Abstract
Autophagy and apoptosis are the two evolutionarily conserved processes regulating the turnover of defective organelles and other contents inside cells and damaged whole cells inside organisms, respectively. Although apoptosis and autophagy function differently, their signaling pathways are interconnected and mediated by a toggle switch that is triggered based on the requirements of a cell and its surroundings. Suppression of apoptosis and autophagy due to uncontrolled stress is thought to be a hallmark of carcinogenesis. In general, autophagy and apoptosis mediate each other through a roller coaster of up- and downregulation of factors; that is, autophagy attenuates apoptosis induction, and caspase-dependent apoptosis turns off the autophagic machinery in cancer cells, with several exceptions. Moreover, in certain scenarios, autophagy or autophagy-associated proteins induce excessive degradation of cytoplasmic components, causing “autophagic cell death.” Autophagy can also rescue cancer cells from apoptosis by modulating stress levels, determining cancer cell fate. However, the molecular signals driving the cell toward either autophagy or apoptosis remain largely unknown. Therefore, in this review, we focus on understanding the complex crossover signaling between autophagy and apoptosis pathways and their modulation in the transformation from benign proliferation to malignant carcinogenesis.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
2.1 Introduction
Cell proliferation and cell death are the two wheels on which life processes roll. The proper organization and construction of a living body depend on the coordination between these two processes. Every living organism has an inherent ability to detect and remove individual damaged cells through a process of regulated cell death (RCD). RCD involves very sophisticated molecular machinery by which cells commit themselves to die after receiving a proper signal of death. The RCD process has multidimensional roles ranging from removal of damaged cells to the proper organization and formation of organs and tissues at specific developmental stages. Different forms of RCD, such as apoptosis, pyroptosis, ferroptosis, and so on, are cellular responses to stress that exceed the limit of tolerance. In the past few years, another guardian of the cellular microenvironment called autophagy was found to be extremely important for the maintenance of cellular homeostasis. Autophagy is a self-digestion process in which unwanted cellular organelles and other cellular components are entrapped within a double-layered vesicular structure known an autophagosome and degraded by the subsequent autophagosome fusion with a lysosome. Hence, the process of autophagy contributes toward the maintenance of cellular homeostasis in a varying range of stress conditions, like nutrient deprivation, an abundance of damaged and misfolded proteins, pathogens, infections, and hypoxia (Klionsky 2005; Mahapatra et al. 2019). The function of autophagy as a cytoprotective process is not limited to scavenging damaged sterile targets as it is also a very effective and regulated way of removing intracellular pathogens through xenophagy, which is the sequestration of pathogen-derived proteins and many other types of invaders (Saha et al. 2018).
The ability of a cell to prevent apoptosis is one of the important hallmarks of cancer (Hanahan and Weinberg 2011). Cancer cells disable the apoptosis mechanism either by mutation of tumor suppressors or by overexpressing anti-apoptotic signals to achieve a malignant state. Conventionally, many cancer therapeutics have been developed to kill cancer cells by promoting apoptosis. These therapeutics mainly target damaged DNA to restore mutated tumor suppressors, such as p53. The strategy of these manipulations is to reduce the uncontrolled proliferation of cancer cells and to eliminate them in a regulated and targeted manner through apoptosis. However, when cancer cells are exposed to any type of stress (such as anticancer therapeutics or nutrient deprivation in the tumor microenvironment), the cellular stress response of autophagy can be activated. Here, the tumor-promoting role of autophagy leads to eradicated cell stress that thus inhibits apoptosis, aiding in tumor growth and progression. Such cancer-cell protective autophagy is one of the main causes of therapeutic resistance shown by different cancers against the majority of anticancer drugs. However, the critical function of the role of autophagy in tumor condition is not simple, as it maintains a critical role in both tumor promotion and suppression during various stages of cancer progression. Therefore, in this review, we shed light on the interaction between the two fundamental processes, that is, autophagy and apoptosis, and discuss every possible angle in their connections to cancer progression and roles in effective cancer treatment.
2.2 Apoptosis in Cancer
Apoptosis (apo-separation, ptosis-falling off) is a highly coordinated process of controlled cell depletion described by morphological features such as a blebbing cell membrane, nuclear fragmentation, condensation of chromatin, and so on, leading to the generation of apoptotic bodies that are successively eliminated by the process of phagocytosis (Kerr et al. 1972). The key intracellular architect of apoptosis is a series of cysteine-dependent aspartate-driven proteases (caspases) (Galluzzi et al. 2008; Garrido and Kroemer 2004), but evidence of caspase-independent cell death has also been presented. Caspases are initially secreted in inactive procaspase form and, as the full name implies, the C-terminus is cleaved at an aspartate residue.
2.2.1 Mechanism of Apoptosis
Depending on the activation of caspases, the apoptotic pathways are primarily induced by either extrinsic mechanism or intrinsic/mitochondrial mechanism. The extrinsic apoptotic signaling is activated through a death receptor (DR), a special type of cell surface receptor in tumor necrosis factor (TNF) superfamily proteins (Bhardwaj and Aggarwal 2003). Signals for the extrinsic apoptotic pathways are transmitted by death ligands that interact with and are activated by DRs. Structurally, TNF family receptors possess a conserved death domain (DD) that undergoes trimerization upon binding with ligands like TRAIL (tumor necrosis factor-related apoptosis-inducing ligand) and that recruits additional DD-containing proteins like FADD (Fas-associated protein with death domain) and TRADD (TNF-R type 1-associated death domain protein) to form the death-inducing signaling complex (DISC), followed by the interaction and induction of caspase 8 and 10. Then the stimulated caspase 8 and 10 cleave caspases 3, 6, and 7, leading to the subsequent cleavage of target peptides (Fig. 2.1) (Mukhopadhyay et al. 2014; Wang and El-Deiry 2003).
Overview of apoptotic pathways. It is mainly regulated through extrinsic and intrinsic way to induce cell death. The extrinsic mode activates the ligands like FasL and TRAIL to bind with receptors and ligand-receptor interaction induces DISC complex consisting of FADD, TRADD, and an initiator caspase-like caspase 8 to form active caspase. The active caspase 8 then activates executioner caspases like caspase 3, 6, and 7 to regulate the apoptotic process. In contrast, the intrinsic mode in response to DNA damage, increased ROS, activate several BH3 only proteins like PUMA, NOXA, Bim, Bid, and Bad to act on mitochondria to release cytochrome c. The cytochrome c then interacts with Apaf-1 and pro-caspase 9 to form a complex called apoptosome to activate the caspase 9. The active caspase then interacts with executioner caspases to regulate the intrinsic apoptosis
The intrinsic apoptotic signaling pathway is triggered by different stresses, but the first and irreversible step in the sequence of events activating this apoptosis is the loss of the mitochondrial membrane potential (MMP) due to outer membrane permeabilization. In this process, the guardians of mitochondrial integrity belong to the Bcl-2 family of proteins containing a Bcl-2 homology (BH) domain, with the BH1–BH4 domains playing important roles. Several proteins belonging to this family have all four BH domains, including Bcl-w, Bcl-xL, Mcl-1, Bcl-B, and A1, which inhibit apoptosis and thus led to their identification as antiapoptotic proteins. The proapoptotic members consist of two groups: BH3-only proteins and BH123 proteins. The BH123 proteins include BCL-2-associated X protein (Bax), BCL-2 antagonist/killer (Bak), and BCL-2 ovarian killer (Bok). The BH3-only protein group includes the p53-upregulated modulator of apoptosis (PUMA), Noxa, BCL2-associated agonist of cell death (Bad), BH3-interacting domain death agonist (Bid), BCL-2-interacting mediator of cell death (Bim), and Harakiri (HRK). The BH3-only proteins perceive apoptosis signalings, such as that induced by DNA damage and ER stress, and are then translocated to the outer mitochondrial membrane, where they activate Bak and Bax or causes inhibition of anti-apoptotic proteins. Activated Bak and Bax oligomerize at the surface of mitochondria and facilitate the formation of pores in the outer membrane of mitochondria, leading to loss of membrane permeability and the release of several apoptosis-inducing proteins, such as SMAC/DIABLO and cytochrome c, from mitochondria to the cytosol. The released cytochrome-c activates with apoptotic protease-activating factor 1 (APAF1) to form the apoptosome complex and induces the activation of caspase-9, which subsequently activates caspases 3, 6, and 7 (Fig. 2.1) (Galluzzi et al. 2012; Kaya-Aksoy et al. 2019; Singh et al. 2019).
