Abstract
The intestines of insects are assumed to be the niche of various microbial groups, and a unique microflora could be formed under environmental conditions different from mammalian intestinal tracts. This chapter describes the bacterial flora formed in the intestines of two dragonfly species, “akatombo” (the red dragonfly; Sympetrum frequens) and “usubaki-tombo” (Pantala flavescens), which fly over a long distance, and carotenoid-producing microorganisms isolated from this flora. C30 carotenoids, which were produced by a bacterium Kurthia gibsonii isolated from S. frequens, were structurally determined.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
The intestines of insects are assumed to be the niche of various microbial groups, and a unique microflora could be formed under environmental conditions different from mammalian intestinal tracts. This chapter describes the bacterial flora formed in the intestines of two dragonfly species, “akatombo” (the red dragonfly; Sympetrum frequens) and “usubaki-tombo” (Pantala flavescens) (Fig. 18.1), and carotenoid-producing microorganisms isolated from this flora. Since these dragonflies fly over a long distance, their intestines can be not only a stable residence for microorganisms but also a means of spreading and moving themselves to a wide range of environments. Recently, Nair and Agashe (2016) examined the gut bacterial flora of eight dragonfly species in southern India, including P. flavescens, through a culture-dependent isolation methodology, and found that its community composition was affected by host species and sampling location and month. Symbiosis of insects and microorganisms in the gastrointestinal tracts is likely to be advantageous for the survival of diverse bacteria other than obligate anaerobic species.
2 Fecal Bacterial Flora of the Two Dragonfly Species
Mature adults of Sympetrum frequens and Pantala flavescens were caught in Koka-shi, Shiga, in September and August–September, respectively. Excrements (feces) were collected from these two dragonfly species (30–50 each). Bacterial genomic DNA was extracted from these feces to amplify the 16S ribosomal RNA gene (rDNA) by PCR (the experimental scheme was indicated in Fig. 18.2). The hypervariable V4 region (approximately 250 bp) was targeted to analyze nucleotide sequence of the gene, and PCR amplicon obtained was sequenced by using next-generation sequencing (NGS) technology with Illumina MiSeq apparatus. Note that feces from multiple dragonflies (30–50 individuals) were mixed together to collect sufficient amounts for the analysis, and thus the result represents the mixed bacterial flora of the multiple individuals (percentage of each taxonomic group represents an average in bacterial flora of multiple dragonflies).
At the phylum level, the fecal flora compositions from the two species were significantly different (Fig. 18.3). Proteobacteria (32% of total bacterial flora) and Firmicutes (66%) accounted for the most dominant bacteria in the flora of S. frequens, whereas Chlamydiae, intracellular parasitic bacteria, was the most dominant in P. flavescens (68%). Except for Chlamydiae, Proteobacteria (9%) and Firmicutes (11%) existed in the feces of P. flavescens, which was similar to the case in S. frequens, and Actinobacteria was additionally present (12%).
At the genus level, it should be noted that Lactococcus (Streptococcaceae) was a major constituent in the fecal bacterial flora of S. frequens (54% of total bacteria), indicating that lactic acid bacteria (LAB) can be dominant in the intestine of this dragonfly (Fig. 18.4). Another LAB genus Lactobacillus also existed at the considerable ratio (11%), also indicative of the lactic acid fermentation occurring in the intestine. Other bacterial groups were identified as multi-taxa belonging to Enterobacteriaceae (Morganella, Serratia, and a genus close to Enterobacter/Klebsiella/Kluyvera/Lelliottia/Leclercia/Erwinia/Pantoea/Buttiauxella), Coxiellaceae (Rickettsiella), and Bartonellaceae (Bartonella). On the other hand, the fecal bacterial flora of P. flavescens was distinctively dominated at the genus level; putative Chlamydiales bacterium occupied 68% of the total flora as mentioned above (Fig. 18.5). The second majority was genus Leifsonia/Cryocola (11%), which belongs to Microbacteriaceae that was also not observed as a major group in the bacterial flora of S. frequens. The LAB genera Vagococcus (6%), Lactococcus (2%), and Lactobacillus (1%) were present as minor constituents.
Thus, it is interesting that bacterial floras are significantly distinct depending on the species of dragonflies, especially for the existence of Chlamydiae. It is also intriguing that general inhabitants in mammal intestinal microflora such as Bacteroidetes were not detectable as a major constituent in dragonfly feces, indicating great difference of gut environments between them.
3 Carotenoid-Producing Microorganisms Isolated from Excrement of Dragonflies S. frequens and P. flavescens
As described in Chaps. 1, 2, 3, 4, and 5, carotenoids are generally found in dragonflies. In S. frequens, carotenoids were distributed in feces (590–910 μg/g: predominant), head (9.4–18.0 μg/g), chest (5.4–19.0 μg/g), and abdomen (10.5–18.0 μg/g), indicating that the intestine is the major place of carotenoid accumulation. Intestinal microorganisms are potential candidates as the producer of their domestic carotenoids. Thus, dragonfly feces could be a fruitful experimental target to isolate strains with carotenoid production ability.
