Abstract
Silver has long time been used as a powerful antimicrobial agent against human pathogens. More recently, incorporating silver nanoparticles in medical tools has become an ultimate strategy to control bacterial biofilm colonization and avoid hospital-acquired diseases in immunocompromised patients. This continued effort has been now reoriented to the potential biomedical applications of the green silver nanoparticles. Besides, a green synthesis appears to be a safer, cheaper, and more effective method to produce a larger amount of silver nanoparticles (AgNPs). In this context, the bacteria-mediated synthesis of AgNPs has been largely used to make green silver nanoparticles (g-AgNPs) owing to the bacterial bioactive biomolecules and the natural capacity of bacteria strains to handle trace metals. We herein exhaustively evaluated the publications describing the production of g-AgNPs mostly using bacteria liquid extracts and their application as antibacterial agents in toxicity assays. Actinobacteria, Proteobacteria, and Firmicutes were the mostly used phyla to synthesize bacteriogenic g-AgNPs. Unfortunately, some technical flaws (undefined nominal concentrations, lack of silver quantification and kinetics of g-AgNP, unawareness of exposure media composition and bacteria biology) jeopardized the g-AgNP final assessment. Henceforth, only a few studies seemed to support a real efficacy of bacteriogenic g-AgNPs to cope on itself with pathogenic bacteria or associated with classical antibiotics bringing the so-called synergistic effect. We further indicated some points to be considered for testing green nanosilver in toxicity assays with bacteria and how g-AgNP toxicity mechanisms could be better assessed in single and multiple exposures.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
10.1 Introduction
Bulk and colloidal silver have culturally a long history of being integrated in the medical practices to fight microbial infections. Back to those times, silver salts were largely used as antibacterial agents against conjunctivitis, gastroenteritis, gonorrhea, and syphilis [1]. Currently, silver (Ag0) has been continuously incorporated as nanoscale particles in everyday life products and medical tools. To secure aseptic practices, medical tools highly susceptible to microbial colonization such as orthopedic implants, synthetic fibrous for sutures, catheters, bandages, surgical meshes, vascular prosthesis, etc. have been successfully fabricated with addition of nanosilver or other silver compounds [2]. As an example, incorporating 0.5% of silver nanoparticles (AgNPs) in acrylic bone cement can significantly reduce biofilm formation of Staphylococcus epidermidis and S. aureus and prevent colonization, proliferation, and infection [3].
Even though recent studies have shown that some Gram-negative bacteria may develop resistance to minimal inhibitory concentrations (MIC) of AgNPs (3.38 mg.L−1) after chronic exposures, they are still viewed as powerful nanoweapons [4]. Thereby AgNP has been also amended with antibacterial molecules. It is postulated that mechanisms of chemically synthesized AgNPs against bacteria involve electrostatic attraction between nanoparticles and the cell wall, production of free radicals, changes in membrane permeability, disturbance of respiratory chain, leakage of intracellular organelles, interaction with thiol groups, further inhibition of protein synthesis, and interaction with phosphorus-containing molecules as DNA [5]. All effects aforementioned might be optimized when AgNPs are coated with biologically active molecules, coming from vascular plants, algae, or microorganisms [6]. In fact, new experiments have been trying to integrate effects of both green and chemically made AgNPs and antibiotics in multiple exposures by functionalizing AgNP surfaces to harm bacteria cells. Lately, the biosynthesis of the so-called green silver nanoparticles (g-AgNPs) represents a new approach in treatment efficiency of microbial diseases [6, 7]. As demonstrated by Divya et al. [8], 30–50 nm g-AgNP-30 μg.mL coating treatment of urinary catheter can significantly prevent biofilm formation by Gram-positive and Gram-negative bacteria known to cause urinary tract infections.
As a rule, a green synthesis of AgNPs by microbial routes basically relies on two approaches: extracellular and intracellular. The intracellular route relies on the ability of cell enzymatic machinery to reduce Ag+ ions accumulated in the intracellular milieu to precipitate Ag0 in situ as nanocrystals. Once the cells reach the optimum time for harvesting, a complementary treatment allows the extraction of the g-AgNPs. On another hand, with the extracellular route, biomolecules released by growing bacteria, for example, are separated from their remaining biomass after centrifugation, and used later on to convert aqueous AgNO3 into nano-colloidal green silver within 24 h. Sometimes the whole bacteria biomass is used as biological matrix to nano-Ag production. In this case, a release of intracellular organics and/or biomolecules on bacteria surface (as amino acids or exopolysaccharides, EPS) may act as reducing agents for some species [9, 10]. Once exposed to Ag, bacteria will depend on their defense mechanisms to control the metal uptake and manage its levels inside the cells. Extracellular polymeric substances produced by proteobacterium Escherichia coli, for example, are proved to reduce surrounding Ag+ into AgNP through hemiacetal and aldehyde groups mitigating thus silver toxicity [11]. Nanoparticles can be also produced by redox reactions inside or outside the cells with involvement of NADH-dependent nitrate reductases, as it happens with marine Streptomyces sp. LK3 [12]. This mechanism has been found with cultures of betaproteobacterium Alcaligenes faecalis as well [8]. Another case is given by Lengke et al. [13] on the biomineralization of Ag by filamentous cyanobacteria Plectonema boryanum from silver (I) nitrate complex. The authors found that 560 mg.L-AgNO3 was precipitated as AgNPs at cell surface (1–200 nm) and within cells (<10 nm) at 25 to 100 °C. In addition, the release of organics from dead cyanobacteria caused further precipitation of AgNPs in solution. In any case, a plethora of biomolecules available in the cell-free supernatant or on bacterial biomass mediate silver reduction and final capping of silver nanoparticles in vitro.
Herein we critically reviewed the studies dealing with the antibacterial properties of eco-friendly AgNPs produced by means of bacteria biomolecules. Experiments were carried out in typical assays for predicting acute toxicity (24–48 h) in agar or broth liquid media at 37 °C. The agar disc or well diffusion, and broth dilution were the mostly used methods to assess antibacterial effects in these studies. Unfortunately, some crucial information for experimental repeatability was often neglected in many reports. Likewise, the lack of chemical data related to soluble Ag and nanosilver forms was another drawback observed. If mechanisms controlling green nanosilver toxicity must be comprehended, the kinetics of g-AgNP dissolution and dispersion in exposure media have to be documented. On another hand, a remarkable outcome of all studies was the diversity of bacteria taxa used for green nano-Ag synthesis. By far, strains of phylum Actinobacteria have been involved for bio-mediated synthesis of AgNPs in about 50% of assays. Actinobacteria have high guanine + cytosine content, and are ubiquitous Gram-positive bacteria present in several ecosystems, especially in the marine environment. Harboring an array of biosynthetic gene clusters, they have an unsurpassed ability to synthesize secondary metabolites of industrial and pharmacological interest [14]. The genus Streptomyces (Actinobacteria) is present in 77% of reviewed publications, followed by Nocardiopsis (15%) and Rhodococcus (8%).
In addition to Actinobacteria, two other bacterial phyla have been used as biological models: Proteobacteria and Firmicutes. Gammaproteobacteria lineage is primarily Gram-negative, as all the others found in Proteobacteria (as Alphaproteobacteria and Betaproteobacteria lineage). Gammaproteobacteria taxon was present at 39% of reported assays, and then Alphaproteobacteria and Betaproteobacteria were used at 15% each. While bacteria species from Gammaproteobacteria lineage are of medical, ecological, and scientific importance like those from genus Pseudomonas; members from Alphaproteobacteria adopt an intracellular lifestyle as plant or animal pathogens, or even as plant mutualists like nonnodulating Bradyrhizobium spp. [15]. Similarly, Betaproteobacteria clade represents a broad variety of habitats and metabolic strategies with members inhabiting oligotrophic groundwater ecosystems and constructed wetlands, or even opportunistically living in human body as Alcaligenes faecalis. Finally, we also registered some bacilli used as medium for AgNP synthesis. Genera Bacillus, Aneurinibacillus, and Lactococcus (Firmicutes) corresponded to 31% of the chosen experimental organisms. Species from Firmicutes have low guanine + cytosine content and mostly show Gram-positive stain. This phylum is known to comprise species largely abundant in soil and aquatic environments, inhabiting normal flora of mammalian intestines, or being pathogenic to plants, to animals; but especially to humans. The species Lactococcus lactis, as an example, has some special biochemical features making them very valuable to the food industry [16].
As previously mentioned, there is scarce chemical data to properly address mechanisms of antibacterial biosynthesized AgNPs. The understanding of toxicity mechanisms of g-AgNPs can get even more complicated when g-AgNPs are used together with antibiotics in order to elucidate their combined effects on bacteria growth. Few researchers have started testing the molecular interactions of g-AgNPs with antibiotics, but much work has yet to be done to clearly understand the complexity of such mechanisms when g-AgNPs are involved. Often times, the wanted synergistic effects of g-AgNP + drug ended up with unexpectedly negative effects and no enhanced inhibition. Therefore, the aim of this chapter is first to briefly review the information available on AgNP biosynthesis with bacteria extracts and mainly explore their toxicity effects against pathogenic bacteria. Then we discuss some mechanisms by which g-AgNPs might be operating against pathogenic bacteria in single exposures or when associated with antibiotics.