2.2.2 Role of Apoptosis in Cancer
Cancer is a complex and variable process with excessive variations in the genetic material that leads to cancer development, with several steps occurring progressively, starting with the initiation of tumorigenesis and eventually leading to metastasis. During cancer development, cells encounter byproducts of the physiological elimination of damaged cells. Therefore, cancer cells must acquire some protective machinery to prevent induced programmed cell death, that is, apoptosis. Cancer cells can regulate apoptotic pathways transcriptionally, translationally, and post-translationally to avoid stress, such as that generated by hypoxia and genomic instability. Moreover, cancer cells may suppress apoptosis by increasing anti-apoptotic genes expression or decreasing proapoptotic genes expression in a context-dependent manner, which results in higher anti-apoptotic protein expression. Generally, apoptotic pathways are restricted in cancer cells to prevent this cellular response. Several reports have indicated that attenuated interactions of the proapoptotic BIM which is a BH3-only protein with the antiapoptotic protein BCL-2 can support the survival and growth of cancer cells (Hübner et al. 2008). Furthermore, the induction of BIM induces oncogenic inactivation and apoptosis in acute lymphoblastic leukemia (Li et al. 2016b). Notably, it was demonstrated that the inhibition of a BH3-only protein or a caspase protein caused a genetic mouse model to develop resistance against certain proapoptotic signals with a commensurate increase in tumor initiation (Parsons et al. 2013). The BH3-only molecules BIM and PUMA are downregulated in breast cancer cells, and overexpression of these molecules induces HER2 inactivation, which induces apoptosis (Bean et al. 2013). Overexpressed BH3-only molecule BIM function as a cytoprotective molecule in cancer cells, and its association through phosphorylation with BCL-xL/MCL-1 block its proapoptotic functions (Gogada et al. 2013). Moreover, caspase-2 deficiency impedes apoptosis and generates genomic instability, resulting in tumorigenesis (Shalini et al. 2016). However, MCL-1 is a vital prosurvival factor in triple-negative breast cancer (TNBC), and its inhibition might be an effective strategy for treating TNBC (Li et al. 2018). Furthermore, the suppression of the tumor suppressor gene p53 mediates cell proliferation, and stabilized p53 phosphorylation activates BAK and BCL-xL, inducing apoptosis (Nieminen et al. 2013). However, suppression of the proapoptotic protein BCL-2 antagonist killer 1 (BAK1) leads to the proliferation of breast cancer cells of various lineages (Zhou et al. 2010), whereas the inhibition of BCL-xL and BCL-2 activates BAX/BAK and induces apoptosis in human myeloid leukemia cells (Rahmani et al. 2013). Thus, it is strongly said that the inhibition of apoptosis has to play a critical role in the case of cancer cell survival and tumor development. Targeting apoptosis induction is a novel strategy for cancer therapy.
2.3 Autophagy in Cancer
Autophagy (in Greek, “auto,” defined as oneself, and “phagy,” defined as to eat) states to a self-cannibalistic mechanism to degrades cytoplasmic components and unwanted organelles through lysosomes to maintain the homeostasis (Levine and Klionsky 2017; Mancias and Kimmelman 2016; Tan et al. 2017). Recent reports have suggested that autophagy significantly modulates various physiological processes, such as growth, development, cell division, and immunity, with any dysfunction in autophagy leading to severe pathophysiological implications, such as neurodegenerative disorders, autoimmune disorders, and cancer.
2.3.1 Mechanism of Autophagy
Autophagy is a complex multistep process regulated through the coordinated action of 42 ATG (AuTophaGy) genes that are sequentially involved in different steps, such as (a) phagophore nucleation and formation, (b) elongation of phagophores, (c) cytoplasmic cargo selection, (d) lysosomal docking and fusion, and (e) cargo degradation (Fig. 2.2) (Mancias and Kimmelman 2016; Wang and El-Deiry 2003; White 2015). Autophagy is initiated upon a shift in the extracellular milieu of a cell, mainly due to different stress signals, such as loss of growth factor signaling, nutrient deprivation, energy depletion, and hypoxia. These stress signals induce the release of reactive oxygen species (ROS) that inhibit the TOR1 (target of rapamycin complex 1)-dependent signaling pathway, which acts as a molecular sensor for autophagy initiation. First, the ATG protein complex is formed as a scaffold of regulatory proteins ATG1, ATG13, ATG17, ATG29, and ATG31, which recruits other ATG proteins to activate downstream targets via phosphorylation and a PAS (phagophore assembly site/pre-autophagosomal structure) (Bhol et al. 2019; Bhutia et al. 2010; Davies et al. 2015; Levine and Klionsky 2004). Then, the cytoplasmic contents, protein aggregates, defective organelles are sequestered through a double-membrane structure known as a phagophore. For phagophore formation, the Ulk1 kinase, in combination with ATG13 and ATG17, is activated at the same time as transmembrane protein ATG9, which extracts phospholipids. Subsequently, the class III PI-3 kinases, particularly Vps34 (vesicular protein sorting) interacts with Beclin1 leading to increase phagophore catalytic activity, in which phosphatidylinositol (PI) is utilized to produce phosphatidylinositol triphosphate (PI3P) for phagophore elongation. The formation of autophagosomes and elongation of vesicles depends on three ubiquitin-like conjugation systems: ATG12 is activated through the utilization of ATP and ubiquitin-activating enzyme E1-like ATG7, which non-covalently binds to ubiquitin-activating enzyme E2-like ATG10. Then, the E2-like action triggers the covalent binding of ATG12 and ATG5, forming the conjugated ATG5-ATG12 complex. This complex then pairs with ATG16L to form the ATG5–ATG12–ATG16L complex, which is essential for the extension of the phagophore and acquisition of the appropriate membrane curvature, and once double-membrane autophagosomes are formed, the complex starts to disintegrate. Thus, the formation of the ATG5-ATG12 conjugated complex does not depend on the activation of autophagy and is a poor marker of autophagy. A second ubiquitin-like system is required for the formation of microtubule-associated protein light chain 3 (LC3), known as the mammalian homolog of ATG8. During autophagy induction, cysteine protease ATG4 cleaves LC3 to form LC3-I. This complex is in turn stimulated in an ATP-dependent way through ATG7, E1-like enzyme. Following this activation stimulated LC3-I is transferred to ATG3 (E2-like) and ATG5-ATG12-ATG16L (E3-ligase complex) before it is conjugated with phosphatidylethanolamine (PE) to generate LC3-II. This lapidated LC3 acts as a receptor for cargo selection in coordination with different adaptor proteins (p62/SQSTM1, NBR1, TAXBP1, etc.) or organelle-specific receptor proteins (PHB2, AMBRA1, Nix, BNIP3L, etc.) contributing to selective molecule uptake and degradation (Bhol et al. 2019; Grumati and Dikic 2018). During docking of autophagosomes to preexisting lysosomes, autolysosomes are formed. In this step, the acidic constituents of the lysosomes digest the selected cargos of the autophagosomes. These organelles migrate along the side of microtubules in a bidirectional manner, with autophagosomes having an affinity for the lysosome-enriched microtubule organizing center (LEMOC). Vesicular docking and fusion are regulated through several proteins, including LAMP-2 (lysosomal membrane protein-2) and GTPases, such as class C Vps proteins, SNARE (soluble N-ethylmaleimide-sensitive factor activating protein receptor), ESCRT (endosomal sorting complex required for transport), and Rabs (Rab7). Any mutation in these proteins halts the progression of autophagosome maturation and fusion. In addition to these proteins, UVRAG, a Beclin1-interacting protein, also plays key roles by maintaining the fusion machinery on autophagosomes and clamping the class C Vps proteins. Thus, Rab7 is activated, which in turn enhances the fusion of lysosomes and late endosomes (Levine and Kroemer 2008; Mathew et al. 2007; Mizushima 2007; Xie and Klionsky 2007).
