Abstract
A program was developed to stitch the cell images with their corresponding spectrophotometer readings. These stitched images will be sent to an artificial intelligence interface to be analysed so as to predict the cell states and suggest ways to maximise the number of cells during the cell manufacturing process. In addition, a CellProfiler pipeline has been made to count the number of cells and predict the amount of mitochondrial activity present in the cell as this would also help to predict the confluency and mitochondrial networks of the cell. This would be significant in ensuring that the cells present in the cell culture are respiring and a larger interconnected network of cells show that there are more cells respiring and that the confluency of the cells present in the cell culture is higher, leading to a better cell culture. These can be predicted using machine learning of biological data in the cell manufacturing process by extracting and analyzing part of the cell culture to aid in growing more healthy cell cultures with higher confluency.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
-
A.
Machine Learning
-
(1)
What is Machine Learning?
Machine Learning is a way to analyse data through automation. It is a branch of artificial intelligence based on the concept of having systems learn from data, identify patterns and making the decisions needed completely free from the restrictions of their human counterparts. In addition, machine learning can be used to reduce the time and errors needed and create through human work, increasing the efficiency of work given to the system.
-
(2)
How does Machine Learning work?
A set of algorithms are used to determine the principles which the rule of the program would follow to learn how to classify and cluster the data according to their similarities. This set of algorithms form a computer model which will be used in the classification of data input. Data input can range from text and images to sound and numbers.
-
(1)
-
B.
Deep Learning
-
(1)
What is Deep Learning?
Deep Learning is a branch of Machine Learning in Artificial Intelligence. Deep Learning has networks which are capable of learning unsupervised from data that is unstructured or unlabeled through learning by example. These networks are more commonly known as neural networks [3]. Deep Learning can be found being used in driverless cars, voice control in devices such as Siri or Google Home and more [5]. In this project, a deep learning model was trained to classify images and the spectrometer data to its different confluencies. Such models are trained through these many hidden layers to be able to classify the numbers, text and images given to them without the need of manual feature extractions. Deep Learning has been shown to be able to give extremely accurate classifications and can sometimes exceed the accuracy of humans [6].
-
(1)
-
C.
Neural Networks
-
(1)
What are Neural Networks?
Inspired by the way the human brain functions, neural networks function in a similar manner. A Neural Network is a set of algorithms which are designed to recognise patterns. They interpret sensory data, similar to a human brain through a kind of machine perception by labelling or clustering the raw input. Neural Networks allow for numerical, images, sounds, text or time series to be input as data into the Neural Network, though they would all be translated into numerical data so that the Neural Network may recognise them since they are only able to recognise the patterns if they are in their numerical form. Neural Networks are used as a way to classify and cluster data according to their similarities among the example inputs which they have been trained on, for easy storage and management. In addition, as the performance of neural networks improves, they grow bigger and are able to work with more and more data, unlike other Machine Learning algorithms which will reach a plateau after a certain point.
-
(2)
Types of Neural Networks
There are many types of neural networks. Each different type of neural network uses a different set of principles to determine their rules which they will follow. Due to this difference in principles and rules, each neural network will have their individual strengths and weaknesses. However, for this research, we have narrowed down to using the convolutional neural networks (CNN) and Artificial Neural Networks (ANN) [2].
-
(1)
-
D.
CellProfiler
-
(1)
What is CellProfiler?
CellProfiler is an open-source software used to quantitatively analyse biological images. It was started in 2003 by Anne E. Carpenter and Thouis (Ray) Jones in the Sabatini Laboratory and Golland Laboratory. CellProfiler is being maintained and improved in the project team in the Carpenter Lab at the Broad Institute of Harvard and MIT. It allows users to construct their own pipelines to extract and manipulate data from the inputted biological images [1].
-
(1)
-
E.
Research Scope and Purpose
It is a known fact that counting and monitoring cells can be very tedious as one needs to check their confluency and be able to predict the different ways as to which the cell culture can be changed so that the confluency of cells and number of cells being cultured will be higher. A higher cell count and better cell quality in the cell culture would be representative of a better cell culture during the cell manufacturing process.
