Abstract
There are more than 200 million cases of lung diseases worldwide. Most commonly they include Obstructive Pulmonary Disease (OPD) like Chronic OPD (COPD), asthma and bronchiectasis. This paper aims to study the causal relationship between the shape of the heart and presence of obstructive pulmonary disease by analyzing HRCT scans which are sensitive and informative as it provides with multiple slices of a patient’s internal structure. A mathematical model to predict disease gives confidence to the radiologists for correct and early diagnosis of the disease. Real life HRCT scans along with the disease information were obtained from the Institute of Pulmocare and Research (IPCR). Using Image Processing techniques we finally obtained the right and left atrium of the heart as individual gray-scale images from the HRCT scans; which were then converted into a gray-scale matrix and finally into a vector. We generated our data-set consisting of 40 patients. For patient diagnosed with Obstructive Pulmonary Disease we assigned the label as +1 and for those who have other disease as −1. Different machine learning algorithms such as kNN, SVM, Random Forest and Naive Bayes were applied to the dataset to find the algorithm with highest accuracy and maximum area under the ROC plot.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Large number of diseases that affect the worldwide population are lung-related. Therefore, research in the field of Pulmonology has great importance in public health studies and focuses mainly on asthma, bronchiectasis and Chronic Obstructive Pulmonary Disease (COPD). The World Health Organization (WHO) estimates that there are 300 million people who suffer from asthma, and that this disease causes around 250 thousand deaths per year worldwide [1]. In addition, WHO estimates that 210 million people have COPD. This disease caused the death of over 300 thousand people in 2005 [2]. Recent studies reveal that COPD is present in the 20 to 45 year-old age bracket, although it is characterized as an over-50-year-old disease. Accordingly, WHO estimates by 2030 COPD will be the third cause of mortality worldwide. For the public health system, the early and correct diagnosis of any pulmonary disease is mandatory for timely treatment and prevents further death. From a clinical standpoint, diagnosis aid tools and systems are of great importance for the specialist and hence for the people’s health [2].
Commonly used diagnosis methods for lung diseases are radiological. A chest X-Ray helps in visualization of the lungs. However, chest X-Rays present blurred images of the lungs and precise observations are not possible with them. In such cases computed tomography (CT) scans are used, which is a more sophisticated and powerful X-Ray that gives a 360\(^{\circ }\) image of the internal organs, spine and the vertebrae. A more sensitive version of CT scan, HRCT (High Resolution CT) scan is used to study the morphological changes associated with certain disease.
Larrey-Ruiz et al. in [3] present an efficient image-driven method for the automatic segmentation of the heart from CT scans. The methodology relies on image processing techniques such as multi-thresholding based on statistical local and global features, mathematical morphology, or image filtering, and it also exploits the available prior knowledge about the cardiac structures involved. The development of such a segmentation system comprises of two major tasks: initially, a pre-processing stage in which the region of interest (ROI) is delimited and the statistical parameters are computed; and next, the segmentation procedure itself, which makes use of the data obtained during the previous stage [3].
HRCTFootnote 1 scanning shows cross sections (slices) through the heart and lungs. For one patient there are approximately 60 slices of HRCT scan. An expert in this field cannot give more than 30 s to each slice. Moreover there are many nuances present in a HRCT scan slice which are hidden from the naked eye. In that case even a close inspection from an expert may not be enough. With a dearth of experts in this field it is absolutely imperative to find a method that can capture as much information from the HRCT scan images and draw quick, efficient and reliable conclusions automatically.
The challenge is to use the HRCT scans and the expertise of a doctor to develop a model that can predict if a patient suffers from obstructive pulmonary disease. However, we are not using the information present in the lungs rather our focus will be to extract the information of the shape of the heart components. In order to draw a causal relationship between the shape of the heart and Obstructive Pulmonary Disease. One of the reasons for this approach is that it takes much less time to examine a heart present in an HRCT scan compared to the lungs.
2 Methodology
Given an HRCT slice of a patient, we propose to extract the portions of the heart which contains the left and right atrium. To the vector obtained from these images we apply different machine learning algorithms to understand which model of which algorithm performs better in terms of accuracy for classification of obstructive pulmonary disease against the background disease.
Our approach is to focus on the heart in order to classify the input to different categories of lung diseases. We do this for multiple reasons:
-
It takes much less time to closely examine the heart than the lungs.