An autophagic mechanism in the mammalian system. It involves four major orchestrated events starting with the initiation of phagophore followed by its elongation, maturation, and fusion with the lysosome. The cellular stress conditions act as inducers of phagophore initiation through activation ULK1/2 associated autophagic initiation complex. The activated ULK1/2 then triggers various ATGs like ATG1, 29, 17, 13, 31, and 9 along with Beclin1-Vps34 complex for initiating phagophore formation by importing lipid molecules from various organelles. The PI to PI3P conversion by the Beclin1-Vps34 complex precedes ATG5–ATG12 conjugation and its interaction with ATG16L. This interaction is essential for elongating phagophore and it is assisted by the processing of LC3 where the LC3 B-II is incorporated into the outer phagophore membrane. This event is followed by the cargo selection for their degradation leading to the formation of a complete phagophore called an autophagosome. The cargo loaded autophagosome then fuses with competent lysosomes to form autolysosome where the lysosomal content is used for the degradation process
2.3.2 Dual Role of Autophagy in Cancer
The actual role of autophagy in cancer is still debatable because of its dual action, functioning as both a survival- and death-promoting mechanism. Survival-promoting autophagy increases the chances of cancer cell survival under adverse conditions by positively regulating the hallmarks of cancer. In contrast, death-promoting autophagy kills cancer cells by limiting mechanisms such as ROS production and the degradation of survival proteins. Therefore, the correct therapeutic approach for cancer treatment, by either promoting or inhibiting autophagy, is still a matter of discussion. Studies have also shown that autophagy is associated with tumor-suppressive function and that inhibition or defective autophagy is associated with tumor induction and malignant transformation. For instance, loss of Beclin1 in mice resulted in the development of hepatocellular carcinoma (Liang et al. 1999). Moreover, heterozygous deletion of Beclin1 was also associated with the development of breast and other human malignancies, suggesting that Beclin1 acts as a tumor suppressor (Qu et al. 2003; Yue et al. 2003). Similarly, the downregulation of ATG5 has been reported to be associated with early-stage cutaneous melanoma pathogenesis (Liu et al. 2013). Although the senescence mechanism by which autophagy acts as a tumor suppressor is controversial, a few studies have suggested that autophagy and senescence can occur simultaneously. Autophagy suppresses melanoma tumorigenesis by inducing senescence (Liu et al. 2014). A recent study in papillary thyroid carcinoma by Liu et al. suggested that BIRC7 induces the epithelial-mesenchymal transition and metastasis by limiting autophagy (Liu et al. 2020). Similarly, ATG7 induces triple-negative breast cancer progression by inhibiting the invasion, migration, and epithelial–mesenchymal transition (Li et al. 2019). In contrast, autophagy has been found to contribute to tumorigenesis by giving the advantage to tumor cells over normal cells under various stress environments and supporting cancer cell aggressiveness. For example, in a Chinese Han population, the expression of ATG12 consequently contributes to the risk of head and neck squamous cell carcinoma (Song et al. 2018). Increased expression of ATG5 induced by HIF1α has been reported to increase tumor size in prostate cancer (Yu et al. 2019). In colon cancer, RACK1-induced protective autophagy triggers cell proliferation and attenuates apoptotic cell death (Xiao et al. 2018). Similarly, in renal cell carcinoma, autophagy is critical for cell survival and the epithelial-mesenchymal transition (Singla and Bhattacharyya 2017). Guo et al. reported an oncogenic role of CCAT1 in hepatocellular carcinoma mediated through ATG7-dependent autophagy induction (Guo et al. 2019). Studies have also reported the association of autophagy with enhanced drug resistance. For example, Wnt3a is reported to promote radioresistance via autophagy in head and neck squamous cell carcinoma (Jing et al. 2019). Similarly, cisplatin resistance and osteosarcoma progression were induced by SNHG16 upregulation of ATG4B expression (Liu et al. 2019).
2.4 The Role of Autophagy and Apoptosis Crosstalk in Cancer Growth and Progression
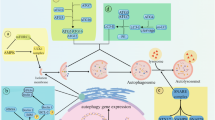
Cells undergo apoptosis under extreme stress, whereas autophagy is well known for its involvement in cellular homeostasis. Hence, the induction of autophagy in cells undergoing stress before the point of no return can protect them from death. However, after cells are destined for death, autophagy can induce a peculiar form of cell death called autophagic cell death (Apel et al. 2009; Wong 2011; Yang et al. 2015). In this context, some reports have shown critical crosstalk between components of two highly complex processes, that is, autophagy and apoptosis (Fig. 2.3).
Autophagy and apoptosis crosstalk during cellular stress. The crosstalk modulates through the interaction of Bcl2/Bcl-xL–Beclin1 and Bcl2/Bcl-xL–proapoptotic BH3 only proteins. Cellular stress like nutrient starvation activates the Ulk1 complex by inhibiting mTOR results in its interaction with the autophagic initiation complex for the onset of autophagy. This is made possible by dissociating Bcl2/Bcl-xL–Beclin1 complex through the phosphorylation of Beclin1 and Bcl2/Bcl-xL. The free Beclin1 with other autophagic proteins activates the elongation complex which directs the cell to employ either autophagy or apoptosis by inhibiting anti-apoptotic proteins through ATG5, ATG12, and by regulating pro-apoptotic proteins through ATG5–FADD interaction and caspase 8 activations through ubiquitinated p62. In contrast, the cell committed to autophagy restricts apoptosis through the lysosomal degradation of caspases. Moreover, stress like DNA damage induces p53 regulates apoptosis through inhibition of anti-apoptotic proteins through BH3 only proteins and through phosphorylating of Bcl2. It also helps in the activation of caspases and proapoptotic proteins like Bax. The p53 also regulates autophagy either by activating it through DRAM mediated mitophagy or by inhibiting through suppression of AMPK and by degrading autophagic proteins like Beclin1, Vps34, ATG3 through activated caspases. Some inhibitor of apoptotic proteins like FLIP regulates apoptosis by inhibiting caspases and autophagy by disrupting the ATG3- ATG8 of the elongation complex
2.4.1 The Intersection of Autophagy and Apoptosis Molecules
One of the most important and well-known points of interaction between the autophagic and apoptotic pathway coincides with the action of antiapoptotic Bcl-2 protein and the conserved autophagic protein Beclin1. Antiapoptotic proteins including Bcl-2 family members (Bcl-2, Bcl-XL, and Mcl-1) with all four BH3 domains interact with the proapoptotic members through BH4 domains (Pattingre et al. 2005). The BH3 binding pocket of Bcl-2 binds with the BH3 domain of Beclin1 to inhibit Beclin1-dependent autophagy. ER-localized Bcl-2 inhibits starvation-induced Beclin1-dependent autophagy by binding with Beclin1 facilitated by NAF-1 (nutrient-deprivation autophagy factor-1) (Chang et al. 2010). The binding affinity of Beclin1 for Bcl-2 is lower than the proapoptotic proteins due to the presence of a polar threonine instead of a hydrophobic amino acid at position 119 (Feng et al. 2007; Oberstein et al. 2007). Therefore, the interaction between Beclin1 and Bcl-2 does not disturb the antiapoptotic property of Bcl-2. During stress-induced autophagy, BH3-only proteins bind to the BH3-binding pocket of Bcl-2 to disrupt their interaction. STKs (Ser/Thr kinases) including JNK (JUN N-terminal kinase), Akt, and DAPK, also make regulatory contributions to the processes of autophagy and apoptosis through their crosstalk with other Bcl-2 family members. c-Jun N-terminal protein kinase 1 (JNK1) controls both autophagy and apoptosis through the phosphorylation of Bcl-2. Under conditions of mild stress, Bcl-2 phosphorylation by JNK-1 causes the dissociation of BCl-2 and Beclin1 but not that of the Bcl-2 and Bax to initiate autophagy; however, under prolonged stress, the Bcl-2-Bax interaction is disrupted, which initiates apoptosis (Wei et al. 2008).
Many studies have shown that active caspases cleave key autophagy-related proteins and allow apoptosis to overtake autophagy. ATG5, a protein required for phagophore elongation, is cleaved by calpain, an active caspase, to form a 24 kDa protein that interacts with Bcl-XL to induce apoptosis through the release of cytochrome c (Yousefi et al. 2006). Furthermore, ATG12, a copartner in the ATG5-ATG12 conjugation system, is thought to act as a proapoptotic protein by supporting the other apoptotic proteins in activating the caspases under various stresses. Moreover, ATG12 can inhibit the anti-apoptotic proteins Bcl-2 and Mcl1 by acting specifically on the BH3-like domain of ATG12. Similarly, in acute lymphoblastic lymphoma (ALL), the cleavage of ATG3 by caspase 8 promotes apoptosis by inhibiting autophagy (Oral et al. 2012). In contrast, caspase 9, which is involved in the intrinsic pathway of apoptosis, is found to lipidated LC3 by interacting with ATG7 to promote autophagy. Hence, apoptosis induction results in the site-specific breakdown of autophagic proteins such as Beclin1, ATG5, and ATG7 to undermine the cytoprotective effects of autophagy (Marquez and Xu 2012). Interestingly, the autophagy adaptor protein p62 displays a critical role in caspase activation to induce apoptosis (Islam et al. 2018; Jung and Oh 2019). The tumor suppressor p53 also regulates crosstalk at various points. It activates autophagy through the AMPK–TSC2–mTOR axis by translocating to the nucleus upon stress, whereas in the cytoplasm, p53 interacts with FIP200 and inhibits autophagy (Vousden and Lane 2007). Cytoplasmic p53 interacts with several Bcl-2 pro-apoptotic family proteins, such as Bax, NOXA, PUMA, and others, to induce apoptosis by regulating mitochondrial outer membrane permeability (MOMP) (Vaseva et al. 2012). Moreover, p53-induced DRAM-mediated autophagy is also associated with apoptosis (Crighton et al. 2006). Furthermore, the anti-apoptotic protein FLICE-like inhibitor protein (FLIP) inhibits apoptosis by inactivating death receptors and is known to regulate autophagy by interacting with autophagic proteins such as ATG3 and LC3. FLIP can also influence both apoptosis and autophagy independently by acting on the plasma membrane to inhibit apoptosis and at the site of autophagosome formation to modulate autophagy, like the action of Bcl-2 (Eisenberg-Lerner et al. 2009). Interestingly, serum starvation was shown to activate antiapoptotic protein cIAP to promote mitochondrial autophagy. It revealed that cIAP1 translocated onto mitochondria to interact with Ulk1, TOM20, and LC3 to stimulate mitophagy through the ubiquitination pathway (Mukhopadhyay et al. 2016).
