosquito cells. Nucleic Acids Res. 2015;43:6545–56.
Zhao S, Gou L-T, Zhang M, Zu L-D, Hua M-M, Hua Y, et al. piRNA-triggered MIWI ubiquitination and removal by APC/C in late spermatogenesis. Dev Cell. 2013;24:13–25.
German MA, Luo S, Schroth G, Meyers BC, Green PJ. Construction of parallel analysis of RNA ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat Protoc. 2009;4:356–62.
Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326:1212–6.
Lécuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–87.
Wolke U, Weidinger G, Köprunner M, Raz E. Multiple levels of posttranscriptional control lead to germ line-specific gene expression in the zebrafish. Curr Biol. 2002;12:289–94.
Lipshitz HD, Smibert CA. Mechanisms of RNA localization and translational regulation. Curr Opin Genet Dev. 2000;10:476–88.
Chartrand P, Meng XH, Huttelmaier S, Donato D, Singer RH. Asymmetric sorting of Ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol Cell. 2002;10:1319–30.
Lewis RA, Kress TL, Cote CA, Gautreau D, Rokop ME, Mowry KL. Conserved and clustered RNA recognition sequences are a critical feature of signals directing RNA localization in Xenopus oocytes. Mech Dev. 2004;121:101–9.
Macdonald PM, Struhl G. Cis-acting sequences responsible for anterior localization of bicoid mRNA in Drosophila embryos. Nature. 1988;336:595–8.
Cenik C, Chua HN, Zhang H, Tarnawsky SP, Akef A, Derti A, et al. Genome analysis reveals interplay between 5′UTR introns and nuclear mRNA export for secretory and mitochondrial genes. PLoS Genet. 2011;7:e1001366.
Palazzo AF, Springer M, Shibata Y, Lee C-S, Dias AP, Rapoport TA. The signal sequence coding region promotes nuclear export of mRNA. PLoS Biol. 2007;5:e322.
Arn EA, Cha BJ, Theurkauf WE, Macdonald PM. Recognition of a bicoid mRNA localization signal by a protein complex containing swallow, nod, and RNA binding proteins. Dev Cell. 2003;4:41–51.
Müller M, Heym RG, Mayer A, Kramer K, Schmid M, Cramer P, et al. A cytoplasmic complex mediates specific mRNA recognition and localization in yeast. PLoS Biol. 2011;9:e1000611.
Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol. 2003;13:1159–68.
Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–56.
Muckenthaler M, Gray NK, Hentze MW. IRP-1 binding to ferritin mRNA prevents the recruitment of the small ribosomal subunit by the cap-binding complex eIF4F. Mol Cell. 1998;2:383–8.
Gebauer F, Grskovic M, Hentze MW. Drosophila sex-lethal inhibits the stable association of the 40S ribosomal subunit with msl-2 mRNA. Mol Cell. 2003;11:1397–404.
Grskovic M, Hentze MW, Gebauer F. A co-repressor assembly nucleated by Sex-lethal in the 3′UTR mediates translational control of Drosophila msl-2 mRNA. EMBO J. 2003;22:5571–81.
Beckmann K, Grskovic M, Gebauer F, Hentze MW. A dual inhibitory mechanism restricts msl-2 mRNA translation for dosage compensation in Drosophila. Cell. 2005;122:529–40.
Nelson MR, Leidal AM, Smibert CA. Drosophila cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. EMBO J. 2004;23:150–9.
Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell. 2004;6:69–78.
Stebbins-Boaz B, Cao Q, de Moor CH, Mendez R, Richter JD. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol Cell. 1999;4:1017–27.
Ostareck DH, Ostareck-Lederer A, Shatsky IN, Hentze MW. Lipoxygenase mRNA silencing in erythroid differentiation: the 3′UTR regulatory complex controls 60S ribosomal subunit joining. Cell. 2001;104:281–90.
Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat Cell Biol. 2010;12:286–93.
Hussey GS, Chaudhury A, Dawson AE, Lindner DJ, Knudsen CR, Wilce MCJ, et al. Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol Cell. 2011;41:419–31.