-
A closer look tells us that the heart and lungs are very closely associated for blood circulation. As a result we try to exploit the proximity between the heart and lungs to study the impact of heart on lung conditions.
-
Aberrations in the volume of the heart chambers can possibly point towards certain abnormalities in the lungs and vice versa.
-
Study the pressure in the two chambers of the heart the Right Atrium (RA) and the Left Atrium (LA) by measuring their volume (which corresponds to area in 2D).
-
Increased pressure in the RA suggests a rise in the Pulmonary artery pressure which goes to the lungs, similarly increased pressure in the pulmonary vein suggests a rise in the pressure in the LA of the heart.
3 Data Collection and Pre-processing
HRCT scans of patients is collected from IPCRFootnote 2, which are available in DICOMFootnote 3 format. A DICOM file consists of a header and image data sets, all packed into a single file [4]. As a result the DICOM files have to be converted to other Image formats such as JPEG, TIFF or PNG for easier visualization and faster analysis through image processing techniques.
Exploiting the mediastinal windowFootnote 4 using one of the many tags present in the DICOM header, we only had to process 3000 image slices instead of 13000, for each of the 50 patients, which reduced the conversion time considerably.
3.1 Contrast Enhancement
Conversion from DICOM to PNG file format, results in loss of contrast. To correct this, histogram equalization was performed on the obtained PNG image.
3.2 Multi-thresholding on CT Scan Image
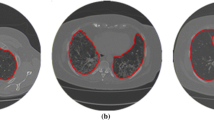
Different parts of a CT scan image have different intensity levels. Exploiting this property, we can extract different portions of the heart by applying different intensity thresholds, which we compute, as per our requirements. This process, we call multi-thresholdingFootnote 5 (Figs. 1 and 2).
3.3 Automatic Selection of the Region of Interest (ROI)
To select only the heart from the image we need to define a Region of Interest (ROI). However, for every patient the size and shape of the heart and lung varies significantly. Moreover the device used to obtain the HRCT scan varies. As a result we cannot fix an ROI, instead the ROI should be selected with respect to each slice of the HRCT scan. To do so, we find the automatic Region of Interest using the algorithm present in [3] with minor parametric tweaks. The algorithm produces a corresponding ROI which only selects the Region of the heart from the entire HRCT scan.
3.4 Algorithm to Extract the Components of Heart from the Background
Step 3 produces an image in which the different sections of the heart are very clearly visible. Step 4 removes the portions of vertebrae from the image.

3.5 Obtaining the Right Atrium and Left Atrium
We can study the ratio (RA:DA) of the area of right atrium (RA) to the descending aorta (DA), and then computing the matrix of the intensity values of finally generating the vectors of the contours of the RA and DA. This approach, however, fails, when the contours of the RA and DA are not clearly distinguishable from other components (Fig. 3).
In this case, we opt for another approach, where we first divide the image obtained after the implementation of the algorithm 1 mentioned above, into four equal parts. This division is supported by the rationale that:
-
the RA will always be present in the top left corner of the image,
-
the DA is in the bottom right corner of the image and
-
the Left Atrium (LA) is in the top right corner of the image.
4 Dataset Generation
Each of the four images obtained after dividing the final image is of dimension 100 \(\times \) 125. We select the image that contains the right atrium. This 100 \(\times \) 125 intensity matrix is converted to a vector of length 12500, which forms 12500 columns in the datasets. There are 40 rows, each corresponding to a patient (Figs. 4 and 5).
Another column, named label is added to the dataset, which is used for supervised learning to classify between Obstructive Pulmonary Disease (OPD) and Background (non-OPD) Disease. The OPD is assigned a label of +1, whereas, the background is assigned a label of −1. OPD consists of COPD and asthma while background diseases include TB, chronic cough and normal cough. This makes our dataset of size: 40 rows \(\times \) 12501 columns.
On close inspection, it was found that the first 2000 columns contain a zero level intensity for each patient, so these columns were omitted, so that they do not act as noise for the machine learning algorithms. The resultant dataset has a size of 40 rows \(\times \) 10501 columns.
5 Machine Learning
Machine Learning, a branch of Artificial Intelligence, relates the problem of learning from data samples to the general concept of inference [4,5,6]. Every learning process consists of two phases: (i) estimation of unknown dependencies in a system from a given dataset and (ii) use of estimated dependencies to predict new outputs of the system. ML has also been proven an interesting area in biomedical research with many applications, where an acceptable generalization is obtained by searching through an n-dimensional space for a given set of biological samples, using different techniques and algorithms [7].