An autophagosome, by itself, modulates apoptosis either through direct sequestration and removal of proapoptotic proteins or through the engulfment and subsequent elimination of damaged cellular molecules, such as those from the mitochondria-dependent apoptotic cell death. In another mechanism, the membrane of autophagosome functions as a platform for processing of apoptotic proteins and thus contributes to the process of apoptosis; for example, in the presence of bortezomib and pan-sphingosine kinase inhibitor, SKI-I, there is autophagy-dependent activation of the extrinsic apoptotic pathway through caspase-8. Caspase 8 in association with FADD is recruited to the membrane of autophagosomes in a p62-dependent manner by interacting with ATG5 (Mukhopadhyay et al. 2014; Young et al. 2012).
2.4.2 The Role of Autophagy and Apoptosis Crosstalk in Cancer
Although the interconnection between apoptosis and autophagy has unique complexities, many studies that explain how apoptosis and autophagy are interlinked and induce cell death or sustain tumor growth and proliferation with common regulators (Table 2.1). For example, p53 was found to modulate autophagy and apoptosis in context-dependent way in different types of cancer. It showed that autophagy degrades p53 to maintain the hepatic cancer stem cells (Liu et al. 2017). Recently, the induction of autophagy by activating the AMPK-ULK1 axis and inhibiting mTOR was found to induce apoptosis through caspase activation, which reduced tumor proliferation in triple-negative breast cancer (Cao et al. 2018). Another study also showed that the activation of autophagy via PI3K/AKT/mTOR signaling reduced the viability of prostate cancer cells by inducing cytotoxicity in conjunction with apoptosis-mediated cell death (Tian et al. 2017). Besides, the autophagy adaptor protein p62 plays central signaling for tumor initiation as well as suppression of tumor progression in the stromal cells (Moscat et al. 2016; Zhang et al. 2013). During cisplatin-mediated ER stress, the induction of cell death occurs through caspase-mediated apoptosis, but Beclin1 mediates autophagy to eliminate excessive stress in human lung cancer cells (Shi et al. 2016). Bax-negative colon cancer cells can undergo TRAIL-induced cell death under compromised autophagy conditions (Li et al. 2016a; Mariño et al. 2014). Moreover, it has been reported that the inhibition of autophagy causes an elevation in NOXA expression, which is a Bcl2 family protein with a BH3-only domain. Generally, NOXA is degraded in the autophagic pathway by sequestration onto autophagosomes through the action of the adaptor protein p62. However, the blockage of autophagy leads to the accumulation of NOXA, which subsequently activates apoptosis and acts as a bridge between autophagy and apoptosis in cancer cells (Wang et al. 2018). In addition, proapoptotic protein PUMA and Bax promote autophagy to contribute to apoptosis (Yee et al. 2009). Interestingly, our study showed that in response to anticancer therapy, mitophagy was induced through PUMA leading apoptosis in glioma cells (Panda et al. 2018; Yee et al. 2009). Moreover, the autophagy protein Ulk1 and ATG14 have found to have an important role in the induction of apoptosis. The upregulation of Ulk1 translocated to mitochondria to inhibit the activity of manganese superoxide dismutase resulting in the production of ROS causing to cell death the cancer cells (Mukhopadhyay et al. 2015, 2017). Furthermore ATG14 along with Ulk1 induced lipophagy, selective autophagy resulting in free fatty acid accumulation leading to ER stress-mediated apoptosis (Mukhopadhyay et al. 2017. On other hand, UVRAG also can act as an antiapoptotic protein by preventing the translocation of Bax to mitochondria, where it initiates mitochondrial apoptosis in response to therapy (Eisenberg-Lerner et al. 2009; Maiuri et al. 2009). Interestingly, UVRAG with truncating mutation displayed higher inflammatory response through NLRP3-inflammasome hyperactivation and exhibited significant spontaneous tumorigenesis through β-catenin stabilization and centrosome amplification (Quach et al. 2019) establishing complex crosstalk between autophagy and apoptosis during tumor growth and progression.
During chemotherapeutic stress, cancer cells trigger autophagy to eradicate stress and support tumor growth and progression. Such protective autophagy is the major cause of therapy resistance that develops against a majority of anticancer drugs used for different cancers. For example, cisplatin-induced autophagy protects cancer cells against drug-induced apoptosis (Harhaji-Trajkovic et al. 2009). Therefore, autophagy inhibitors in combination with apoptosis-inducing drugs might increase the efficacy of anticancer therapy. Chloroquine, a potent autophagy inhibitor, is reported to enhance existing chemotherapeutics without inducing toxicity in cells (Amaravadi et al. 2007). Hence, the use of chloroquine and its analog hydroxychloroquine for inhibiting autophagy following anticancer therapy is widely accepted and is currently under clinical trial (Cudjoe et al. 2019). Docetaxel-induced autophagy (Zhang et al. 2019) and paclitaxel-induced autophagy (Kim et al. 2013) play tumor protective roles in cancer cells, leading to treatment failure. Pretreatment with 3-MA along with paclitaxel significantly enhances cytochrome C release and the subsequent induction of the mitochondrial apoptotic pathway (Xi et al. 2011). Furthermore, the expression of FOXO3, an autophagic transcription factor, is increased after autophagy is blocked. Then, FOXO3 at increased levels can directly bind to the promoter of the proapoptotic protein PUMA and hence trigger the apoptotic pathway in osteosarcoma and other cancer cells (Fitzwalter and Thorburn 2018; Fitzwalter et al. 2018; Jiang et al. 2017). Collectively, these studies implicate the antiapoptotic function of protective autophagy against therapy-induced apoptosis in cancer cell lines that leads to therapy resistance. However, contradictory evidence is presented for cases of cancer cells induced toward death upon radiation treatment, which attenuates apoptosis by knocking out proapoptotic proteins (such as Bax and Bak), and the treatment-associated cell death is not attributed to apoptosis but type-II programmed cell death, that is, autophagy (Kim et al. 2006).
2.5 Modulation of Apoptosis and Autophagy in Potential Cancer Therapeutics
The present era of targeted cancer therapy is rationally based on specific signaling pathways that have high expression in specific tumor types. The paradigm suggests that target-specific therapeutic agents sensitize cancer cells toward chemotherapy through a closed circuit of signals shuffled between apoptosis and autophagy components (Table 2.2).