10.2 Antibacterial Effects of Green Nanosilver Produced by Extracts of Actinobacteria
From cell-free extracts of marine actinobacterium Nocardiopsis sp. (MBRC-1) isolated from marine sediment, Manivasagan et al. [17] synthesized mostly spherical and aggregated 30–90 nm g-AgNPs. These nanoparticles were enrobed by different biomolecules whose chemical profiles were obtained by Fourier transform infrared (FT-IR) spectroscopy analysis. Spectra indicated the presence of -OH stretches, C-H groups related to alkanes, -C=C- from alkenes, and H:C-H bend of the alkyne group. After exposing Escherichia coli, Bacillus subtilis, Enterococcus hirae, Pseudomonas aeruginosa, Shigella flexneri, and Staphylococcus aureus to g-AgNPs (10–50 μg.mL) at 35 °C up to 24 h, growth inhibition ocurred in a concentration-dependent trend. The highest antibacterial activity was detected against B. subtilis and P. aeruginosa. Interestingly, growth inhibition zones by all microorganisms tested were only wider than those induced by amoxicillin (positive control, 30 μg.mL) once g-AgNPs exceeded 30 μg.mL. As a consequence, the minimum inhibitory concentration of g-AgNPs was always ~1.1-fold stronger for each bacteria tested than those used with a classic antibiotic molecule. Similarly, Rajivgandhi et al. [18] exposed a strain of methicillin-resistant coagulase-negative Staphylococcus sp. to well dispersed and roughly spherical 20–50 nm g-AgNPs (5–100 μg.mL) obtained from 20 g biomass of marine Nocardiopsis sp. GRG1 (KT235640) challenged with 1 mM (170 µg.mL) AgNO3 at 28 °C within 6 days. These Nocardiopsis AgNPs showed N-H vibration motions of amides and amines and C-Cl moieties in alkyl halides by FT-IR. Only medium to large concentrations (50–100 μg.mL) of g-AgNPs prevented Staphylococcus sp. from growing at ~18 mm (50 μg.mL) and ~15 mm (100 μg.mL) within 24 h at 37 °C, contrarily to ceftazidime (positive control, 30 μg.mL).
Using 2 isolates of Streptomyces rochei (MHM13) from marine sediment, Abd-Elnaby et al. [19] obtained spherical 22–85 nm g-AgNPs. To reduce Ag+ to nanosilver, 1 mM (170 µg.mL) of AgNO3 (50 mL) was mixed with actinomycete supernatants (50 mL) at pH 8.5. After synthesis, capping agents of g-AgNPs held IR stretches for primary amines (N-H), alkanes (C-H), carbon dioxide (O=C=O), aliphatic ethers (C-O), halo compounds (C-Cl or C-Br), and bending for alkanes (C-H). From the stock solution of g-AgNPs obtained, 50 μL aliquots were injected to exposure media with growing bacteria. Unexpectedly, g-AgNPs of isolate 13 induced the maximum zones of inhibition (16–19 mm) against all bacteria tested, but especially against Vibrio fluvialis; while g-AgNPs of isolate 38 caused moderate zones of inhibition (13–18 mm) (see Adb-Elnaby et al. [19] in Table 10.1). E. coli and P. aeruginosa were not affected by isolate 38 g-AgNPs. Later on, the authors combined isolate 13 g-AgNPs with antibiotics (Table 10.1) to evaluate mixed chemical effects on V. fluvialis, Vibrio damsel, Salmonella typhimurium, and Escherichia coli at 37 °C overnight. As a result, the antimicrobial effects were in some way enhanced by all chemicals associated with nanosilver (for a more detailed discussion between g-AgNPs and antibacterial drug interactions, see Sect. 10.5). Notwithstanding, the newly produced stock solution of g-AgNP was used without any dialysis process to isolate nanoparticles from unreacted biomolecules and residual soluble Ag, which is an active antimicrobial metal. Furthermore, due to dilution effect caused by mixture of both silver stock solution and actinomycete liquid extract, it is difficult (even impossible) to estimate what were the final nominal concentrations of g-AgNPs actually tested.
Along similar lines, 100 μg.mL of nearly spherical 30 nm g-AgNPs produced from marine Streptomyces violaceus (MM72) extracts caused strong antibacterial activity against Escherichia coli and Pseudomonas aeruginosa (with ~16 mm of inhibition zone for each), whereas Bacillus subtilis and Staphylococcus aureus were less affected. Both showed ~10 and ~ 4 mm of growth inhibition at 28 °C within 24 h, respectively [27]. In this study, Gram-negative bacteria had higher sensitivity than the Gram-positive ones. With Streptomyces violaceus (MM72)-g-AgNPs, α-D-galactopyranose units as well as sugar backbone components (such as glucose and galactose residues) and rhamnose residues were depicted by proton nuclear magnetic resonance (1HNMR) analysis. Reducing sugars are known to be actively involved in AgNP mineralization in bacteria. After reaction with dissolved silver, the bands corresponding to rhamnose and pyranose structures normally become much weaker in the FT-IR spectra, while the bands of carboxyl groups get much sharper and stronger in exopolysaccharides of Escherichia coli. It means that aldehyde groups in these sugars were oxidized to carboxyl groups by Ag+ [11].
Smaller and dispersed 15 nm g-AgNPs have been also produced from marine Streptomyces strains [29]. After mixing 50 mL of 1 mM (170 µg.mL) AgNO3 with 50 mL of cell filtrated of Streptomyces olivaceus (MSU3) at 28 °C in dark; spherical and monodispersed ~12.3 nm g-AgNPs were produced. Five major functional groups were found in FT-IR: alkynes (C=C-H), alcohol (-OH), oxime (C=N-OH), alkene (C=C), carboxyl (-COOH), and amine (-NH2) with alkynes and alcohol being predominant. By testing several nominal concentrations of g-AgNPs (0.313–20 μg.mL) within 24 h at 37 °C, a strong antibacterial activity was confirmed against Streptococcus pneumoniae with 0.625 μg.mL (MIC, minimum inhibitory concentration) and ≤ 2.50 μg.mL (MBC, minimum bactericidal concentration) in a concentration-dependent manner. Other bacteria such as Streptococcus mutans (MIC 0.625 and MBC ≤ 5.00 μg.mL), Klebsiella pneumoniae (MIC 2.50 and MBC ≥ 5.00 μg.mL), Escherichia coli (MIC 1.25 and MBC ≤ 5.00 μg.mL), and Enterobacter faecalis (MIC 1.25 and MBC ≤ 2.50 μg.mL) were also affected. More evidences supporting that small g-AgNPs might have a potential to act as broad-spectrum agents against different bacteria species came out from Al-Dhabi et al. [28] experiments. Using Streptomyces sp. (Al-Dhabi-87) supernatant and 1–5 mM AgNO3 suspension, slightly polydispersed 9.7–17.25 nm g-AgNPs were prepared. By high-resolution scanning electron microscope (HRSEM) analysis, these g-AgNPs looked rather clustered-like and united by the biomolecular coating. Using concentrations at mg.mL level, biocide efficiency of 9.7–17.25 nm g-AgNPs was registered with Gram-negative bacteria models (such as Enterobacter faecalis and Staphylococcus aureus) showing a MIC of 0.039 mg.mL each. Among drug-resistant pathogens as Escherichia coli, Acinetobacter baumannii, and Proteus mirabilis, g-AgNPs were also efficient with MIC values reaching 18.0 µg.mL. A second drug-tolerant strain of A. baumannii showed higher MIC values (39, 156, and 312 µg.mL) though. Against Pseudomonas aeruginosa drug-resistant strains, MIC value reached 39 µg.mL, whereas a 312 µg.mL g-AgNPs treatment prevented Enterococcus faecium strains from growing. According to the authors, MIC values were comparatively lower than streptomycin concentration for Gram-positive bacteria.
Microbe-assisted synthesis of g-AgNPs has been equally performed with supernatants of Streptomyces spp. isolated from mangrove sediments [20, 21]. While spherical and agglomerated S. albogriseolus 16.2 ± 1.6 nm g-AgNPs exhibited N-H groups, C-O ester groups, C=O, and C=C=C (polyenes) on their organic coating, Streptomyces sp. (BDUKAS10) 21–48 nm g-AgNPs held free N-H and -C=C-, groups aparently related to heterocyclic compounds present in proteins [20, 21]. These nanoparticles were visually clustered-like. Pathogenic bacteria treated with Streptomyces sp. (BDUKAS10) g-AgNPs went through some growth limitation compared to treatments of pure actinobacteria extracts using disc diffusion method on agar plates up to 14 h at 37 °C [21]. For instance, Bacillus cereus was negatively affected by pure cell-free Streptomyces sp. (BDUKAS10) extracts at 1.25-fold increase compared to 5% diluted cell-free extracts. Once treated with g-AgNPs, only 1.3-fold increase of growth inhibition was detected if compared to pure cell-free supernatant. Correspondingly, 1.7-fold increase of antibacterial effect was observed against Staphylococcus aureus when treated with non-diluted extracts in comparison to 5% diluted fractions. After g-AgNPs exposure, Staphylococcus aureus had 1.1-fold increase of growth disruption. The same effect was observed with Pseudomonas aeruginosa. Because no additional positive controls (as Ag+ ions, chemically produced AgNPs, or antibiotics) were evaluated, it is challenging to estimate at which degree these g-AgNPs might effectively prevent bacterial growth. Without quantification of remaining free Ag and/or free antibacterial biomolecules in raw g-AgNP stock solutions after reduction process (1) or final silver concentrations measured in the exposure media (2), it is tricky to unequivocally attribute the toxicological effects of g-AgNPs against bacteria to the green nanosilver itself. By investigating Streptomyces albogriseolus-made 16.2 ± 1.6 nm g-AgNPs’ antibacterial properties against Bacillus cereus, Escherichia coli, and Staphylococcus aureus using agar well diffusion methods, Samundeeswari et al. [20] showed that g-AgNPs were able to display antimicrobial activity against S. aureus at ~18.5 mm, followed by E. coli at ~16.5 mm and B. cereus at ~14 mm. Compared to positive controls (free Ag and pure supernatant extract of S. albogriseolus), zones of inhibition created by g-AgNPs appeared to be slightly wider than those induced by dissolved Ag and cell-free supernatant, so no significant differences were observed. Both controls showed considerable antimicrobial effects against bacterial strains, especially for S. aureus. It is interesting to note that after biosynthesis of g-AgNPs, the yield of nanosilver obtained by atomic absorption spectrophotometer (AAS) was about 73%. Hence, ~27% of dissolved silver likely remained in the stock solution or converted to silver oxides or silver sulfides with unknown antimicrobial activities.