As cells are living beings and their growth and reaction of their growth mediums can be unpredictable, this would provide cell manufacturers with a better way to consistently grow cell cultures through understanding the confluency and possible ways to improve cell growth through non-invasive means of analysing the cells in the cell culture, thereby reducing the waste which invasive methods of analysing cells.
-
F.
Real Life Applications
Currently, in cell manufacturing it is difficult to ensure and maintain the consistent quality and quantity of cells grown in a cell culture. This is due to the fact that cells may respond differently to the same medium, making it difficult for manufacturers to predict the response of the cell culture to a medium and properly determine if the medium is best for the cell to grow in.
Therefore, through this project, we hope to be able to find a way to aid in analysing the cell culture through non-invasive methods like spectrophotometry, before suggesting changes to improve the medium of the cell culture to aid in encouraging better quality and quantity of the cells manufactured.
-
G.
Other Similar Technologies
As of now, there are neural networks which are able to count cells. However, no model is currently available to form the correlation of the confluency of the cells, which is related to the number of cells in the cell culture, to the health of the cells. Therefore, this machine learning would be the first of its kind.
In addition, current CellProfiler pipelines monitoring cell activity are not mainly focused on mitochondrial interconnected networks or activity in the cells. Although there has been a pipeline [4] made concerning analyzing mitochondrial activity, it measures this by average intensity per square pixel. This is extremely inaccurate as the average can be vastly affected by bright spots present in certain cells if some cells have higher activity than others or if only one cell is alive. Thus, this machine learning would provide a better and more accurate way to measure the interconnectedness and intensity of the mitochondrial networks and mitochondrial activity of a cell respectively.
2 Hypothesis
As previous research has shown that making such a pipeline and stitching images together is possible, we hypothesise that it is possible to construct the pipeline and stitch the images together. In addition, we also hypothesise that the greater the fluorescence of the cytoplasm, the higher the confluency of the cell culture.
3 Materials and Methods
-
A.
Materials and Software Used
-
1.
Python version 2.7.
-
2.
CellProfiler 3.
-
3.
Cell images obtained from an Olympus microscope.
-
4.
Spectrophotometer data from the cell culture.
-
1.
-
B.
Procedure for Stitching Images
-
1.
Import numpy, pandas, matplotlib.pyplot, cv2, os, sys, Image from PIL, splittext os.path, Counter from collections, tkinter, filedialog from tkinter for the Python operating system version 2.7.
-
2.
Checking the filenames in the directory, extract the files with the spectrophotometer data using their file extensions.
-
3.
Using matplotlib.pyplot, plot a graph of the reading against the wavelength on a black background and black axes.
-
4.
Save the graph using the same filename as the spectrophotometer data.
-
5.
Using the filename of the data, search the relevant directory for the filename for the microscope image of the cell culture.
-
6.
Stitch the two images together by plotting the two images using matplotlib.pyplot and cv2.
-
7.
Save the image under desired name in the specified directory.
-
1.
-
C.
Procedure for CellProfiler Pipeline Construction
-
1.
Set up a new pipeline for CellProfiler.
-
2.
Under Metadata, ensure that Metadata is not extracted from the images.
-
3.
Under Names and Types, assign a name to images matching the file extension of the images. They can be assigned the names OrigDAPI, OrigMito, OrigActin, IllumDAPI, IllumMito and IllumActin.
-
4.
Select all image types to Grayscale Image and do not process them as 3D.
-
5.
Set all intensity ranges to be from Image Metadata.
-
6.
Set image set matching method to Order.
-
7.
Under Groups, ensure that the inputted images are not grouped.
-
8.
Add a EnhanceorSuppressFeatures module, enhancing the speckles from the input image OrigMito with feature size of 20 and fast speed and accuracy.
-
9.
Add two modules of IdentifyPrimaryObjects and input the images OrigDAPI and OrigActin to identify the nuclei and fluorescence reading of the cells.
-
10.
Add a module of IdentifySecondaryObject to identify the cells present in the image.
-
11.
Add a module of IdentifyTertiaryObjects to identify the cytoplasm of the cells around the nuclei.
-
12.
Add a module of Threshold, using the OrigMito image, change the correction factor to 0,9 and bounds on threshold to 0.0–1.0.