2.5.1 Distinguishing Autophagic Cell Death from Apoptosis for Cancer Therapeutics
Since cancer cells can block apoptosis and increase their resistance to chemotherapeutic agents, targeting autophagy as an alternative cell death pathway is an attractive strategy for anticancer therapy (Jain et al. 2013). Arsenic trioxide and EB1089 (a vitamin D analog) have been reported to induce dynamic changes in lysosomal activity that provoke Beclin1-mediated autophagic cell death (Lo-Coco et al. 2013; Qian et al. 2007). Several chemotherapeutic agents, such as dexamethasone with fenretinide or etoposide, along with key dietary phytochemicals, such as resveratrol and fisetin, have also been reported to induce autophagic cell death in a type of non-apoptotic programmed cell death mediated by Bcl2-dependent autophagy genes (Fazi et al. 2008; Jain et al. 2013; Laane et al. 2009). Sodium selenite, a mitophagy inducer, has also emerged as a therapeutic agent against malignant glioma cells that subsequently promotes cell death through superoxide-mediated mitochondrial damage (Kim et al. 2007). Imatinib and cannabinoid have also exhibited autophagic cell death in glioma cells through the inhibition of autophagy, leading to the stimulation of ER stress (Salazar et al. 2009). Spautin-1, a potent autophagy inhibitor, triggers the inhibition of autophagy in chronic myeloid leukemia cells to induce autophagic cell death through the inhibition of class III PI3K and targeting the deubiquitination of USP10- and USP13-mediated degradation of Beclin1 (Liu et al. 2011; Shao et al. 2014; Wilde et al. 2018). Moreover, β-lapachone and elisidepsin also induce marked levels of autophagic cell death in lung cancer. Tamoxifen, bortezomib, trastuzumab, and sulforaphane have been used against breast cancer and provoke cell death by inhibiting autophagy. Bortezomib, 5-FU, and sulforaphane have also been used in colorectal cancer treatment. Temozolomide, 4-HPR, imatinib, rapamycin, and PI-103 are effective in the treatment of colorectal cancer. The inhibition of autophagy at the early stage attenuates the cytotoxicity induced by chemotherapeutic drugs, and this effect is augmented when autophagy is inhibited at later stages. Chronic myeloid leukemia responds to treatment with SAHA and OSI-027 in an autophagy-dependent cell death manner through the epigenetic modulation of histone proteins (Choi 2012). ADI-PEG20 and saracatinib have been reported to be effective against prostate cancer. AZD2014, another small-molecule autophagy inhibitor, has shown promise in the clinical treatment of breast cancer and acute lymphoblastic leukemia through the modulation of PI3K/AKT/mTORC2 signaling (Tabe et al. 2013). A dietary phytochemical, quercetin also displays potent anticancer efficacy in gastric cancer. Voacamine, a bisindole alkaloid, has been shown to induce autophagic cell death in an apoptosis-independent manner (Panda et al. 2015). In preclinical trials with patients diagnosed with melanoma, pancreatic adenocarcinoma or bladder cancer, CQ and HCQ, as single agents, have been shown to have potent anti-cancer properties, as exhibited through the inhibition of autophagy. Furthermore, treatments based on CQ or HCQ combined with metabolic stressors have been found to potentiate autophagy-mediated cell death (Amaravadi et al. 2011; Jain et al. 2013; Panda et al. 2015; Wilde et al. 2018).
2.5.2 Autophagic Facilitation of Apoptosis in Cancer Therapeutics
Autophagy-mediated facilitation of apoptosis in several cancer cells displays a remarkable therapeutic avenue against cancer. Both induction and inhibition of autophagy drive cellular mechanisms toward apoptosis induction. Recent studies have shown that the gallic acid in Terminalia bellirica extracts inhibits autophagy and can be used to fuel the induction of apoptosis in a ROS-dependent manner in oral squamous cell carcinoma (Patra et al. 2020). Verteporfin, a benzoporphyrin derivative, in conjunction with gemcitabine, inhibits autophagy and promotes apoptosis in acute promyelocytic leukemia and exhibits clinical potency against glioma, prostate, and colon cancer (Donohue et al. 2011, 2013). Furthermore, elaiophylin has also been reported to inhibit the autophagic flux that is associated with the endoplasmic reticulum stress to promote apoptosis in ovarian cancer (Zhao et al. 2015). Deguelin, another autophagy inhibitor, triggers apoptosis and enhances the chemosensitization of several types of cancer cells to doxorubicin (Xu et al. 2017). Also, withaferin A and ginsenoside have been reported to induce apoptosis after autophagy inhibition in breast and esophageal cancer cells (Muniraj et al. 2019; Zheng et al. 2016). Similarly, liensinine, an isoquinoline alkaloid, has also been reported to inhibit autophagosome-lysosome fusion to provoke the induction of mitochondrial fission and apoptosis in triple-negative breast cancer (Zhou et al. 2015).
2.5.3 Apoptosis-Autophagy Links in Cancer Therapeutics
Apoptosis and autophagy may induce cell death during chemotherapy in a parallel or sequential manner. The cooperation of apoptotic and autophagic machinery is required for the induction of cell death in the mature tumor environment (Jain et al. 2013). Arsenic trioxide in combination with all-trans retinoic acid has evolved as a potent drug against acute promyelocytic leukemia by modulating both apoptotic and autophagic pathways (Lo-Coco et al. 2013; Qian et al. 2007). Furthermore, plumbagin (a naphthoquinone derivative) has also been reported to induce apoptosis and autophagy with two long synthetic MAPK inhibitors, SB202190 and SB203580 (Li et al. 2014). Several dietary phytochemicals, including resveratrol, curcumin, quercetin, lutein, lycopene, catechin, and β-carotene, have demonstrated proapoptotic and autophagic potential owing to their antioxidant properties in several cancer cells (Choi 2012). Recently, different plant lectins, such as Abrus agglutinin, a lectin from Abrus precatorius, was implicated in the onset of autophagy and is being considered as a means to induce apoptotic cell death in prostate cancer and oral squamous cell carcinoma (Panda et al. 2020; Panigrahi et al. 2020). Another lectin, concanavalin A, has also been reported to downregulate PI3K/Akt/mTOR signaling, thus contributing to autophagic cell death (Roy et al. 2014). Finally, concanavalin A has also induced caspase-dependent apoptosis in human melanoma cells (Liu et al. 2009).
2.6 Conclusion and Future Perspective
Being a double-edged sword, autophagy regulates other modes of cell death in cancer cells. Several of its key regulators act as tumor suppressors or promoters, depending on the threshold level of the cellular or tumor microenvironmental stress in the cancer cell milieu. Moreover, the inhibition of autophagy disrupts cancer cell metabolism, interferes with differentiation, and destabilizes anticancer immunosurveillance. At later stages of tumorigenesis, restoration of autophagy leads to the development of chemo- and/or radioresistance in cancer cells. As explained earlier, the majority of research has focused on understanding the coordinated regulation of autophagy and apoptosis, which sensitizes cancer cells toward death. However, several issues remain unresolved, such as the mechanism by which apoptotic activation of effector caspases turns off the autophagic machinery and the identities of key autophagy proteins that drive cells from being in a pro-autophagic state to acquiring a proapoptotic phenotype during this period, causing various pathophysiological consequences. Several clinical trials using autophagy inhibitors, such as chloroquine and hydroxychloroquine, have also been used for targeted cancer therapy. Besides, several new anticancer drugs have been formulated to modulate both autophagy and apoptosis. However, altering only autophagy in cancer cells might not be an ideal approach; although it is beneficial at low levels, overactive autophagy becomes detrimental, leading to tumor development. Hence, anticancer or antidegenerative drugs modulating autophagy and targeting apoptosis may work more effectively in the clinic when combined. Altogether, it will be interesting to reveal the mechanism and thereby understand autophagy and apoptosis in cancer, which will help to leverage their functional interrelation for developing new targets for the possible effective therapeutic intervention of cancer therapy.