Silver reduction by terrestrial Streptomyces kasugaensis (NH28/M338-M1) liquid extracts produced spherical and polydispersed ~13 nm g-AgNPs likely holding proteins on their organic shell [25]. Concentrations of raw solution of green nano-Ag (1.25–200 μg.mL) were tested against bacteria at 1 x 106 CFU.mL (CFU, colony formation unity) final concentrations per well. The strongest antibacterial activity of g-AgNPs given by MIC was against Staphylococcus aureus (1.25 μg.mL), Klebsiella pneumoniae (1.25 μg.mL), Proteus mirabilis (1.25 μg.mL), and Escherichia coli (1.25 μg.mL) followed by Bacillus subtilis (2.5 μg.mL), Salmonella infantis (10 μg.mL), and Pseudomonas aeruginosa (10 μg.mL) up to 24 h at 37 °C. The lowest MIC value (1.25 μg.mL) for growth inhibition was found against S. aureus (48% of inhibition), K. pneumoniae (29%), P. mirabilis (23%), and E. coli (14%). The MIC for B. subtilis 13 nm g-AgNP-treated was 2.5 μg.ml, whereas for P. aeruginosa and S. infantis 10 μg.mL inhibited bacteria at 8%. Expectedly, Iniyan et al. [26] recently found that rod-shaped, smaller, and highly aggregated g-AgNPs (3–12 nm)) synthesized from Streptomyces exfoliatus ICN25 were greatly functional against methicillin-resistant and methicillin-sensitive Staphylococcus aureus (ATCC33591 and ATCC29213, respectively), E. coli, P. aeruginosa, and K. pneumoniae at 105–106 CFU.mL cells. These 3–12 nm g-AgNPs also taken directly from newly prepared raw solution led to a much lower MIC (1 μg.mL; ~15.5 mm of inhibition zone) against methicillin-resistant S. aureus compared to methicillin-sensitive S. aureus (2 μg.mL; ~6.33 mm), E. coli (2 μg.mL; ~5.83 mm), P. aeruginosa (2 μg.mL; ~12.5 mm), and K. pneumoniae (2 μg.mL; ~4.33 mm). Antimicrobial effects were dose-dependent. As it turned out, g-AgNPs’ impact over bacterial strains was twofold higher than dissolved ion counterparts [26]. Culture supernatant itself exhibited considerable antibacterial properties with inhibition zones being only about 2 mm smaller than those zones cleared by g-AgNP treatment. This is quite predictable owing to the fact that Streptomyces exfoliatus is able to produce a β-lactamase inhibitory protein of molecular mass estimated to 17.5 kDa [30] and β-Lactam antibiotics are normally applied to inhibit the synthesis of the bacterial peptidoglycan layer. As for K. pneumoniae, no differences were detected. Conversely, rounded ~5–50 nm g-AgNPs (1000–10000 µg.mL) capped by biomolecules extracted from Streptomyces naganishii (MA7) mycelium showed some inhibition effect against biofilm of several strains of pathogenic bacteria growing in Luria-Bertani (LB) media well plates [22]. Such effects were not significantly higher compared to either dissolved silver or classic antibiotics throughout 24 h at 37 °C. Staphylococcus aureus was the most affected species with >12 mm of inhibition due to g-AgNP effect. Overall, silver toxicity arose in a concentration-dependent manner. In this experiment, g-AgNPs were previously separated by ultracentrifugation after synthesis and weighted per 100 mL. Main features of g-AgNP synthesis and exposure conditions of toxicity assays reported in papers reviewed in this section are summarized in chronological order in Table 10.1.
10.3 Antibacterial Effects of Green Nanosilver Produced by Extracts of Proteobacteria and Firmicutes
Bacterial species of Firmicutes and Proteobacteria have been isolated from marine ecosystem, plant tissues, soils, chemical effluents, and even milk; then used to fabricate g-AgNPs. A wide range of pathogenic bacteria was exposed to lower concentrations of green nano-Ag compared to the experiments conducted with actinobacteria-made g-AgNPs. A summary of AgNP synthesis methods and exposure conditions for papers reviewed in this section is given in Table 10.2.
After a 24 h-mixing process of 100 mL cell-free supernatant of marine seaweed epibiont Bacillus vallismortis (SS7) (Firmicutes) with 1 mM AgNO3 final concentration at 37 °C, partially polydispersed ~22.76 nm g-AgNPs emerged in the stock solution [40]. FT-IR spectrum of g-AgNPs showed biomolecules harboring primary and secondary amines, C-H, N-O, and halogen-alkanes (C-F and C-Cl) of methyl (–CH3) and methylene (=CH2) groups of proteins. Throughout a 48 h exposure at 37 °C, Pantoea agglomerans was the most affected biofilm-forming bacteria whose zones of inhibition ranged from ~5 mm (0.25 nM) to ~14 mm (1.25 nM). For testing, initial cell concentrations in each well were ~ 2 x 108 cells.mL. Then Pseudomonas aeruginosa (~5–12 mm), Vibrio alginolyticus (~4–12 mm), Escherichia coli (~3–11 mm), Serratia marcescens (~3–10 mm), and Aeromonas hydrophila (~3–8 mm) were inhibited in a wide range of low nominal concentrations (0.027–0.16 µg.mL of green nano-Ag) as well. In terms of minimum inhibition concentration (MIC) of green nano-Ag, bacteriostatic effects were reached with 0.027 µg.mL in P. agglomerans, P. aeruginosa, and V. alginolyticus cultures, whereas for the other species tested MIC corresponded 0.054 µg.mL. The bactericidal effect was reached at higher doses only (0.054–0.82 µg.mL). In comparison, roughly spherical 35–58 nm g-AgNPs bioreduced by liquid and solid extracts of marine bacillus Ochrobactrum anthropi (CB2/JQ435714) were able to disrupt growth of Staphylococcus aureus, Salmonella typhi, Salmonella paratyphi, and Vibrio cholerae at ~15, ~14, ~15, and ~ 16 mm, respectively [35]. The authors mentioned that 108 µg.mL of soluble Ag (1 mM) was used as positive control and did not stop bacteria growth. It should be also noted that bacteria were exposed to both cell-free supernatant and mycelia-synthesized g-AgNPs in a single exposure with no final concentrations established.
Bacterial endophytes are known to be plant symbionts inhabiting plant tissues for the majority of their life cycle without detrimental interactions between symbiont and host [43]. For instance, they can create an endophytic microbiome in the roots mostly dominated by Proteobacteria, Actinobacteria, and in a lesser extent Firmicutes [44]. Isolated from Coffea arabica L. (Rubiaceae) tissues, the endophytic Pseudomonas fluorescens gave rise to multi-shaped 5–50 nm g-AgNPs by reduction of Ag+ ions in bacteria cell-free extracts at different concentrations (0.5–2 mM) and temperatures (20–90 °C) [36]. Before synthesis, P. fluorescens strain was pre-screened twice by culturing it on media supplemented with AgNO3. After a 72 h incubation, culture broth was centrifuged to obtain a supernatant challenged with 1 mM AgNO3. With initial cell concentrations reaching 5 x 105 CFU.mL in test chambers, pathogenic bacteria had colony formation decreased as a function of nominal concentrations up to 100 μg.mL within 24 h and at 37 °C. Alone both chemically prepared AgNPs and g-AgNPs showed similar results inhibiting bacteria growth. Minimum inhibitory concentrations varied from 31.25 to 250 μg.mL, and it seemed to be more effective against Klebsiella pneumoniae and less effective against Escherichia coli. Antibiotics kanamycin (a) and tetracycline were stronger than AgNPs and g-AgNPs for all bacteria tested. In fact, AgNO3 treatment and supernatant single exposures did not succeed to prevent bacterial growth. Only Staphylococcus aureus growth was affected at ~6 mm by Ag+ ion exposures with concentrations not precisely determined. Having both kanamycin (1 μg.mL) and g-AgNPs (10 μg.disc) in the same treatment led to combined effects defined as synergistic by Syed et al. [36] (for a more detailed discussion about such interactions, see the Sect. 10.5).
These same authors synthesized ~24 nm g-AgNPs from supernatant of the endophytic bacterium Aneurinibacillus migulanus 141 isolated from shameplant Mimosa pudica L. (Fabaceae) tissues [37]. Differently from previous methods used with gammaproteobacterium P. fluorescens, endophytic A. migulanus (Firmicutes) was pre-screened (1x) with a nutrient media previously amended with 170 µg.mL AgNO3 at 37 °C. Then colonies went through a 72 h fermentation process before being centrifuged to obtain the cell-free extracts. Once again, the supernatant was mixed with 170 µg.mL AgNO3 in order to obtain the g-AgNPs. The pathogenic bacteria were concentrated ~5 × 106 CFU.mL at time 0 when challenged with a 25–100 μg.mL g-AgNPs range. Antibacterial activity, given by inhibition growth (in mm and MIC), was primarily found with Pseudomonas aeruginosa (~21 mm, 12.5 μg.mL), then Escherichia coli (~18 mm, 12.5 μg.mL), Staphylococcus aureus (~16 mm, 12.5 μg.mL), Bacillus subtilis (~19 mm, 12.5 μg.mL), and Klebsiella pneumoniae (~17 mm, 25 μg.mL). The positive control gentamicin (1 mg.mL) showed similar or stronger effects compared to g-AgNPs against all bacteria tested, except for P. aeruginosa. With a different approach, Sunkar and Nachiyar [32] managed to reduce 1 mM of dissolved silver solution into slightly aggregated and spherical 20–40 nm g-AgNPs from biomass of the endophytic bacterium Bacillus cereus GX1 isolated from leafs of yellow mangosteen Garcinia xanthochymus (Guttiferae). After bacteriogenic nanosilver treatment, only S. aureus and E. coli clearly showed a better growth inhibition (~22 mm and ~ 18 mm, respectively) compared to all antibiotics used. Other bacteria tested such as P. aeruginosa, K. pneumoniae, and Salmonella typhi showed growth inhibition as well, but often similar to at least one of the antibiotics tested (positive controls).