-
13.
Add two modules of MeasureObjectIntensity to measure the intensity of the cytoplasm and nuclei of the cells in the image.
-
14.
Add a module of MeasureObjectSizeShape to measure the cytoplasm while ensuring that the Zernike features are not being calculated.
-
15.
Measure the average intensity per square pixel and the standard deviation using CalculateMath and MeasureImageIntensity modules.
-
16.
Export the data to a spreadsheet using the ExportToSpreadsheet module.
-
1.
-
K.
Procedure to test Hypothesis of relationship between mitochondrial networks and confluency
-
1.
From the spreadsheet obtaind from the CellProfiler pipeline, extract the data of the intensity per square pixel of the cells’ cytoplasm
-
2.
Draw a graph of average intensity per square pixel against confluency of the cell.
-
1.
5 Discussion
-
A.
Analysis
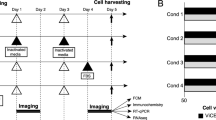
It has to be noted that there are no axes on the graphs generated, such as in Fig. 1, since the presence of extra features in the graph would cause the neural network model to be incorrectly trained in recognising the keywords in the axes instead of the plot itself. This is due to the fact that features in white, like the axes headings, would be read by the neural network as having a reading of 255, causing it to disrupt the reading of the graph since computers read by taking the colour values of the pixels. However, the x-axis is wavelength (nm) and the y-axis is the fluorescence intensity. All cell images, like Fig. 2, are images of fibroblast cells from different cell cultures with different confluencies taken from the Olympus Microscope by connecting the laptop to the microscope. For all stitched images generated, such as Fig. 3, it was generated uniformly with the graph generated from the spectrophotometer data (Fig. 1) on the right and the cell image on the left (Fig. 2).
The stitching of images causes no significant difference in the accuracy of the results, the results stay between 90 and 95%. This means that the stitching of the images alone does not provide an aid to the neural networks to increase the accuracy of the classification of confluencies of cell cultures. Despite this, the hypothesis that the images could be stitched together through a Python code was confirmed.
However, since no research has been done on the effect of the stitching of images on improving the accuracy of neural networks, there are unfortunately no studies to compare this against.
The CellProfiler pipeline has confirmed the hypothesis that the higher the average intensity of the cytoplasm fluorescence of the cells present in the microscope image, the higher the confluence of the cell culture. It has done this by showing a positive trend between confluency and intensity of cytoplasm fluorescence, as seen in Fig. 4. Detailed values of the average intensity of the fluorescence of the cells’ cytoplasmis in the Appendix (Table 1). This indicates that the higher the confluency of the cell culture, the healthier the cells in the cell culture. These mitochondrial networks arise from mitochondrial fusion and fission [8]. Mitochondrial fusion is extremely important in living cells as it allows the spreading of metabolites, enzymes and mitrochondrial gene products through the mitochondrial network. It can optimise mitochondrial function and counteract the accumulation of mitochondrial mutations due to aging. Fragmented mitochondrial networks are more commonly found in resting cells. However, mitochondrial fission aids in the removal of damaged organelles [7]. Thus, it can be said that the intensity of the cytoplasm fluorescence of cell cultures with a higher confluence is indeed expected since it would indicate that the cells have a high respiration activity and are alive. This would therefore provide a more accurate insight on the interconnectedness, activity and quality of the cell culture, thereby making it more able to provide better feedback on changing the medium to suit the cell culture and manufacture better cells. This can be done since it has been found that healthy mitochondria and thus healthy cell cultures have mitochondrial networks between being highly fragmented and completely fused.
-
B.
Conclusion
Stitching the spectrum together with the microscopy cell image does improve the classification accuracy of the machine learning method. Although this was not a significant improvement because of the already high accuracy of the spectrum-only machine learning method, it does indicate a potential in using both spectrum and image for classification. However, the CellProfiler pipeline would be able to serve as another way to predict and classify the confluence of the cell while checking the activity levels of the mitochondria to ensure that the cells within the culture are healthy and alive.
-
C.