References
Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB (2007) Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest 117:326–336
Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E (2011) Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res 17:654–666
Apel A, Zentgraf H, Buchler MW, Herr I (2009) Autophagy—a double-edged sword in oncology. Int J Cancer 125:991–995
Bean GR, Ganesan YT, Dong Y, Takeda S, Liu H, Chan PM, Huang Y, Chodosh LA, Zambetti GP, Hsieh JJD, Cheng EHY (2013) PUMA and BIM are required for oncogene inactivation-induced apoptosis. Sci Signal 6:ra20
Bhardwaj A, Aggarwal BB (2003) Receptor-mediated choreography of life and death. J Clin Immunol 23:317–332
Bhol CS, Panigrahi DP, Praharaj PP, Mahapatra KK, Patra S, Mishra SR, Behera BP, Bhutia SK (2019) Epigenetic modifications of autophagy in cancer and cancer therapeutics. Semin Cancer Biol. https://doi.org/10.1016/j.semcancer.2019.05.020
Bhutia SK, Dash R, Das SK, Azab B, Su ZZ, Lee SG, Grant S, Yacoub A, Dent P, Curiel DT, Sarkar D, Fisher PB (2010) Mechanism of autophagy to apoptosis switch triggered in prostate cancer cells by antitumor cytokine melanoma differentiation-associated gene 7/interleukin-24. Cancer Res 70:3667–3676
Cao C, Huang W, Zhang N, Wu F, Xu T, Pan X, Peng C, Han B (2018) Narciclasine induces autophagy-dependent apoptosis in triple-negative breast cancer cells by regulating the AMPK-ULK1 axis. Cell Prolif 51:e12518
Chang NC, Nguyen M, Germain M, Shore GC (2010) Antagonism of Beclin 1-dependent autophagy by BCL-2 at the endoplasmic reticulum requires NAF-1. EMBO J 29:606–618
Choi KS (2012) Autophagy and cancer. Exp Mol Med 44:109–120
Crighton D, Wilkinson S, O’Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM (2006) DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 126:121–134
Cudjoe EK, Lauren Kyte S, Saleh T, Landry JW, Gewirtz DA (2019) Chapter 12: Autophagy inhibition and chemosensitization in cancer therapy. In: Johnson DE (ed) Targeting cell survival pathways to enhance response to chemotherapy. Academic Press, London, pp 259–273
Davies CW, Stjepanovic G, Hurley JH (2015) How the ATG1 complex assembles to initiate autophagy. Autophagy 11:185–186
Donohue E, Tovey A, Vogl AW, Arns S, Sternberg E, Young RN, Roberge M (2011) Inhibition of autophagosome formation by the benzoporphyrin derivative verteporfin. J Biol Chem 286:7290–7300
Donohue E, Thomas A, Maurer N, Manisali I, Zeisser-Labouebe M, Zisman N, Anderson HJ, Ng SS, Webb M, Bally M, Roberge M (2013) The autophagy inhibitor verteporfin moderately enhances the antitumor activity of gemcitabine in a pancreatic ductal adenocarcinoma model. J Cancer 4:585–596
Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A (2009) Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ 16:966–975
Fazi B, Bursch W, Fimia GM, Nardacci R, Piacentini M, Di Sano F, Piredda L (2008) Fenretinide induces autophagic cell death in caspase-defective breast cancer cells. Autophagy 4:435–441
Feng W, Huang S, Wu H, Zhang M (2007) Molecular basis of Bcl-xL’s target recognition versatility revealed by the structure of Bcl-xL in complex with the BH3 domain of Beclin1. J Mol Biol 372:223–235
Fitzwalter BE, Thorburn A (2018) FOXO3 links autophagy to apoptosis. Autophagy 14:1467–1468
Fitzwalter BE, Towers CG, Sullivan KD, Andrysik Z, Hoh M, Ludwig M, O’Prey J, Ryan KM, Espinosa JM, Morgan MJ, Thorburn A (2018) Autophagy inhibition mediates apoptosis sensitization in cancer therapy by relieving FOXO3a turnover. Dev Cell 44:555–565.e553
Galluzzi L, Joza N, Tasdemir E, Maiuri MC, Hengartner M, Abrams JM, Tavernarakis N, Penninger J, Madeo F, Kroemer G (2008) No death without life: vital functions of apoptotic effectors. Cell Death Differ 15:1113–1123
Galluzzi L, Kepp O, Kroemer G (2012) Mitochondria: master regulators of danger signalling. Nat Rev Mol Cell Biol 13:780–788
Garrido C, Kroemer G (2004) Life’s smile, death’s grin: vital functions of apoptosis-executing proteins. Curr Opin Cell Biol 16:639–646
Gogada R, Yadav N, Liu J, Tang S, Zhang D, Schneider A, Seshadri A, Sun L, Aldaz CM, Tang DG, Chandra D (2013) Bim, a proapoptotic protein, up-regulated via transcription factor E2F1-dependent mechanism, functions as a prosurvival molecule in cancer. J Biol Chem 288:368–381
Grumati P, Dikic I (2018) Ubiquitin signaling and autophagy. J Biol Chem 293:5404–5413
Guo J, Ma Y, Peng X, Jin H, Liu J (2019) LncRNA CCAT1 promotes autophagy via regulating ATG7 by sponging miR-181 in hepatocellular carcinoma. J Cell Biochem 120:17975–17983
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674
Harhaji-Trajkovic L, Vilimanovich U, Kravic-Stevovic T, Bumbasirevic V, Trajkovic V (2009) AMPK-mediated autophagy inhibits apoptosis in cisplatin-treated tumour cells. J Cell Mol Med 13:3644–3654
Hübner A, Barrett T, Flavell RA, Davis RJ (2008) Multisite phosphorylation regulates Bim stability and apoptotic activity. Mol Cell 30:415–425
Islam MA, Sooro MA, Zhang P (2018) Autophagic regulation of p62 is critical for cancer therapy. Int J Mol Sci 19:1405
Jain MV, Paczulla AM, Klonisch T, Dimgba FN, Rao SB, Roberg K, Schweizer F, Lengerke C, Davoodpour P, Palicharla VR, Maddika S, Los M (2013) Interconnections between apoptotic, autophagic and necrotic pathways: implications for cancer therapy development. J Cell Mol Med 17:12–29
Jiang K, Zhang C, Yu B, Chen B, Liu Z, Hou C, Wang F, Shen H, Chen Z (2017) Autophagic degradation of FOXO3a represses the expression of PUMA to block cell apoptosis in cisplatin-resistant osteosarcoma cells. Am J Cancer Res 7:1407–1422
Jing Q, Li G, Chen X, Liu C, Lu S, Zheng H, Ma H, Qin Y, Zhang D, Zhang S, Ren S, Huang D, Tan P, Chen J, Qiu Y, Liu Y (2019) Wnt3a promotes radioresistance via autophagy in squamous cell carcinoma of the head and neck. J Cell Mol Med 23:4711–4722
Jung K-T, Oh S-H (2019) Polyubiquitination of p62/SQSTM1 is a prerequisite for Fas/CD95 aggregation to promote caspase-dependent apoptosis in cadmium-exposed mouse monocyte RAW264.7 cells. Sci Rep 9:12240
Kaya-Aksoy E, Cingoz A, Senbabaoglu F, Seker F, Sur-Erdem I, Kayabolen A, Lokumcu T, Sahin GN, Karahuseyinoglu S, Bagci-Onder T (2019) The pro-apoptotic Bcl-2 family member Harakiri (HRK) induces cell death in glioblastoma multiforme. Cell Death Discov 5:64
Kerr JF, Wyllie AH, Currie AR (1972) Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer 26:239–257
Kim KW, Mutter RW, Cao C, Albert JM, Freeman M, Hallahan DE, Lu B (2006) Autophagy for cancer therapy through inhibition of pro-apoptotic proteins and mammalian target of rapamycin signaling. J Biol Chem 281:36883–36890
Kim EH, Sohn S, Kwon HJ, Kim SU, Kim MJ, Lee SJ, Choi KS (2007) Sodium selenite induces superoxide-mediated mitochondrial damage and subsequent autophagic cell death in malignant glioma cells. Cancer Res 67:6314–6324
Kim HJ, Lee SG, Kim YJ, Park JE, Lee KY, Yoo YH, Kim JM (2013) Cytoprotective role of autophagy during paclitaxel-induced apoptosis in Saos-2 osteosarcoma cells. Int J Oncol 42:1985–1992
Klionsky DJ (2005) The molecular machinery of autophagy: unanswered questions. J Cell Sci 118:7–18
Laane E, Tamm KP, Buentke E, Ito K, Kharaziha P, Oscarsson J, Corcoran M, Bjorklund AC, Hultenby K, Lundin J, Heyman M, Soderhall S, Mazur J, Porwit A, Pandolfi PP, Zhivotovsky B, Panaretakis T, Grander D (2009) Cell death induced by dexamethasone in lymphoid leukemia is mediated through initiation of autophagy. Cell Death Differ 16:1018–1029
Levine B, Klionsky DJ (2004) Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 6:463–477