According to Rasulov et al. [38], legume root-nodulating nitrogen-fixing gammaproteobacterium Bradyrhizobium japonicum SDK276 isolated from the soil makes high and low molecular mass exopolysaccharides (EPS). Dissolved silver (10 M) was then reduced in a solution of EPS (100 mL) and stored for 2 months at ambient temperature before producing rod and oval-shaped ~5–50 nm g-AgNPs. No attempt to separate the nanoparticles from the remaining reducing solution seemed to be applied. While high molecular mass-EPS g-AgNPs (HMM-g-AgNPs) ranged from ~20 to 50 nm, low molecular mass-EPS g-AgNPs (LMM-g-AgNPs) had ~5–20 nm. Toxicological testing of green nanosilver (0.10–2.5 μg.mL) took place at 37 °C throughout 24 h against Gram-negative Escherichia coli and Gram-positive Staphylococcus aureus. Inhibition of bacterial growth occurred as a function of nominal concentrations used. LMM-g-AgNPs were more effective to reduce bacterial cell viability and growth though. With lower doses (1–2.5 μg.mL), inhibition zones generally reached ~16–32 mm with LMM-g-AgNPs, whereas for HMM-g-AgNPs ~6–22 mm was found for both species. S. aureus was the most sensitive bacteria, and it had 70–75% of cells dead after 12 h in 2.0–2.5 μg.mL g-AgNP testing media. When exposed to 10–50 μg.mL g-AgNPs, E. coli cells started sharply dropping in number after 6 h. Using only high doses of LMM-g-AgNPs (10–50 μg.mL), cell viability of bacteria started to drop in a shorter period of time right after exposure (~2 h). In this case, the most effective concentration was 30 μg.mL. The best results found with LMM-g-AgNPs can possibly be explained by differences of solubility of both g-AgNPs in the exposure media [38].
Bacteria isolated from chemically polluted soils or industrial effluents have been also used to biofabricate AgNPs [31, 34]. Liquid and solid extracts of betaproteobacterium Alcaligenes faecalis inhabiting metal-rich dump sites near industrial areas were used to obtain spherical and monodispersed ~11 nm g-AgNPs [31]. In their study, increasing concentrations of g-AgNPs (2.16–216 µg.mL) were more efficient to block bacteria growth in culture media than Ag ions. Lowest concentrations of silver only stopped bacteria growing in g-AgNP exposure media. Escherichia coli growth was reduced in ~3.2 mm. A dose of 2.16 µg.mL AgNP also inhibited Pseudomonas aeruginosa growing at ~2.0 mm, Bacillus cereus at ~2.2 mm, and Staphylococcus aureus at ~1.5 mm. Additionally, 21.6 µg.mL nominal concentrations for both g-AgNPs and Ag+ ions inhibited E. coli at ~12.5 mm/1.2 mm ratio for g-AgNPs/Ag+, ~9.2/0.8 for P. aeruginosa, ~9.8/1.3 for B. cereus, and ~ 9.9/1.8 for S. aureus. Similar effects likely emerged with spherical 4 nm g-AgNPs synthesized from gammaproteobacterium Serratia nematodiphila liquid extracts [34]. Green nanosilver showed high antimicrobial effects against P. aeruginosa by preventing its growth at ~18 mm and Bacillus subtilis at ~17 mm within 24 h at 37 °C. Lower effects were found against Klebsiella planticola, with ~14 mm of inhibition zone detected. In both studies, raw 24–72 h preincubated ~108 µg.mL g-AgNP stock solutions were used.
Finally, cell-free supernatant of lactic acid bacterium Lactococcus lactis (Firmicutes) isolated from milk products and mixed with 170 µg.mL AgNO3 formed spherical silver nanoparticles of 5–50 nm (19 nm in average) with antibacterial activity [39]. This fermentative bacterium can almost totally convert its carbon source into L-lactate from pyruvate through lactate dehydrogenase, which dominates the maximum enzymatic activity once L. lactis is exposed to high doses of sugar [16]. So the preparation of the Ag-reducing media involved a preincubation of inoculate in MH (Mueller–Hinton) broth medium in a bioreactor for 7 days at 26 °C. Following a centrifugation process, a supernatant was combined with dissolved silver and incubated again at 26 °C for another 7 days in the dark. After this process, the residual silver ions and biomolecules were washed out from silver biocolloids in a 3-day dialysis. L. lactis is a natural factory of many aromatic acetylated products such as diacetyl, acetaldehyde, and acetate that could likely have acted as Ag reducers. The newly made g-AgNPs exhibited a large amount of functional groups (see Table 10.2) and were poorly dispersible, as depicted by TEM micrographs. The most effective g-AgNP concentration to avoid pathogenic bacteria growth was 15 μg per well with inhibition zones varying from ~11 to 16 mm at 35 °C within 24 h. Proteus mirabilis was the least sensitive, whereas Staphylococcus epidermidis was the most affected. The MIC for Staphylococcus aureus, S. epidermidis, and P. mirabilis was calculated to be ~3.125 μg.mL, and then 6.25 μg.mL for P. aeruginosa and 12.5 μg.mL for methicillin-sensitive S. aureus (MSSA) at 35 °C within 18 h. Surprisingly, with 200 μg.mL, S. aureus was resistant to nano-Ag and showed ~70.2 in cell density (CD) compared to less than ~9 CD in absorbance measurements. Unfortunately, with no positive controls, there was no estimation of toxicity compared to silver ions or antibiotics.
10.4 Assessing g-AgNPs Effects on Bacteria Models: Accomplishments, Pitfalls, and Challenges
10.4.1 Nominal Concentrations and Stock Solution Preparation
So far, the research conducted to biosynthesize green silver nanoparticles and evaluate their potential as antimicrobial agents was successful in obtaining a great variety of isolates of bacterial strains (Tables 10.1 and 10.2). Promising results regarding antibacterial properties of green nanosilver were obtained with pathogenic bacteria known to cause hospital-acquired infections. Notwithstanding, the lack of information about final nominal concentrations for the antimicrobial assays can be considered as a major drawback in several studies dealing with g-AgNP toxicity. The nominal concentration refers to the calculated amount of a chemical to be introduced in a test chamber, while the measured concentration is referring to the real amount of the test substance analyzed in the exposure media at a given time. More than 30% of the reviewed papers have inaccurately reported the g-AgNP nominal concentration as a volume (μL) taken from the stock solution, which is considered as the initial concentration, without any additional analysis. One must take into consideration that after reduction process of silver nitrate (usually ~1 mM of AgNO3; or 170 µg.mL) by liquid or solid extracts of bacteria, the main stock solution of newly produced g-AgNPs will doubtlessly retain an unknown proportion of unreacted dissolved silver and metabolites/exudates. In addition, this unknown proportion will be changing with reaction time and incubation temperature. Sampling μL aliquots from the raw g-AgNP stock solution to be injected into the exposure media implies exposing bacteria to residual Ag+ ions, g-AgNPs, and unreacted and unknown biomolecules all together. Therefore, without a final dialysis of reaction fluid to wash out dissolved ions and free reactive biomolecules from the colloidal nano-Ag, the final testing concentrations of g-AgNP will be likely biased, and it becomes impossible to compare claimed antibacterial activity of such mixture (or protocol) proposed by different authors.
10.4.2 The Green AgNP Morphology and its Toxicity Mechanisms
After isolation of bacterial strains, the two principal matrices for AgNP mineralization (supernatant and biomass) are separated as schematically illustrated in Fig. 10.1a–c. Generally, the multiple biomolecules in a cell-free liquid extract drive the production of silver nanoparticles in a rapid process (~24 to 48 h) (Fig. 10.1d–f, Fig. 10.2). In this context, AgNP size is likely related to the incubation time, the ability of functional groups of biomolecules to catch Ag+ ions along the process, and increasing AgNO3 concentrations mixed with bacteria exudates [45]. Henceforth, FT-IR or 1HNMR spectral analysis of g-AgNPs has been performed in the majority of publications we analyzed. What has been revealed is that amines, amides, carbonyl, hydroxyl, and alcohol functions, as well as different hydrocarbons, are found in the coating of g-AgNPs. Even though bacteria extracellular synthesis has been the more frequently used technique to reduce silver, cell-fabricated g-AgNPs have an unexplored potential to be used as antibacterial agents in toxicity testing [24] (see Table 10.1 for a summary).
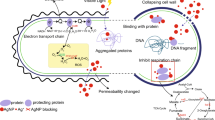
Schematic representation of procedures for green synthesis of silver nanoparticles. A- After isolation from the environment, bacteria are cultured in laboratory at around 30 °C. B- Centrifugation of broth cultures is a crucial step to obtain the biological matrix for green synthesis of AgNP. The matrix may be the bacterial biomass or the cell-free supernatant (presented in D-F ). Nucleation of dispersed ions may occur on the bacteria surface (when the biomass is resuspended into a new solution) or by means of free exudates dispersed in the solution (for cell-free supernatants). C- A cell-free extract is often used to mineralize soluble Ag into AgNP. Biomolecules of cultured bacteria are then studied by FT-IR or 1HNMR spectral analysis to depict functional groups likely responsible for reducing silver and lately capping the newly produced nanoparticles. D, E, and F- In many studies, 170 µg.mL AgNO3 (1 mM) is mixed with bacteria supernatant in a wide range of temperatures (20–37 °C), and at different reaction times (1 h to ~6 days) to biomineralize Ag. Unreacted products (silver ions and biomolecules) likely remain in the final g-AgNP solution if a separation process is not applied
Silver reduction in a cell-free bacterial supernatant after mixing both AgNO3 and bacterial supernatant (following Fig. 10.1). A- The visible color change in the AgNO3 + bacteria supernatant solution is an indicator of the reduction of soluble silver into g-AgNP. It is known that several factors govern this process such as organic function of free biomolecules in solution, pH, composition of medium, metallic salt concentration, and temperature. B and C- In solution, reducing agents such as NADH, NADH-dependent reductases, proteins, exopolysaccharides (EPS), and other molecules can mediate Ag+ silver loading in AgNP. D- EPS with reducing sugars can chelate, reduce, and stabilize metal ions by several functional groups. Polyanionic groups such as hydroxyl, carboxyl, phosphoric groups, and amino end groups have been suggested as main sites for soluble Ag reduction into nano-Ag
It has been postulated that several factors are actually governing the toxicity of chemically synthesized AgNPs towards bacteria including shape, concentration, size, zeta potential, pH of exposure media, presence of capping agents, and electrical charge of organic ligands coating AgNP surface. In fact, much of the AgNP identity is related to the corona composition and the way it interacts with cells and surrounding molecules dispersed in the exposure media. As a consequence, the molecular/protein composition of the corona is strongly influencing nanoparticle-cell interactions, its biological fate, and final functions as well [46]. Therefore, more studies focusing the corona identity (i.e., chemical characterization) of green nanosilver with experimental media and biological fluids are needed to first reveal which functional groups could optimize g-AgNP attachment to Gram-positive and Gram-negative bacteria and how drug delivery would target pathogenic bacteria with these particles. We can suggest that g-AgNPs will behave like chemically made organic-coated AgNPs. Nonetheless, not only soluble silver and nano-Ag form (as a single particle and/or aggregates) will impair bacteria cell, but also the byproducts of bacteria used as capping shell as well. For instance, Ferreyra Maillard et al. [47] showed that green nano-Ag produced by plant extracts had a high zeta potential and seemed to adhere to bacteria cell wall in tryptic soy broth (TSB) media at 37 °C up to 18 h. Green AgNPs disrupted bacterial envelope just 1 h after exposure. Overall, an increase in surface roughness, membrane disruption, and leakage of intracellular contents was mainly observed with Gram-negative Escherichia coli, whereas Gram-positive Staphylococcus aureus appeared to be less affected. The authors stated that aromatic/hydrophobic moieties from the main phenolic compounds in plant extract adsorbed on AgNPs would likely allow interactions with hydrocarbon chain of lipids on bacteria surface.