Culturable bacteria and yeasts were grown on six media, tryptone soya agar (TSA) (Eiken, Tokyo, Japan), Gifu anaerobic medium (GAM) agar (Nissui, Tokyo, Japan), de Man, Rogosa, and Sharpe (MRS) medium agar (Becton Dickinson, Franklin Lakes, NJ, USA), nutrient agar (NA) (Nissui), standard method agar (SA) (Nissui), and potato dextrose medium agar (PDA, containing 0.01% chloramphenicol) (Eiken). The medium plates were used to isolate aerobic bacteria with TSA, NA, and SA, anaerobic bacteria with GAM, lactic acid bacteria with MRS, and yeasts with PDA. For both dragonflies P. flavescens and S. frequens, viable counts were approximately 106 cfu per gram feces both for aerobic and anaerobic bacteria, and yeast counts were around 103–104 cfu per gram feces (Table 18.1).
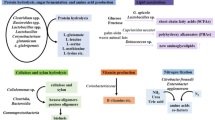
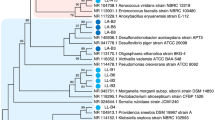
Among microorganisms isolated, yellowish, pinkish, and red colonies were picked and streaked on the medium plates (Fig. 18.6), and their genomic DNA was extracted to analyze nucleotide sequence of the 16S ribosomal RNA gene (rDNA). The hypervariable V1–V3 regions were covered to identify species in the analysis. In the excrement of S. frequens, the most frequently isolated yellow-pigmented microorganism was a bacterial species very close to Kurthia gibsonii that belongs to the Firmicutes phylum (Kg in Fig. 18.6). Thus, we classified this strain as K. gibsonii. Secondary frequent isolates were identified as Bacillus sp. (Bs in Fig. 18.6); some species of genus Bacillus have been reported to be C30 carotenoid producers (Khaneja et al. 2010; Steiger et al. 2012a). Other three yellowish strains were identified as Enterobacter sp. (light-yellow colony, Es in Fig. 18.6), Stenotrophomonas sp. (light-yellow colony, Ss in Fig. 18.6), and Pseudomonas sp. (thick-yellow colony, Ps in Fig. 18.6). This Pseudomonas sp. was found to produce β-carotene and zeaxanthin by Fukaya et al. (2018). One strain indicating sharp red color was isolated and identified as Serratia marcescens (Sm in Fig. 18.6), which has been known as a producer of non-carotenoid pigment prodigiosin (Williams 1973). Interestingly, K. gibsonii was also isolated from the excrement of P. flavescens (Fig. 18.6), suggesting that this bacterium may be commonly detectable from dragonfly intestines. Abovementioned bacteria, however, were not the species dominating the fecal bacterial flora as seen in Figs. 18.3, 18.4, and 18.5; K. gibsonii was detected at low existing ratio (0.8% of total bacteria) in the feces of P. flavescens and was not detected in that of S. frequens possibly due to an extremely low population. Bacillus was also quite low population (<1%) both in the fecal flora of S. frequens and P. flavescens. Thus, it was predicted that the carotenoid producers do not occupy majority in the intestines of these two dragonflies. Finally, one pinkish strain isolated from the feces of P. flavescens was identified as a basidiomycete yeast close to the genus Pseudozyma by analyzing the ITS1 and ITS2 sequences (P in Fig. 18.6).
Bacterial strains belonging to phylum Firmicutes generally produce short-chain C30 carotenoids as seen in Staphylococcus aureus (Pelz et al. 2005) and Planococcus maritimus (Shindo et al. 2008b; Shindo and Misawa 2014), while bacteria belonging to phylum Proteobacteria generally produce C40 carotenoids such as the Pseudomonas sp. described above and Erwinia uredovora (reclassified as Pantoea ananatis) that was elucidated to possess the biosynthetic pathway producing zeaxanthin and its glycosides (Choi et al. 2013; Misawa et al. 1990). A broad range of microorganisms, i.e., yellowish bacterial strains of both phyla described above and the yeast strain producing pinkish pigments, were isolated from the excrements of the dragonflies, indicating that dragonfly feces could be a potential source for the isolation of carotenoid-synthesizing strains.
4 Structural Determination of C30 Carotenoids Produced by Fecal Bacteria of Dragonflies
As described, one strain of Pseudomonas sp. producing β-carotene and zeaxanthin, C40 carotenoid, was isolated from the excrement of S. frequens (Fukaya et al. 2018). Another yellow-pigmented bacterium K. gibsonii, belonging to Firmicutes, was here analyzed for its carotenoids. Through UPLC, UV/VIS, MS, and NMR analyses, a major carotenoid produced by this species was proved to be a novel C30 carotenoid 5,6-dihydro-4,4′-diapolycopene-5-ol 1-glucoside fatty acid ester (Fig. 18.7) (our unpublished data). We also identified 4,4′-diapolycopene, 5,6-dihydro-4,4′-diapolycopene-5-ol and 5,6-dihydro-4,4′-diapolycopene-5-ol 1-glucoside as the minor components (Fig. 18.8).