Suggestions for Future Work
Perhaps it could be future researched to find out if other features indicating the health of the cell can be found so that it would be possible to rely on more than one feature to determine the health of the cells in the cell culture. In addition, as it is currently not fully known how fluorescence intensity of a cell’s cytoplasm relates to the health of a cell, it could also be further researched upon in order to give a better understanding and narrowing of the range of values possible for a cell culture to be considered healthy.
Additionally, it is possible that by stitching the graph of the spectrophotometer data and corresponding cell images together in addition to using the average intensity per square pixel of the cell’s cytoplasm in the cell image through the Python code and CellProfiler pipeline, it would be possible to aid in increasing the accuracy of the neural network closer to 100% accuracy. This would be extremely useful in cell manufacturing to give feedback, analyse and predict the confluencies of the cells as well as to aid in suggesting adjustments in the medium to ensure better manufactured cells.
-
D.
Contriution to Cell Manufacturing through Machine Learning
Machine Learning is able to classify features without manually extracting them, allowing for a hands-off classification of images, text and sounds. In addition, machine learning allows for extremely complex features to be recognized by the computer models allowing for classification of much higher accuracy than that of their human counterparts.
Cell Manufacturing is made up of many components. These include cell counting and checking if the cells in the cell culture vessel are healthy. These are labour-intensive tasks which generate a lot of data for which labour can be reduced if machine learning was used in cell manufacturing by feeding the Neural Network images of the cells in the cell culture vessel to measure the number of cells and the health of the cells in the cell culture vessel so that proper adjustments to the cell culture vessel can be suggested to improve the health and growth of the cells in the cell culture vessel. In this case, to reduce the manpower and time needed for the monitoring of cells in the cell culture vessels during cell manufacturing, machine learning is essential.
References
CellProfiler. (2003). Retrieved from https://www.cellprofiler.org/.
Chauhan, N. S. (2019, October 10). Introduction to Artificial Neural Networks(ANN). Retrieved from https://towardsdatascience.com/introduction-to-artificial-neural-networks-ann-1aea15775ef9.
Hargrave, M. (2019, November 18). Deep Learning. Retrieved from https://www.investopedia.com/terms/d/deep-learning.asp.
Kitami, T., Logan, D. J., Negri, J., Hasaka, T., Tolliday, N. J., Carpenter, A. E., … Mootha, V. K. (2012). A Chemical Screen Probing the Relationship between Mitochondrial Content and Cell Size. PLoS ONE, 7(3). https://doi.org/10.1371/journal.pone.0033755.
MathWorks. (n.d.). What Is Deep Learning?: How It Works, Techniques & Applications. Retrieved from https://www.mathworks.com/discovery/deep-learning.html.
pathmind. (n.d.). A Beginner’s Guide to Neural Networks and Deep Learning. Retrieved from https://pathmind.com/wiki/neural-network.
Westermann, B. (2012). Bioenergetic role of mitochondrial fusion and fission. Biochimica Et Biophysica Acta (BBA) - Bioenergetics, 1817(10), 1833–1838. https://doi.org/10.1016/j.bbabio.2012.02.033.
Zamponi, N., Zamponi, E., Cannas, S. A., Billoni, O. V., Helguera, P. R., & Chialvo, D. R. (2018). Mitochondrial network complexity emerges from fission/fusion dynamics. Scientific Reports, 8(1). https://doi.org/10.1038/s41598-017-18351-5.
Acknowledgements
I would like to thank my teacher-mentor, Dr Derrick Yong for his help, advice and feedback throughout the duration of the project, Singapore Institution of Manufacturing Technologies for being supportive of this project, Afif for working on the neural networks to analyse and predict the confluency of the cell culture from the images presented.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Appendix
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Suan, E., Yong, D. (2021). Machine Learning of Biological Data in Cell Manufacturing. In: Guo, H., Ren, H., Kim, N. (eds) IRC-SET 2020. Springer, Singapore. https://doi.org/10.1007/978-981-15-9472-4_10
Download citation
DOI: https://doi.org/10.1007/978-981-15-9472-4_10
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-9471-7
Online ISBN: 978-981-15-9472-4
eBook Packages: Physics and AstronomyPhysics and Astronomy (R0)