Final g-AgNP morphology will depend on the way chemical groups of bacterial exudates will handle dissolved ions during the reducing process (Fig. 10.2). Nonetheless, there is no information about the molecular weight of biomolecules capping these nanoparticles. In fact, capping biomolecules might play a crucial role on g-AgNPs toxicity considering (1) the interaction with exposure media (and its constituents like Cl− ions) and bacterial cell envelope, and (2) the kinetics involving leaching Ag+ ions out of g-AgNP surface. It has been hypothesized that once exposed in liquid media, high molecular weight (65,000 Da) poly-allylamine-coated AgNPs released in a few hours a first layer of Ag+ ions already entrapped by capping polymers [48]. As time goes on, the dissolution of Ag core slowly takes place under mediation of AgNP organic layer. Another example is given by the nanofibrillated cellulose used as antimicrobial agent in association with g-AgNPs where Ag+ ions’ release time is prolonged due to efficient in situ immobilization of nanosilver by carboxyl groups [49]. As demonstrated by the authors, silver nanoparticles were reduced by dialdehyde nanofibrillated celluloses by means of silver mirror reaction between the aldehyde groups and [Ag(NH3)2]+, generating three carboxyl groups per unit in the molecular chain. As it follows, the reduced AgNPs were then firmly anchored by the carboxylate groups on the surface of fibers. Experiments carried out by Baygar et al. [50] with surgical sutures coated by Streptomyces griseorubens-made 30–50 nm g-AgNPs released an increasing amount of silver up to 2.93 μg.L after 21 days in phosphate-buffered saline (PBS). The Ag-coated material limited Escherichia coli and Staphylococcus aureus growing in agar, but no clear quantification was presented.
Bacteriogenic silver nanoparticles have been reported to be in a wide range size (4 to 85 nm, or even large particules reaching ~220 nm), and mostly spherical. However, the shape of all g-AgNP-types can vary from rod-shaped, oval, spherical, roughly spherical, or less frequently quadrangular (Tables 10.1 and 10.2). As it has been pointed out by Vetchinkina et al. [51], when increasing incubation time, the ultrasmall g-AgNPs tend to disappear, and the larger ones will rise in number. As a general rule, the longer the incubation time, the larger will likely be the g-AgNPs, which is an expected observation. Accordingly, sticking g-AgNPs and formation of agglomerates also increase with reaction time [51]. Because a large part of these capping exudates could be amino-polysaccharides, they might have a crucial role in keeping AgNPs aggregated and/or partially dispersed after synthesis. The way small g-AgNPs combined with large aggregates will provoke toxicity in bacterial cells remains a mechanism poorly discussed so far. Moreover, AgNPs with different shapes might exhibit different antimicrobial properties probably because they give rise to many degrees of nanoparticle-cell membrane interactions leading to cell damage [6]. However, how exactly AgNP shape would affect toxicity towards microbial cells is still a matter of debate.
10.4.3 The Influence of the Exposure Media Features on g-AgNP Toxicity
Finding out how g-AgNPs behave in agar or liquid broth is another piece of the puzzle to understand their toxicity. Work by Oves et al. [33] has scrutinized the release kinetics of cubic and well-dispersed 93 nm g-AgNPs obtained from gammaproteobacterium Stenotrophomonas maltophilia OS4 liquid extracts. Their assays were performed with a dialysis bag suspended in a HEPES buffer solution over a period of 48 h. A slow release of Ag (~10%) was detected within a few hours; but after 12 h, ~70% of nano-Ag was already released; and at 48 h, 81% of g-AgNPs was dispersed in the medium. Interestingly, the smaller g-AgNPs were more rapidly dispersed than the bigger ones. Some variables such as oxygen diffusion and nano-Ag dispersion should have a significant impact on the results. First of all, dissolved oxygen is known to leach Ag+ ions from nanosilver surface. Thus, antibacterial properties of nano-Ag will strongly depend on optimally displayed oxidized surfaces found in well-dispersed suspensions [44]. A limited diffusion rate of both g-AgNPs and free silver in the solid medium diminishes the amount of silver to readily react with bacteria [52]. As stated by Schumacher et al. [53], the handling of disc diffusion methods is quite simple and straightforward, but it requires appropriate diffusion. In other words, the density of the medium (which is defined by solubility, gelling agent’s concentration, as well as the degree of polymerization), the molecular weight of capping agents; and the silver concentration (in terms of nominal concentrations and measured concentrations) will define how effective the g-AgNPs could act against bacteria.
The effect of Cl− ions must also be considered for green nanosilver toxicity assays with bacteria. The majority of antibacterial tests we reviewed were carried out within 24–48 h in agar diffusion methods (with discs or well plates), and sometimes in broth liquid media. As observed by Lok et al. [44], 9 nm AgNPs synthesized by borohydride reduction normally exhibit antibacterial effects in water or in buffered solutions, but they aggregate in commonly used culture media and biological buffers having chloride and/or phosphate in their contents. As an example, Luria-Bertani liquid broth media (or lysogeny broth medium) contain high concentrations of Cl− ions (~ 5 g.L for Lennox or Luria medium and ~ 10 g.L for Miller formulation). In turn, chloride concentration in agar is around 0.5%. As a rule, Cl− ions can clearly interfere with silver Ag+ concentration to form less soluble chloro complexes (AgClx(x-1))− in the solution, affecting thus AgNP toxicity. Therefore, bactericidal activity interpreted as zone diameter or optical density measurement can be misinterpreted due to the presence of chloride in the medium. Working on silver fate in MH (Mueller-Hinton) agar, Tuncer and Seker [54] revealed that a clear local deposition of AgCl happened in a MIC antibacterial test of nanocomposites amended with dissolved silver (containing 29% Ag-TiO2; titanium dioxide nanoparticles). This deposition was later involved by the bacteria-free zone, as depicted by a reflective optical microscope. In any case, the Cl/Ag ratio is considered as a governing factor of AgNP toxicity in exposure media with bacteria [55]. The rate of dissolution of AgNP will be strongly related to the solid AgCl/ AgClx(x-1)- ratio: as dissolution rate increases, AgCl/AgClx(x-1)- decreases. Using Escherichia coli as biological model, the reaction of Cl− ions on AgNP surfaces begins within 2h of exposure [55]. It results either in precipitation of AgCl or formation and dispersion of AgClx(x-1)--soluble forms. As Cl/Ag ratio increases, AgClx(x-1)--soluble species begin to dominate compared to solid AgCl. After 24 h of exposure, with lower initial concentrations of Ag+ or AgNPs (~1.10–6 or 1.10–5 M) in 0.01–0.1 M NaCl-containing media, density of E. coli living cells remains close to 100%. Hence, it seems unlikely that high concentrations of dissolved silver (~34 to 170 μg.mL), used as positive controls in some experiments with g-AgNPs, failed to stop bacteria growth if we bear in mind that chloride ions have this critical influence on silver ion-mediated acute toxicity.
10.4.4 The Bacterial Model Used in the Toxicity Assays
According to Gordienko et al. [52], there is also another important limitation to be aware of in order to properly evaluate g-AgNP antibacterial activity: the growth phase of testing bacteria. A visible change in bacteria metabolism and cell structure in exponential and stationary phases is known to influence toxicity. Cultures of Bacillus cereus, Pseudomonas aeruginosa, and Staphylococcus aureus are proven to be more resistant to silver during exponential phase at 37 °C within 24 h. To date, it looks like the bacterial growth-stage aspect has been neglected in a majority of the studies reviewed in this report. Indeed, the combination of low to middle g-AgNP nominal concentrations and growth phases can show surprising results in bacteria species. Rhodococcus sp. 10–50 nm intracellularly produced-AgNP (10 µg.mL) seemed to be efficient to reduce Klebsiella pneumoniae activity only until 48 h of exposure [24]. The same concentration affected log phase of Escherichia coli, Enterococcus faecalis, Staphylococcus aureus, Pseudomonas aeruginosa, and Proteus vulgaris only delaying growth to ~20–30 h compared to controls. Later on, a 10 μg.mL g-AgNP treatment caused bacteriostatic effects in Enterococcus faecalis and Staphylococcus aureus reaching to the stationary phase earlier than controls. Even with a 30 μg.mL exposure, some of these pathogens continued to grow after 50 h of incubation.
Noteworthy, some bacteria exhibit a natural resistance against silver nanoparticles; so the best exposure conditions (single or combined exposure, temperature range, acute or chronic treatment, and nominal concentration) should be pre-screened before testing. Thus, the biology of the species must be taken into account for toxicity assays. For instance, Rafińska et al. [56] observed that Bacillus subtilis displays different responses to silver in a short 24 h exposure at 37 °C. Briefly, four conditions were chosen as a function of silver chemical form and tetracycline combination: 1) 4–45 nm actinomycete-produced AgNPs [57] (6.25–200 μg.mL); 2) 4–45 nm g-AgNP + tetracycline (6.25–200 μg.mL and 0.25–128 μg.mL, respectively); 3) 4–45 nm g-AgNP functionalized with tetracycline (6.25–200 μg.mL); and 4) soluble silver (0.25–128 μg.mL). It was noted that MIC value for g-AgNPs was very high (200 μg.mL), while for functionalized g-AgNP with tetracycline, the value dropped to 50 μg.mL. At concentration of 12.5 µg.mL, silver ions seemed to provoke cell lysis. However, toxicity of ionic silver in low concentrations could be alleviated by interaction with compounds produced by B. subtilis cells. The mixture AgNP + tetracycline and the drug itself were the most effective treatments to induce intracellular reactive oxygen species production and impair B. subtilis growth. It was suggested that during incubation of B. subtilis culture with nanosilver, bacterial exudates expelled into the culture might have changed AgNP zeta potential, causing thus aggregation [56].