Levine B, Klionsky DJ (2017) Autophagy wins the 2016 Nobel Prize in Physiology or Medicine: breakthroughs in baker’s yeast fuel advances in biomedical research. Proc Natl Acad Sci U S A 114:201–205
Levine B, Kroemer G (2008) Autophagy in the pathogenesis of disease. Cell 132:27–42
Li YC, He SM, He ZX, Li M, Yang Y, Pang JX, Zhang X, Chow K, Zhou Q, Duan W, Zhou ZW, Yang T, Huang GH, Liu A, Qiu JX, Liu JP, Zhou SF (2014) Plumbagin induces apoptotic and autophagic cell death through inhibition of the PI3K/Akt/mTOR pathway in human non-small cell lung cancer cells. Cancer Lett 344:239–259
Li F, Zheng X, Liu Y, Li P, Liu X, Ye F, Zhao T, Wu Q, Jin X, Li Q (2016a) Different roles of CHOP and JNK in mediating radiation-induced autophagy and apoptosis in breast cancer cells. Radiat Res 185:539–548, 510
Li Y, Deutzmann A, Choi PS, Fan AC, Felsher DW (2016b) BIM mediates oncogene inactivation-induced apoptosis in multiple transgenic mouse models of acute lymphoblastic leukemia. Oncotarget 7:26926–26934
Li H, Liu L, Chang H, Zou Z, Xing D (2018) Downregulation of MCL-1 and upregulation of PUMA using mTOR inhibitors enhance antitumor efficacy of BH3 mimetics in triple-negative breast cancer. Cell Death Dis 9:137–137
Li M, Liu J, Li S, Feng Y, Yi F, Wang L, Wei S, Cao L (2019) Autophagy-related 7 modulates tumor progression in triple-negative breast cancer. Lab Investig 99:1266–1274
Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402:672–676
Liu B, Min MW, Bao JK (2009) Induction of apoptosis by Concanavalin A and its molecular mechanisms in cancer cells. Autophagy 5:432–433
Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, Cai Y, Norberg HV, Zhang T, Furuya T, Jin M, Zhu Z, Wang H, Yu J, Li Y, Hao Y, Choi A, Ke H, Ma D, Yuan J (2011) Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 147:223–234
Liu H, He Z, von Rutte T, Yousefi S, Hunger RE, Simon HU (2013) Down-regulation of autophagy-related protein 5 (ATG5) contributes to the pathogenesis of early-stage cutaneous melanoma. Sci Transl Med 5:202ra123
Liu H, He Z, Simon HU (2014) Autophagy suppresses melanoma tumorigenesis by inducing senescence. Autophagy 10:372–373
Liu K, Lee J, Kim JY, Wang L, Tian Y, Chan ST, Cho C, Machida K, Chen D, Ou JJ (2017) Mitophagy controls the activities of tumor suppressor p53 to regulate hepatic cancer stem cells. Mol Cell 68:281–292.e285
Liu Y, Gu S, Li H, Wang J, Wei C, Liu Q (2019) SNHG16 promotes osteosarcoma progression and enhances cisplatin resistance by sponging miR-16 to upregulate ATG4B expression. Biochem Biophys Res Commun 518:127–133
Liu K, Yu Q, Li H, Xie C, Wu Y, Ma D, Sheng P, Dai W, Jiang H (2020) BIRC7 promotes epithelial-mesenchymal transition and metastasis in papillary thyroid carcinoma through restraining autophagy. Am J Cancer Res 10:78–94
Lo-Coco F, Avvisati G, Vignetti M, Thiede C, Orlando SM, Iacobelli S, Ferrara F, Fazi P, Cicconi L, Di Bona E, Specchia G, Sica S, Divona M, Levis A, Fiedler W, Cerqui E, Breccia M, Fioritoni G, Salih HR, Cazzola M, Melillo L, Carella AM, Brandts CH, Morra E, von Lilienfeld-Toal M, Hertenstein B, Wattad M, Lubbert M, Hanel M, Schmitz N, Link H, Kropp MG, Rambaldi A, La Nasa G, Luppi M, Ciceri F, Finizio O, Venditti A, Fabbiano F, Dohner K, Sauer M, Ganser A, Amadori S, Mandelli F, Dohner H, Ehninger G, Schlenk RF, Platzbecker U (2013) Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. N Engl J Med 369:111–121
Mahapatra KK, Panigrahi DP, Praharaj PP, Bhol CS, Patra S, Mishra SR, Behera BP, Bhutia SK (2019) Molecular interplay of autophagy and endocytosis in human health and diseases. Biol Rev Camb Philos Soc 94:1576–1590
Maiuri MC, Tasdemir E, Criollo A, Morselli E, Vicencio JM, Carnuccio R, Kroemer G (2009) Control of autophagy by oncogenes and tumor suppressor genes. Cell Death Differ 16:87–93
Mancias JD, Kimmelman AC (2016) Mechanisms of selective autophagy in normal physiology and cancer. J Mol Biol 428:1659–1680
Mariño G, Niso-Santano M, Baehrecke EH, Kroemer G (2014) Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol 15:81–94
Marquez RT, Xu L (2012) Bcl-2:Beclin 1 complex: multiple, mechanisms regulating autophagy/apoptosis toggle switch. Am J Cancer Res 2:214–221
Mathew R, Karantza-Wadsworth V, White E (2007) Role of autophagy in cancer. Nat Rev Cancer 7:961–967
Mizushima N (2007) Autophagy: process and function. Genes Dev 21:2861–2873
Moscat J, Karin M, Diaz-Meco MT (2016) p62 in cancer: signaling adaptor beyond autophagy. Cell 167:606–609
Mukhopadhyay S, Panda PK, Sinha N, Das DN, Bhutia SK (2014) Autophagy and apoptosis: where do they meet? Apoptosis 19:555–566
Mukhopadhyay S, Das DN, Panda PK, Sinha N, Naik PP, Bissoyi A, Pramanik K, Bhutia SK (2015) Autophagy protein Ulk1 promotes mitochondrial apoptosis through reactive oxygen species. Free Radic Biol Med 89:311–321
Mukhopadhyay S, Naik PP, Panda PK, Sinha N, Das DN, Bhutia SK (2016) Serum starvation induces anti-apoptotic cIAP1 to promote mitophagy through ubiquitination. Biochem Biophys Res Commun 479:940–946
Mukhopadhyay S, Schlaepfer IR, Bergman BC, Panda PK, Praharaj PP, Naik PP, Agarwal R, Bhutia SK (2017) ATG14 facilitated lipophagy in cancer cells induce ER stress mediated mitoptosis through a ROS dependent pathway. Free Radic Biol Med 104:199–213
Muniraj N, Siddharth S, Nagalingam A, Walker A, Woo J, Gyorffy B, Gabrielson E, Saxena NK, Sharma D (2019) Withaferin A inhibits lysosomal activity to block autophagic flux and induces apoptosis via energetic impairment in breast cancer cells. Carcinogenesis. https://doi.org/10.1093/carcin/bgz015
Nieminen AI, Eskelinen VM, Haikala HM, Tervonen TA, Yan Y, Partanen JI, Klefström J (2013) Myc-induced AMPK-phospho p53 pathway activates Bak to sensitize mitochondrial apoptosis. Proc Natl Acad Sci 110:E1839
Oberstein A, Jeffrey PD, Shi Y (2007) Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem 282:13123–13132
Oral O, Oz-Arslan D, Itah Z, Naghavi A, Deveci R, Karacali S, Gozuacik D (2012) Cleavage of ATG3 protein by caspase-8 regulates autophagy during receptor-activated cell death. Apoptosis 17:810–820
Panda PK, Mukhopadhyay S, Das DN, Sinha N, Naik PP, Bhutia SK (2015) Mechanism of autophagic regulation in carcinogenesis and cancer therapeutics. Semin Cell Dev Biol 39:43–55
Panda PK, Naik PP, Meher BR, Das DN, Mukhopadhyay S, Praharaj PP, Maiti TK, Bhutia SK (2018) PUMA dependent mitophagy by Abrus agglutinin contributes to apoptosis through ceramide generation. Biochim Biophys Acta Mol Cell Res 1865:480–495
Panda PK, Patra S, Naik PP, Praharaj PP, Mukhopadhyay S, Meher BR, Gupta PK, Verma RS, Maiti TK, Bhutia SK (2020) Deacetylation of LAMP1 drives lipophagy-dependent generation of free fatty acids by Abrus agglutinin to promote senescence in prostate cancer. J Cell Physiol 235:2776–2791
Panigrahi DP, Bhol CS, Nivetha R, Nagini S, Patil S, Maiti TK, Bhutia SK (2020) Abrus agglutinin inhibits oral carcinogenesis through inactivation of NRF2 signaling pathway. Int J Biol Macromol 155:1123–1132
Parsons MJ, McCormick L, Janke L, Howard A, Bouchier-Hayes L, Green DR (2013) Genetic deletion of caspase-2 accelerates MMTV/c-neu-driven mammary carcinogenesis in mice. Cell Death Differ 20:1174–1182
Patra S, Panda PK, Naik PP, Panigrahi DP, Praharaj PP, Bhol CS, Mahapatra KK, Padhi P, Jena M, Patil S, Patra SK, Bhutia SK (2020) Terminalia bellirica extract induces anticancer activity through modulation of apoptosis and autophagy in oral squamous cell carcinoma. Food Chem Toxicol 136:111073
Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B (2005) Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122:927–939
Qian W, Liu J, Jin J, Ni W, Xu W (2007) Arsenic trioxide induces not only apoptosis but also autophagic cell death in leukemia cell lines via up-regulation of Beclin1. Leuk Res 31:329–339
Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B (2003) Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 112:1809–1820
Quach C, Song Y, Guo H, Li S, Maazi H, Fung M, Sands N, O’Connell D, Restrepo-Vassalli S, Chai B, Nemecio D, Punj V, Akbari O, Idos GE, Mumenthaler SM, Wu N, Martin SE, Hagiya A, Hicks J, Cui H, Liang C (2019) A truncating mutation in the autophagy gene UVRAG drives inflammation and tumorigenesis in mice. Nat Commun 10:5681
Rahmani M, Aust MM, Attkisson E, Williams DC Jr, Ferreira-Gonzalez A, Grant S (2013) Dual inhibition of Bcl-2 and Bcl-xL strikingly enhances PI3K inhibition-induced apoptosis in human myeloid leukemia cells through a GSK3- and Bim-dependent mechanism. Cancer Res 73:1340–1351
Roy B, Pattanaik AK, Das J, Bhutia SK, Behera B, Singh P, Maiti TK (2014) Role of PI3K/Akt/mTOR and MEK/ERK pathway in Concanavalin A induced autophagy in HeLa cells. Chem Biol Interact 210:96–102
Saha S, Panigrahi DP, Patil S, Bhutia SK (2018) Autophagy in health and disease: a comprehensive review. Biomed Pharmacother 104:485–495
Salazar M, Carracedo A, Salanueva IJ, Hernandez-Tiedra S, Lorente M, Egia A, Vazquez P, Blazquez C, Torres S, Garcia S, Nowak J, Fimia GM, Piacentini M, Cecconi F, Pandolfi PP, Gonzalez-Feria L, Iovanna JL, Guzman M, Boya P, Velasco G (2009) Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest 119:1359–1372
Shalini S, Nikolic A, Wilson CH, Puccini J, Sladojevic N, Finnie J, Dorstyn L, Kumar S (2016) Caspase-2 deficiency accelerates chemically induced liver cancer in mice. Cell Death Differ 23:1727–1736
Shao S, Li S, Qin Y, Wang X, Yang Y, Bai H, Zhou L, Zhao C, Wang C (2014) Spautin-1, a novel autophagy inhibitor, enhances imatinib-induced apoptosis in chronic myeloid leukemia. Int J Oncol 44:1661–1668
Shi S, Tan P, Yan B, Gao R, Zhao J, Wang J, Guo J, Li N, Ma Z (2016) ER stress and autophagy are involved in the apoptosis induced by cisplatin in human lung cancer cells. Oncol Rep 35:2606–2614
Singh R, Letai A, Sarosiek K (2019) Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat Rev Mol Cell Biol 20:175–193
Singla M, Bhattacharyya S (2017) Autophagy as a potential therapeutic target during epithelial to mesenchymal transition in renal cell carcinoma: an in vitro study. Biomed Pharmacother 94:332–340
Song X, Yuan Z, Yuan H, Wang L, Ji P, Jin G, Dai J, Ma H (2018) ATG12 expression quantitative trait loci associated with head and neck squamous cell carcinoma risk in a Chinese Han population. Mol Carcinog 57:1030–1037
Tabe Y, Jin L, Iwanami H, Matsushita H, Kazuno S, Fujimura T, Ueno T, Miida T, Weinstock DM, Thomas DA, Andreeff M, Konopleva M (2013) Efficacy and mechanisms of the mTOR inhibitor AZD2014 combined with L-Asparaginase or JAK2 inhibitor TG101348 in ALL. Blood 122:1282–1282
Tan YQ, Zhang J, Zhou G (2017) Autophagy and its implication in human oral diseases. Autophagy 13:225–236
Tian X, Song HS, Cho YM, Park B, Song Y-J, Jang S, Kang SC (2017) Anticancer effect of Saussurea lappa extract via dual control of apoptosis and autophagy in prostate cancer cells. Medicine 96:e7606
Vaseva AV, Marchenko ND, Ji K, Tsirka SE, Holzmann S, Moll UM (2012) p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell 149:1536–1548
Vousden KH, Lane DP (2007) p53 in health and disease. Nat Rev Mol Cell Biol 8:275–283
Wang S, El-Deiry WS (2003) TRAIL and apoptosis induction by TNF-family death receptors. Oncogene 22:8628–8633
Wang J, Cui D, Gu S, Chen X, Bi Y, Xiong X, Zhao Y (2018) Autophagy regulates apoptosis by targeting NOXA for degradation. Biochim Biophys Acta Mol Cell Res 1865:1105–1113
Wei Y, Sinha S, Levine B (2008) Dual role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation. Autophagy 4:949–951
White E (2015) The role for autophagy in cancer. J Clin Invest 125:42–46
Wilde L, Tanson K, Curry J, Martinez-Outschoorn U (2018) Autophagy in cancer: a complex relationship. Biochem J 475:1939–1954
Wong RS (2011) Apoptosis in cancer: from pathogenesis to treatment. J Exp Clin Cancer Res 30:87
Xi G, Hu X, Wu B, Jiang H, Young CYF, Pang Y, Yuan H (2011) Autophagy inhibition promotes paclitaxel-induced apoptosis in cancer cells. Cancer Lett 307:141–148
Xiao T, Zhu W, Huang W, Lu SS, Li XH, Xiao ZQ, Yi H (2018) RACK1 promotes tumorigenicity of colon cancer by inducing cell autophagy. Cell Death Dis 9:1148
Xie Z, Klionsky DJ (2007) Autophagosome formation: core machinery and adaptations. Nat Cell Biol 9:1102–1109
Xu XD, Zhao Y, Zhang M, He RZ, Shi XH, Guo XJ, Shi CJ, Peng F, Wang M, Shen M, Wang X, Li X, Qin RY (2017) Inhibition of autophagy by Deguelin sensitizes pancreatic cancer cells to doxorubicin. Int J Mol Sci 18:370
Yang X, Yu DD, Yan F, Jing YY, Han ZP, Sun K, Liang L, Hou J, Wei LX (2015) The role of autophagy induced by tumor microenvironment in different cells and stages of cancer. Cell Biosci 5:14
Yee KS, Wilkinson S, James J, Ryan KM, Vousden KH (2009) PUMA- and Bax-induced autophagy contributes to apoptosis. Cell Death Differ 16:1135–1145
Young MM, Takahashi Y, Khan O, Park S, Hori T, Yun J, Sharma AK, Amin S, Hu C-D, Zhang J, Kester M, Wang H-G (2012) Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis. J Biol Chem 287:12455–12468
Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon H-U (2006) Calpain-mediated cleavage of ATG5 switches autophagy to apoptosis. Nat Cell Biol 8:1124–1132
Yu K, Xiang L, Li S, Wang S, Chen C, Mu H (2019) HIF1alpha promotes prostate cancer progression by increasing ATG5 expression. Anim Cells Syst (Seoul) 23:326–334
Yue Z, Jin S, Yang C, Levine AJ, Heintz N (2003) Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A 100:15077–15082
Zhang YB, Gong JL, Xing TY, Zheng SP, Ding W (2013) Autophagy protein p62/SQSTM1 is involved in HAMLET-induced cell death by modulating apoptosis in U87MG cells. Cell Death Dis 4:e550–e550
Zhang M, Zhang W, Tang G, Wang H, Wu M, Yu W, Zhou Z, Mou Y, Liu X (2019) Targeted codelivery of docetaxel and ATG7 siRNA for autophagy inhibition and pancreatic cancer treatment. ACS Appl Bio Mater 2:1168–1176
Zhao X, Fang Y, Yang Y, Qin Y, Wu P, Wang T, Lai H, Meng L, Wang D, Zheng Z, Lu X, Zhang H, Gao Q, Zhou J, Ma D (2015) Elaiophylin, a novel autophagy inhibitor, exerts antitumor activity as a single agent in ovarian cancer cells. Autophagy 11:1849–1863
Zheng K, Li Y, Wang S, Wang X, Liao C, Hu X, Fan L, Kang Q, Zeng Y, Wu X, Wu H, Zhang J, Wang Y, He Z (2016) Inhibition of autophagosome-lysosome fusion by ginsenoside Ro via the ESR2-NCF1-ROS pathway sensitizes esophageal cancer cells to 5-fluorouracil-induced cell death via the CHEK1-mediated DNA damage checkpoint. Autophagy 12:1593–1613
Zhou M, Liu Z, Zhao Y, Ding Y, Liu H, Xi Y, Xiong W, Li G, Lu J, Fodstad O, Riker AI, Tan M (2010) MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J Biol Chem 285:21496–21507
Zhou J, Li G, Zheng Y, Shen HM, Hu X, Ming QL, Huang C, Li P, Gao N (2015) A novel autophagy/mitophagy inhibitor liensinine sensitizes breast cancer cells to chemotherapy through DNM1L-mediated mitochondrial fission. Autophagy 11:1259–1279
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Haragannavar, V.C. et al. (2020). Autophagy: An Agonist and Antagonist with an Interlink of Apoptosis in Cancer. In: Bhutia, S.K. (eds) Autophagy in tumor and tumor microenvironment . Springer, Singapore. https://doi.org/10.1007/978-981-15-6930-2_2
Download citation
DOI: https://doi.org/10.1007/978-981-15-6930-2_2
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-6929-6
Online ISBN: 978-981-15-6930-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)