Past several reports described that some bacteria produced carotenoids with the C30 skeleton of 4,4′-diapolycopene similar to our results (Steiger et al. 2012a, b, 2015; Shindo et al. 2007, 2008a, b), but their chemical structures are different from the major carotenoids of K. gibsonii. It is likely that various carotenoids are produced by gut microbes in dragonfly intestines, suggesting that they may have some role to contribute to the balance of antioxidative chemical substances in the host intestines. On the other hand, either of S. frequens and P. flavescens did not accumulate the C30 carotenoids in the feces and any other body parts as major components (majority was C40 carotenoids, β,β-carotene 14.4%, β,γ-carotene 9.6%, β-zeacarotene 16.2%, and β,φ-carotene 34.0%; our unpublished results), so that further study should be needed to reveal whether, if any, C30 carotenoids could play some role in the dragonfly body or not.
References
Choi S–K, Osawa A, Maoka T, Hattan J, Ito K, Uchiyama A, Suzuki M, Shindo K, Misawa N (2013) 3-β-Glucosyl-3′-β-quinovosyl zeaxanthin, a novel carotenoid glycoside synthesized by Escherichia coli cells expressing the Pantoea ananatis carotenoid biosynthesis gene cluster. Appl Microbiol Biotechnol 97:8479–8486
Fukaya Y, Takemura M, Koyanagi T, Maoka T, Shindo K, Misawa N (2018) Structural and functional analysis of the carotenoid biosynthesis genes of a Pseudomonas strain isolated from the excrement of Autumn Darter. Biosci Biotechnol Biochem 82:1043–1052
Khaneja R, Perez-Fons L, Fakhry S, Baccigalupi L, Steiger S, To E, Sandmann G, Dong TC, Ricca E, Fraser PD, Cutting SM (2010) Carotenoids found in Bacillus. J Appl Microbiol 108:1889–1902
Misawa N, Nakagawa M, Kobayashi K, Yamano S, Izawa Y, Nakamura K, Harashima K (1990) Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J Bacteriol 172:6704–6712
Nair A, Agashe D (2016) Host-specific spatial and temporal variation in culturable gut bacterial communities of dragonflies. Curr Sci 110:1513–1523
Pelz A, Wieland KP, Putzbach K, Hentschel P, Albert K, Götz F (2005) Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus. J Biol Chem 280:32493–32498
Shindo K, Misawa N (2014) New and rare carotenoids isolated from marine bacteria and their antioxidant activities. Mar Drugs 12:1690–1698
Shindo K, Mikami K, Tamesada E, Takaichi S, Adachi k MN, Maoka T (2007) Diapolycopenedioic acid xylosyl ester, a novel glyco-C30-carotenoic acid produced by a new marine bacterium Rubritalea squalenifaciens. Tetrahedron Lett 48:2725–2727
Shindo K, Asagi E, Sano A, Hotta E, Minemura N, Mikami K, Tamesada E, Misawa N, Maoka T (2008a) Diapolycopenedioic acid xylosyl esters A, B, and C, novel antioxidative glyco-C30-carotenoic acids produced by a new marine bacterium Rubritalea squalenifaciens. J Antibiot 61:185–191
Shindo K, Endo M, Miyake Y, Wakasugi K, Morritt D, Bramley PM, Fraser PD, Kasai H, Misawa N (2008b) Methyl 5-glucosyl-5,6-dihydro-apo-4,4′-lycopenoate, a novel antioxidative glyco-C30-carotenoic acid produced by a marine bacterium Planococcus maritimus. J Antibiot (Tokyo) 61:729–735
Steiger S, Perez-Fons L, Fraser PD, Sandmann G (2012a) Biosynthesis of a novel C30 carotenoid in Bacillus firmus isolates. J Appl Microbiol 113:888–895
Steiger S, Perez-Fons L, Fraser PD, Sandmann G (2012b) The biosynthetic pathway to a novel derivative of 4,4′-diapolycopene-4,4′-oate in a red strain of Sporosarcina aquimarina. Arch Microbiol 194:779–784
Steiger S, Perez-Fons L, Cutting SM, Fraser PD, Sandmann G (2015) Annotation and functional assignment of the genes for the C30 carotenoid pathways from the genomes of two bacteria: Bacillus indicus and Bacillus firmus. Microbiology 161:194–202
Williams RP (1973) Biosynthesis of prodigiosin, a secondary metabolite of Serratia marcescens. Appl Microbiol 25:396–402
Acknowledgment
We thank Masato Koshino, Ayaka Mochinaga, Miho Nagao, and Nagisa Matsumura for their contributions to isolation, identification, and microbial evaluation of the carotenoid-producing strains.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Koyanagi, T., Maoka, T., Misawa, N. (2021). Fecal Microflora from Dragonflies and Its Microorganisms Producing Carotenoids. In: Misawa, N. (eds) Carotenoids: Biosynthetic and Biofunctional Approaches. Advances in Experimental Medicine and Biology, vol 1261. Springer, Singapore. https://doi.org/10.1007/978-981-15-7360-6_18
Download citation
DOI: https://doi.org/10.1007/978-981-15-7360-6_18
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-7359-0
Online ISBN: 978-981-15-7360-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)