10.5 Bacteria-Green Nanosilver and Antibiotics Working Together: What Is the Deal?
There are current efforts to combine the effects of green nanosilver produced by bacterial extracts and classical drugs in an attempt to provide an alternative to cope with pathogenic bacteria that are very often resistant to antibiotics. Numerous cellular processes in bacteria are targeted by conventional antibiotics: ATP biosynthesis and e− transport, cell envelope structure, DNA replication and transcription, inhibition of folic acid metabolism, and disruption of 50S and 30S subunits of ribosomes. Chiefly, antibiotics basically change the metabolic state of bacteria, which leads to either death or stasis [58]. Moreover, once the bacterial metabolic state is changed by antibiotics, bacteria will be more susceptible to other chemicals (as nano-Ag or even other drugs).
As expected, some microorganisms appear to be more sensitive to g-AgNP + antibacterial drug combinations than others. Green nanosilver (~8–16 nm) obtained with liquid extracts of Aeromonas sp. THG-FG1.2 inhibited alone Pseudomonas aeruginosa (~16 mm), Vibrio parahaemolyticus (~16 mm), Staphylococcus aureus (~15.5 mm), Bacillus cereus (~13.5 mm), Bacillus subtilis (~13 mm), Escherichia coli (~13 mm), and Salmonella enterica (~11 mm) up to 24 h at 28 °C [41]. Combined effects of green nano-Ag and antibiotics (erythromycin and lincomycin) were especially observed with bacteria V. parahaemolyticus. Accordingly, with erythromycin + g-AgNPs (15 μg + 15 μg per disc) or lincomycin + g-AgNPs (15 μg + 15 μg per disc); the bacteriostatic effects became onefold stronger to those coming out from the antibiotic acting alone. On the contrary, novobiocin or penicillin G + g-AgNP effects were diminished by the nanoparticles from ~29.5 to ~12.5 mm and ~ 45.5 to ~10.5 mm of V. parahaemolyticus inhibition zones, respectively. Drug-resistant S. enterica, P. aeruginosa, and E. coli displayed similar inhibition zones once treated with 6 different antibiotics and green nano-Ag, and in a g-AgNP single condition. Comparatively, ~228.7 nm-Pseudomonas sp.-made-green Ag composites associated with the same 6 antibiotics (erythromycin, novobiocin, vancomycin, lincomycin, penicillin G and oleandomycin) unleashed contrasting effects on B. cereus, S. aureus, C. tropicalis, V. parahaemolyticus, E. coli, P. aeruginosa, and S. enterica during a 24 h treatment at 28 °C [42]. The colloidal Ag solution, synthesized through Ag+ reduction in supernatant of Pseudomonas sp. THG-LS1.4, was composed by larger Ag particles (~228.7 nm) and smaller (10–40 nm) Ag nanoparticles. Briefly, the protocol was as follows: small paper discs were soaked up with 500 μg.mL (500 ppm in 30 μL) of the green-made silver yield and then impregnated again with different antibiotics (10–30 μg.disc each) before being placed in the agar plates. Later on, clinical bacteria P. aeruginosa, E. coli, and S. enterica highly resistant strains to the 6 commercial antibiotics were sensitive to green AgNPs and exhibited inhibition zones reaching ~14.5 mm, ~13 mm, and ~ 11 mm, respectively. After a new set of treatments, it was suggested that synergistic effects possibly emerged when green nano-Ag slightly enhanced bacterial sensitivity to some of the antibiotics (see Table 10.2).
In this particular case as in many others we reviewed, it is questionable to consider such interactions emerging from g-AgNPs and antibiotics as a direct association. First, with no g-AgNP-cleaning up procedures after synthesis, both soluble and nano-Ag remained in solution. Secondly, silver concentration was extremely high (500 μg.mL). Thirdly, the inhibition zones from g-AgNP single exposure and antibiotic + g-AgNP treatments were very much alike and nonsignificant. As another example, a twofold increase against methicillin-resistant Staphylococcus aureus was documented by Manikprabhu and Lingappa [23] with 1 μg of oxacillin in agar, and freshly produced 28–50 nm g-AgNPs reduced from blue pigments of Streptomyces coelicolor. The synergistic effect might have been the result of multiple interacting factors as Ag+, free pigments, active biomolecules, and green nanosilver. With equimolar concentrations of cyanobacterium Oscillatoria limnetica-made 3–18 nm g-AgNP 10 mg.mL and antibiotics (tetracycline or cefaxone at 10 mg.mL), Hamouda et al. [59] evaluated their antibacterial properties against Escherichia coli and Bacillus cereus by measuring inhibition growth in culture optical density with Luria-Bertani medium, and in a classical disc-diffusion method. By spectrophotometry, no significant differences were found with g-AgNP + cefaxone treatment compared to cefaxone itself or soluble Ag against both bacteria species. With the diffusion test, an antibacterial effect appeared, but neither g-AgNP + tetracycline nor g-AgNP + cefaxone conditions showed significant inhibition to be called synergistic.
Working with combined exposures of g-AgNP and antibiotics requires a very accurate experimental setup with objectives clearly formulated, and possible sources of errors comprehensively stated when comes time to interpret and discuss the results. So far, the studies dealing with g-AgNP produced by means of bacteria bioactive molecules and antibiotic combination employed ineffective (and often obsolete methods) to properly test their combined antibacterial proprieties. These studies also failed to fully explain their final results. Before such exposures, it is mandatory to anticipate (or in some cases to model) how the molecular structure of drugs would interact with silver and which testing doses should be used to assess bacterial sensitivity avoiding thus technical difficulties (as nanoparticle aggregation and precipitation). Often, a diffusion method with agar is used with antibiotic disc papers (5–30 μg) soaked up with raw solution of g-AgNPs. By using this simplistic exposition method, it is impossible to carefully calibrate the optimal dosage of drugs and AgNPs that cause real toxic effects in bacterial cells. Most probably, small changes in drug concentration and proportion can optimize or disrupt AgNP activity [56, 60, 61]. Very complex chemical and biochemical interactions between cell membranes, particulate/dissolved silver, and antibiotics are expected; and it gets even more complicated if no purification of the newly synthesized g-AgNP solution is performed before testing. In such a case, soluble silver, g-AgNPs, antibiotics, and unknown free bioactive molecules in solution will likely all interact with each other and bacteria in same time or in a stepwise chain of reactions (Fig. 10.3 A-C). With chloride and phosphate ions frequently found in agar or broth media composition, the equation achieves a new level of complexity.
Schematic drawing of toxicity mechanisms of green silver nanoparticles (associated with antibiotic discs); and chemically produced tetracycline-conjugated AgNP against bacteria [60]. A- With an unfiltered solution of freshly produced g-AgNPs, a bacteria model will be exposed to soluble Ag, nanosilver, and bacteria exudates. By using antibiotic discs soaked with g-AgNP raw solution, an additional stressor will complexify green silver toxicity against bacteria. Considering the high levels of Cl− ions in agar (as in many other media), Ag+ may precipitate as AgCl, which affects silver toxicity. B- Free bioactive bacterial biomolecules (previously separated from isolated strains) are an auxiliary agent to provoke joint toxicity with g-AgNPs on bacteria walls. C- Green nanosilver must behave like organic-coated AgNPs and easily penetrate in cells. By doing so, both interaction of the particle with cell structures and dissolution of Ag core will lead to nanotoxicity. D - A tetracycline-conjugated AgNP previously stabilized with citrate allows an increase of Ag+ release. E and F- The presence of small amounts of tetracycline facilitates AgNP attachment to bacteria. Synergistic interactions will mainly occur following a principal pathway where a tetracycline + AgNP complex binds to bacteria and releases dissolved silver more than AgNP alone. G- Considered as a secondary and less likely pathway, AgNP alone would provoke toxicity by binding to bacteria and releasing Ag+. The antibiotic molecule itself is not considered as a third pathway due to bacterial resistance to tetracycline
From our exhaustive examination of published literature, we observed that several experiments seeking to obtain a synergistic effect with a co-exposure of g-AgNP + antibiotics failed to prove that such an effect was real. Many times, an antibiotic single treatment is first tested on bacteria as a “stand-alone drug control” using microbial strains known to be resistant to the given drug. Then, authors observe moderate or no effects, and confirm the resistance of strains. In the following step, the results of the exposure to the mixtures (antibiotic + g-AgNP) are described, and the synergistic effects are claimed only based on onefold (or less) increase of growth inhibition comparing to the antibiotic-alone condition in a basic ratio estimation. Oddly, g-AgNP antibacterial synergistic effect is estimated with b-a/a x 100 formula (wherein a is the drug effect and b referred to g-AgNP + drug condition) that falls short to confirm such effects owing to the fact that it could just be an additive effect. A significant synergy in pharmacology is accepted when there is more than twofold deviation from concentration addition (dose addition) effects [62]. In other words, synergy rises up when mixtures obtain a minimum twofold difference between observed versus predicted effect concentrations, and a concentration addition as a reference model [62, 63]. Furthermore, synergism (also called superadditivity) for a drug pair not only relies on the agonist drug pair (herein g-AgNP and antibiotic), it depends on the ratio of the doses as well [63]. Hence, finding a range of dose ratios of two agonists is an additional step to bring synergistic interactions in a chemical exposure. These relationships can be easily illustrated with a theoretical isobologram between g-AgNPs and an antibiotic causing low to higher effects in bacteria (Fig. 10.4).