Villalba A, Coll O, Gebauer F. Cytoplasmic polyadenylation and translational control. Curr Opin Genet Dev. 2011;21:452–7.
Reed R, Hurt E. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell. 2002;108:523–31.
LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–24.
Hwang J, Sato H, Tang Y, Matsuda D, Maquat LE. UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol Cell. 2010;39:396–409.
Ruiz-Echevarría MJ, Peltz SW. The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell. 2000;101:741–51.
Chester A, Somasekaram A, Tzimina M, Jarmuz A, Gisbourne J, O’Keefe R, et al. The apolipoprotein B mRNA editing complex performs a multifunctional cycle and suppresses nonsense-mediated decay. EMBO J. 2003;22:3971–82.
Inada T, Aiba H. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J. 2005;24:1584–95.
Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208.
Kaygun H, Marzluff WF. Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol Cell Biol. 2005;25:6879–88.
Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–10.
Ray D, Kazan H, Chan ET, Castillo LP, Chaudhry S, Talukder S, et al. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat Biotechnol. 2009;27:667–70.
Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1:302–7.
Kishore S, Jaskiewicz L, Burger L, Hausser J, Khorshid M, Zavolan M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods. 2011;8:559–64.
Corcoran DL, Georgiev S, Mukherjee N, Gottwein E, Skalsky RL, Keene JD, et al. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011;12:R79.
Butter F, Scheibe M, Mörl M, Mann M. Unbiased RNA-protein interaction screen by quantitative proteomics. Proc Natl Acad Sci U S A. 2009;106:10626–31.
Khorshid M, Rodak C, Zavolan M. CLIPZ: a database and analysis environment for experimentally determined binding sites of RNA-binding proteins. Nucleic Acids Res. 2011;39:D245–52.
Allers J, Shamoo Y. Structure-based analysis of protein-RNA interactions using the program ENTANGLE. J Mol Biol. 2001;311:75–86.
Sigrist CJA, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, et al. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38:D161–6.
Sarver M, Zirbel CL, Stombaugh J, Mokdad A, Leontis NB. FR3D: finding local and composite recurrent structural motifs in RNA 3D structures. J Math Biol. 2008;56:215–52.
Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–90.
Hiller M, Pudimat R, Busch A, Backofen R. Using RNA secondary structures to guide sequence motif finding towards single-stranded regions. Nucleic Acids Res. 2006;34:e117.
Pavesi G, Mauri G, Stefani M, Pesole G. RNAProfile: an algorithm for finding conserved secondary structure motifs in unaligned RNA sequences. Nucleic Acids Res. 2004;32:3258–69.
Simultaneous SD. Solution of the RNA folding, alignment and protosequence problems. SIAM J Appl Math. 1985;45:810–25.
Yao Z, Weinberg Z, Ruzzo WL. CMfinder—a covariance model based RNA motif finding algorithm. Bioinformatics. 2006;22:445–52.
Rabani M, Kertesz M, Segal E. Computational prediction of RNA structural motifs involved in posttranscriptional regulatory processes. Proc Natl Acad Sci U S A. 2008;105:14885–90.
Foat BC, Houshmandi SS, Olivas WM, Bussemaker HJ. Profiling condition-specific, genome-wide regulation of mRNA stability in yeast. Proc Natl Acad Sci U S A. 2005;102:17675–80.
Foat BC, Stormo GD. Discovering structural cis-regulatory elements by modeling the behaviors of mRNAs. Mol Syst Biol. 2009;5:268.
Kazan H, Ray D, Chan ET, Hughes TR, Morris Q. RNAcontext: a new method for learning the sequence and structure binding preferences of RNA-binding proteins. PLoS Comput Biol. 2010;6:e1000832.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Ethics declarations
None
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive licence to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Rathore, S.K., Pati, P. (2021). RNA–Protein Interaction Analysis. In: Gupta, M.K., Behera, L. (eds) Bioinformatics in Rice Research. Springer, Singapore. https://doi.org/10.1007/978-981-16-3993-7_16
Download citation
DOI: https://doi.org/10.1007/978-981-16-3993-7_16
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-3992-0
Online ISBN: 978-981-16-3993-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)