Synergistic interactions between nanosilver and antibiotics can be understood by a model predicting real synergism between two drugs (figure modified from [63]). To achieve a synergistic effect AgNP-antibiotic, it is necessary to optimize the drug combination dose ratio. Each axe represents the dose combinations of AgNPs and antibacterial drugs (dose ratios). The optimal synergistic dose will be found between a range of synergistic and subadditive doses. Synergism for a desired effect against bacteria will be determined for dose ratios between radial lines C and D (indicated by black arrowheads), while the subadditive effect is placed within the dose ratios between A and B (indicated by blue arrowheads). The intersection of these areas (orange triangle) represents the optimal set of ratios to reach a synergistic effect with no undesired side effects [63]
Henceforth, in order to properly investigate the optimal synergism AgNP-drug ratio, an appropriate choice of dose combinations and the bacteria models must be made as shown hereafter. Chemically produced citrate-stabilized 29.8 nm AgNPs (50 μM or 5.4 µg.mL) combined with tetracycline, enoxacin, kanamycin, and neomycin at different ratios (0.1–100 μM antibiotic/Ag) showed strong synergistic effects against drug-resistant Salmonella sp. [60]. The same outcome was not observed with ampicillin and penicillin. Even at lower doses of tetracycline, enoxacin, or kanamycin (~0.5 μM) in antibiotic + AgNP treatments, the inhibition was still observed. With 8–16 μM tetracycline + AgNPs, the inhibition reached nearly 100%. Quite the opposite, the AgNP itself was only able to inhibit 10% of Salmonella sp. growth in single treatments. The ratio of nano-Ag particles to Salmonella cells was ~73:1 in 0.108 µg.mL (1 μM) AgNP medium with ~1 x 107 CFU.mL of bacteria cells. Testing 1 μM AgNPs for Ag binding to Salmonella cells (~1 × 107 CFU.mL), the binding was 42.2% in the presence of 10 μM tetracycline; and it was not different when Salmonella cells increased to 4 × 107 CFU.mL. Most strikingly, 50 μM AgNPs +1 μM tetracycline eliminated 90% of bacteria. Moreover, with a higher dose of tetracycline (10 μM), Ag-bacteria binding falls to 50%, but inhibition was still reaching 95%. Tetracycline molecules increased Ag+ release from AgNPs to 14.5%, and once bacteria cells were present in AgNPs +10 μM tetracycline conditions, 18.2% of Ag+ ions were leached away. In the presence of Salmonella only, Ag+ is released at 12.1% (25% more than without bacteria cells). It can be concluded that some molecules such as tetracycline facilitate Ag+ release [60] (Fig. 10.3 D-G). Noteworthy, aggregation of AgNPs emerged at low antibiotic/AgNP molar ratios except for tetracycline. For a better comparison of mechanisms describing antibacterial effects of g-AgNP in exposure medium enriched with antibiotics (by disk diffusion method), and those observed with chemically synthesized AgNP + tetracycline molar combination (in aqueous solution); see Fig. 10.3.
Using the disc diffusion method, Railean-Pluragu et al. [61] tested a series of combinations with green synthesized AgNPs and classical antibiotics (kanamycin 5 μg, ampicillin 25 μg, neomycin 30 μg, streptomycin 25 μg) against Gram-negative Salmonella infantis, Pseudomonas aeruginosa, Klebsiella pneumoniae, and Staphylococcus aureus. The antibiotic disc was impregnated with 100 μg.mL g-AgNPs and placed into trypticase soy agar (TSA) plates. The 4–45 nm actinomycete-produced AgNPs were first properly separated from unreacted silver and bacterial exudates in a 3-day dialysis. A detailed study using flow cytometry showed that Pseudomonas aeruginosa was the most affected species by both g-AgNP single treatment (50 and 100 μg.mL) and streptomycin, neomycin, or kanamycin + g-AgNPs (12.5 μg.mL) exposures. After treatments, concentration of living cells was significantly lower than negative or positive controls. Such combinations were even less effective than g-AgNP single treatment in the K. pneumoniae case. Reduced doses of g-AgNPs (12.5 μg.mL) in single exposures were enough to significantly inhibit other bacteria such as S. aureus and P. mirabilis.
At the molecular level, g-AgNP and tetracycline combination seem to impair Bacillus subtilis metabolism [56] (Fig. 10.5). After a 2 h incubation in g-AgNP + tetracycline liquid medium, a sporulation process of Bacillus subtilis started. This condition was also triggered by g-AgNPs in single treatments. Bacillus subtilis is a rod-shaped bacteria known to go through sporulation process under inhospitable and lethal conditions to its normal vegetative forms. The coiling of bacterial DNA in g-AgNP single treatments after 6 h indicated the beginning of sporulation, with two cell populations present in the medium: a modified cell and a normal cell similar to the controls. Tetracycline-alone condition did not cause sporulation, but rather disintegrated cell cytoplasm after 2 h. In g-AgNP + tetracycline medium, cells were unable to complete the sporulation, which seems to be mostly due to tetracycline. Later on (4 h), cell-damaging condition was visible and it appeared in g-AgNP + tetracycline medium as well. Likewise, dissolved silver seems to contribute to toxicity: 12.5 μg.mL Ag+-treated cells were severely damaged just after 2 h. With such combination, the sporulation is only induced by g-AgNPs, but tetracycline simultaneously avoided its completion.
Schematic drawing of toxicity mechanisms of green silver nanoparticles combined with tetracycline (in solution) against Bacillus subtilis, and assessed in Mueller-Hinton broth medium [53]. A- The experiment began with a 2 h incubation in g-AgNP + tetracycline medium. B and C- The DNA coiling process indicated sporulation (from normal cell on the left to shrunken cell on the right; the spores below them). In g-AgNP single exposures, the same two cell types were observed. Sporulation began after 2h of exposure in the g-AgNP + tetracycline medium and in the g-AgNp single exposure. D and E- Tetracycline alone disintegrated cell cytoplasm. With g-AgNP mixed with tetracycline, cells do not complete the sporulation and started dying 4 h later. Dotted arrows represent possible pathways taken by tetracycline and g-AgNP. Due to dissolution of Ag core, soluble silver is also represented in the figure
Above published papers have shown promising results when g-AgNPs and antibiotics are added together to induce antibacterial effects, but some results are conflicting. Sometimes a green nano-Ag + antibiotic exposure can bring synergistic effects and detrimental effects to bacteria, but in other assays, these interactions may lead to nano-Ag aggregation and a reduced toxicity as well [60]. In order to accurately evaluate satisfactory synergistic effects of green nano-Ag and antibiotics in vitro, the following guidelines should be considered: (1) careful selection of the growing phase of bacteria cells [52]; (2) accurate preparation of g-AgNP inoculates including efficient dialysis of stock solution and determination of actual concentrations of g-AgNPs); (3) running pre-screened low nominal concentrations of both green nano-Ag and antibiotics before testing [60]; (4) identification by FT-IR and NMR techniques of the main functional groups present in antibiotic molecules and green nano-Ag surfaces; and the exposure media constituents.
10.6 Final Thoughts
To date, most of the research dealing with green silver nanoparticles obtained by bacteria byproducts aimed to primarily describe the AgNP biosynthesis methods using a wide range of bacteria taxa and then eventually test the antibacterial potential of g-AgNPs against pathogenic strains. The lack of analytical data, dialysis of green nano-Ag-stock solution, and g-AgNP kinetic experiments in so many reports is, however, a major setback. The pre-definition of final nominal concentrations is another crucial point to be taken into consideration if precision and accuracy are intended for such assays. Overall, FT-IR or 1HNMR spectral analysis provided a helpful dataset to progress forward new investigations of g-AgNP toxicity mechanisms. We suggest that g-AgNPs likely behave as chemically produced organic-coated AgNPs. By doing so, the g-AgNP-coating layer could have an important role in green nanosilver toxicity by mediating Ag+ release and/or attaching nanoparticles to bacteria cells. But what about the capping biomolecules obtained from actinobacteria residues, for example? Do they just act as facilitators of Ag toxicity or directly interfere with bacteria metabolism as well? And which free biomolecules in bacteria supernatant interfere with g-AgNP solubility? Only by elucidating the complex interactions among silver forms, bacteria-capping agents, and antibiotics can the way be paved to a deeper understanding whether or not g-AgNPs might be really useful to treat bacterial infections in humans. There is no doubt these interactions are very complex, and progress of the knowledge at the molecular level would require the contribution of a number of sub-disciplines such as structural biochemistry, surface nanochemistry, and toxicokinetic modeling, among others.
References
Medici S et al (2019) Medical uses of silver: history, myths, and scientific evidence. J Med Chem 62:5923–5943
Cohen M et al (2007) In vitro analysis of a nanocrystalline silver-coated surgical mesh. Surgical Infections (Larchmt) 8:397–403
Slane J et al (2015) Mechanical, material, and antimicrobial properties of acrylic bone cement impregnated with silver nanoparticles. Mater Sci Eng C 48:188–196
Panáček A et al (2018) Bacterial resistance to silver nanoparticles and how to overcome it. Nat Nanotechnol 13:65–71
Durán N et al (2015) Silver nanoparticles: a new view on mechanistic aspects on antimicrobial activity. Nanomedicine 12(3):789–799
Roy A et al (2019) Green synthesis of silver nanoparticles: biomolecule-nanoparticle organizations targeting antimicrobial activity. RSC Adv 9:2673–2702
Jelinkova P et al (2019) Nanoparticle-drug conjugates treating bacterial infections. J Control Release 307:166–185
Divya M et al (2019) Biogenic synthesis and effect of silver nanoparticles (AgNPs) to combat catheter-related urinary tract infections. Biocatal Agric Biotechnol 18:101037. https://doi.org/10.1016/j.bcab.2019.101037
Gahlawat G, Choudhury R (2019) A review on the biosynthesis of metal and metal salt nanoparticles by microbes. RSC Adv 9(12944):12944
Khan Z, Al-Thabaiti S (2019) Biogenic silver nanoparticles: green synthesis, encapsulation, thermal stability and antimicrobial activities. J Mol Liq 289:111102. https://doi.org/10.1016/j.molliq.2019.111102
Kang F et al (2013) Microbial extracellular polymeric substances reduce Ag+ to silver nanoparticles and antagonize bactericidal activity. Environ Sci Technol 48:316–322
Karthik L et al (2014) Streptomyces sp. LK3 mediated synthesis of silver nanoparticles and its biomedical application. Bioprocess Biosyst Eng 37:261–267
Lengke M et al (2007) Biosynthesis of silver nanoparticles by filamentous cyanobacteria from a silver (I) nitrate complex. Langmuir 23:2694–2699
Subramani R, Aalbersberg W (2012) Marine actinomycetes: an ongoing source of novel bioactive metabolites. Microbiol Res 167:571–580
Gano-Cohen K et al (2016) Nonnodulating Bradyrhizobium spp. modulate the benefits of legume-rhizobium mutualism. Appl Environ Microbiol 82:17. https://doi.org/10.1128/AEM.01116-16
Song AA-L et al (2017) A review on Lactococcus lactis: from food to factory. Microb Cell Factories 16:55. https://doi.org/10.1186/s12934-017-0669-x
Manivasagan P et al (2013) Biosynthesis, antimicrobial and cytotoxic effect of silver nanoparticles using a novel Nocardiopsis sp. MBRC-1. BioMed Res Int 2013: 287638. https://doi.org/10.1155/2013/287638
Rajivgandhi G et al (2019) Biosynthesized silver nanoparticles for inhibition of antibacterial resistance and biofilm formation of methicillin-resistant coagulase negative staphylococci. Bioorg Chem 89:103008
Abd-Elnaby HM et al (2016) Antibacterial and anticancer activity of extracellular synthesized silver nanoparticles from marine Streptomyces rochei MHM13. Egypt J Aquat Res 42:301–312
Samundeeswari A et al (2012) Biosynthesis of silver nanoparticles using actinobacterium Streptomyces albogriseolus and its antibacterial activity. Biotechnol Appl Biochem 59:503–507
Sivalingam P et al (2012) Mangrove Streptomyces sp. BDUKAS10 as nanofactory for fabrication of bactericidal silver nanoparticles. Colloids Surf B: Biointerfaces 98:12–17
Shanmugasundaram T et al (2013) A study of the bacterial, anti-fouling, cytotoxic and antioxidant properties of actinobacterially synthesized silver nanoparticles. Colloids Surf B: Biointerfaces 111:680–687
Manikprabhu D, Lingappa K (2013) Antibacterial activity of silver nanoparticles against methicillin-resistant Staphylococcus aureus synthesized using model Streptomyces sp. pigment by photo-irradiation method. J Pharm Res 6:255–260
Otari SV et al (2015) Intracellular synthesis of silver nanoparticle by actinobacteria and its antimicrobial activity. Spectrochim Acta A Mol Biomol Spectrosc 136:1175–1180
Skladlanowski M et al (2016) Evaluation of cytotoxicity, immune compatibility and antibacterial activity of biogenic silver nanoparticles. Med Microbiol Immunol 205:603–613
Iniyan AM et al (2017) In vivo safety evaluation of antibacterial silver chloride nanoparticles from Streptomyces exfoliatus ICN25 in zebrafish embryos. Microb Pathog 112:76–82
Sivasankar P et al (2018) Characterization, antimicrobial and antioxidant property of exopolysaccharide mediated silver nanoparticles synthesized by Streptomyces violaceus MM72. Carbohydr Polym 181:752–759
Al-Dhabi NA et al (2018) Characterization of silver nanomaterials derived from marine Streptomyces sp. Al-Dhabi-87 and its in vitro application against multidrug resistant and extent-spectrum beta-lactamase clinical pathogens. Nanomaterials 8:279
Sanjivkumar M et al (2019) Investigation on characterization and biomedical properties of silver nanoparticles synthesized by an actinobacterium Streptomyces olivaceus (MSU3). Biocatal Agric Biotechnol 17:151–159
Kang SG et al (2000) New β-lactamase inhibitory protein (BLIP-I) from Streptomyces exfoliatus SMF19 and its roles on the morphological differentiation. J Biol Chem 22:16851–16856
El-Deeb B, Mostafa NY, Altalhi A, Gherbawy Y (2013) Extracellular biosynthesis of silver nanoparticles by bacteria Alcaligenes faecalis with highly efficient anti-microbial property. Int J Chem Eng 30:1137–1144
Sunkar S, Nachiyar CV (2012) Biogenesis of antibacterial silver nanoparticles using the endophytic bacterium Bacillus cereus isolated from Garcinia xanthochymus. Asian Pac J Trop Biomed 2:953–959
Oves M et al (2013) Antibacterial and cytotoxic efficacy of extracellular silver nanoparticles biofabricated from chromium reducing novel O64 strain of Stenotrophomonas maltophilia. PLoS One 8:e59140
Malarkodi C et al (2013) Bactericidal activity of bio-mediated silver nanoparticles synthesized by Serratia nematodiphila. Drug Invention Today 5:119–125
Thomas R et al (2014) Antibacterial properties of silver nanoparticles synthesized by marine Ochrobactrum sp. Braz J Microbiol 45:1221–1227
Syed B et al (2016) Synthesis of silver nanoparticles by endosymbiont Pseudomonas fluorescens CA417 and their bacterial activity. Enzym Microb Technol 95:128–136
Syed B et al (2019) Synthesis and characterization of silver nanobactericides produced by Aneurinibacillus migulanus 141, a novel endophyte inhabiting Mimosa pudica L. Arab J Chem. 12(8) 3743-3752. https://doi.org/10.1016/j.arabjc.2016.01.005
Rasulov B et al (2016) Synthesis of silver nanoparticles on the basis of low and high molar mass exopolysaccharides of Bradyrhizobium japonicum 36 and its antimicrobial activity against some pathogens. Folia Microbiology 61:283–293
Railean-Plugaru V et al (2017) Lactococcus lactis as a safe and inexpensive source of bioactive silver composites. Appl Microbiol Biotechnol 101:7141–7153
Ramasubburayan R et al (2017) Synthesis of nano silver by a marine epibiotic bacterium Bacillus vallismortis and its potent ecofriendly antifouling properties. Environmental Nanotechnology, Monitoring and Management 8:112–120
Singh H et al (2017) Biosynthesis of silver nanoparticles using Aeromonas sp. THG-FG1.2 and its antibacterial activity against pathogenic microbes. Artificial cells, Nanomedicine, and Biotechnology 45:584–590
Singh H et al (2018) Extracellular synthesis of silver nanoparticles by Pseudomonas sp. THG-LS1.4 and their microbial application. Journal of Pharmaceutical Analysis 8:258–264
Kandel S et al (2017) Bacterial endophyte colonization and distribution within plants. Microorganisms 5:77. https://doi.org/10.3390/microorganisms5040077
Lok C-N et al (2017) Silver nanoparticles: partial oxidation and antibacterial activities. J Biol Inorg Chem 12:527–534
Htwe YZN et al (2019) Effect of silver nitrate concentration on the production of silver nanoparticles by green method. Mater Today Proc 17(3):568–573
Gunawan C et al (2014) Nanoparticle-protein corona complexes govern the biological fates and functions of nanoparticles. J Mater Chem B 2:2060–2083
Ferreyra Maillard A et al (2019) Studies on interactions of green silver nanoparticles with whole bacteria by surface characterization techniques. BBA-Biomembranes 1861:1086–1092
Bardaxoglou G et al (2017) High stability and very slow dissolution of bare and polymer coated silver nanoparticles dispersed in river and coastal waters. J Aquatic Pollut Toxicol 12:15
Li J et al (2019) Controlled release and long-term antibacterial activity of dialdehyde nanofibrillated cellulose/silver nanoparticle composites. ACS Sustainable Chemistry Engineering 7:1146–1158
Baygar T et al (2019) Antimicrobial characteristics and biocompatibility of the surgical sutures coated with biosynthesized silver nanoparticles. Bioorg Chem 86:254–258
Vetchinkina E et al (2019) Shape and size diversity of gold, selenium, and silica nanoparticles prepared by green synthesis using fungi and bacteria. Industrial & Engineering Chemistry Research DOI 58:17207. https://doi.org/10.1021/acs.iecr.9b03345
Gordienko MG et al (2019) Antimicrobial activity of silver salt and silver nanoparticles in different forms against microorganisms of different taxonomic groups. J Hazard Mater 378:120754
Schumacher A et al (2018) In vitro antimicrobial susceptibility testing methods: agar dilution to 3D tissue-engineered models. Eur J Clin Microbiol Infect Dis 37:187–208. https://doi.org/10.1007/s10096-017-3089-2
Tuncer M, Seker E (2011) Single step sol-gel made silver chloride on Titania xerogels to inhibit E. coli bacteria growth: effect of preparation and chloride ion on bactericidal activity. J Sol Gel Sci Technol 59:304–310
Levard C et al (2013) Effect of chloride on the dissolution rate of silver nanoparticles and toxicity to E. coli. Environ Sci Technol 47:5738–5745
Rafińska K et al (2019) Study of Bacillus subtilis response to different forms of silver. Science of Total Environment 661:120–129
Railean-Pluragu V et al (2016) Study of silver nanoparticles synthesized by acidophilic strain of Actinobacteria isolated from the Picea sitchensis forest soil. J Appl Microbiol 120:1250–1263
Stokes JM et al (2019) Bacterial metabolism and antibiotic efficacy. Cell Metab 30(6):251–259
Hamouda RA et al (2019) Characterization of silver nanoparticles derived from the cyanobacterium Oscillatoria limnetica. Scientific Rep 9:13071. https://doi.org/10.1038/s41598-019-49444-y
Deng H et al (2016) Mechanistic study of the synergistic antibacterial activity of combined silver nanoparticles and common antibiotics. Environ Sci Technol 50:8840–8848
Railean-Pluragu V et al (2016) Antimicrobial properties of biosynthesized silver nanoparticles studied by flow cytometry and related techniques. Electrophoresis 37:752–761
Cedergreen N (2014) Quantifying synergy: a systematic review of mixture toxicity studies within environmental toxicology. PLoS One 9(5):e96580
Tallarida RJ (2011) Quantitative methods for assessing drug synergism. Genes Cancer 2:1003–1008
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Magesky, A., Pelletier, É. (2020). Toxicity of Green-made Bacteriogenic Silver Nanoparticles Against Bacterial Pathogens: A Critical Review. In: Saquib, Q., Faisal, M., Al-Khedhairy, A.A., Alatar, A.A. (eds) Green Synthesis of Nanoparticles: Applications and Prospects. Springer, Singapore. https://doi.org/10.1007/978-981-15-5179-6_10
Download citation
DOI: https://doi.org/10.1007/978-981-15-5179-6_10
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-5178-9
Online ISBN: 978-981-15-5179-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)