Spathis et al. [8] attempted to choose a representative portion used in the literature comprising of multiple categories of classifiers including linear, non-linear, kernel-based, trees and probabilistic algorithms for the diagnosis of asthma and chronic obstructive pulmonary disease. For similar types of problems Decision Trees were used by Metting et al. [9] Mohktar et al. [10], Prosperi et al. [11] and Prasad et al. [12] and Kernel-based methods such as SVMs were used by Dexheimer et al. [13]. In addition to this Random Forest was also examined by Leidy et al. [14]as well as Prosperi et al. [11]. We have used the method of k-fold cross validation [15], i.e., we created a bunch of train/test splits, calculated the testing accuracy for each, and averaged the results together. We have deployed machine learning algorithms such as kNN, SVM, Random Forest and Naive Bayes on our dataset for different values of \(k \in \{5,7,10 \}\), i.e., 5-cross validation, 7-cross validation, 10-cross validation as well as Jack-Knife Validation. The Python library ‘Sklearn’ [16] was used for performing the ML classifications.
6 Results
In this section, we show a comparison among the performance of different machine learning algorithms for the different sets of data. We present the threshold-dependent plots and threshold-independent ROCFootnote 6 plots, which serve as standard performance metric of the algorithms.
6.1 Testing Set
K-fold cross validation and Jack-Knife validation is done on the Testing set.
6.2 Blind Set
From a total of 40 observations in our dataset, we kept aside a total 6 observations as the blind set. 2 observations with label \(+1\) and 4 observations with label \(-1\). The blind set is disjoint form the training/test set in order to avoid biased prediction (Figs. 6 and 7).
6.3 Best Model on the Blind Set
The confusion matrix generated for the kNN algorithm using 5-,7-,10-cross gives a 100% accuracy whereas it gives an accuracy of 83% using the Jack Knife validation techniques.
7 Discussion
Based upon the threshold-dependent plots and the Receiver operating Characteristic plots shown in the Results section we can say that the k-Nearest Neighbour Classifier (k = 1) performs best on our data-set giving a 77% accuracy and an area under the curve (AUC) of 0.66 for the ROC plots, using 5-cross validation technique on the training/testing data; and giving a 78% accuracy and an area under the curve (AUC) of 0.64 for the ROC plots, using 7-cross validation technique on the training/testing data. For, kNN, the blind set used for the validation of the model gives an accuracy of 100% with a 100% sensitivity using 5-cross, 7-cross and 10-cross validation techniques. Our data-set contains over 10,000 columns, making our data-set complex. The reason why K nearest neighbors performs well in our case can be because this algorithm classifies new cases based on similarity measures, based upon the cases it stored earlier (Figs. 8, 9 and 10).
Random Forest Classifier has also performed well giving an accuracy of 74% and an area under the curve (AUC) = 0.62 for the ROC plots, using 7-cross validation technique on the training/testing data.
Since the data-set contains a vector of image pixels, where the vector length is greater than 10,000; the data-set is a very complex one and Naive Bayes being a simple and crude algorithm could not process the complexity of the data and performed poorly.
8 Conclusion
Based upon our results we can conclude that k-Nearest Neighbor (k = 1) performs better for complex data. Random Forest Classifier has also performed well giving an accuracy of 74%. Naive Bayes performed the worst comparatively and Support Vector Machines was a significant improvement over Naive Bayes.
9 Future Scope
In future, we plan to expand this problem to a multi-class classification problem, which will enable us to predict across a range of pulmonary diseases such as COPD, asthma, tuberculosis, ILD, DPLD, chronic cough, etc. We can also use Deep Neural Networks (DNNs) for image-based classification, owing to the complex nature of the available dataset. However, application of DNN demands a significantly larger sample size which will be possible with the availability of more patient data in the near future.
Notes
- 1.
High-resolution computerized tomography.
- 2.
Institute of Pulmocare & Research, Kolkata, India.
- 3.
Digital Imaging and Communications in Medicine.
- 4.
Part of the chest that lies between the sternum and the spinal column.
- 5.
It will help us obtain the different components belonging to the heart only.
- 6.
Receiver Operating Characteristic, is a graphical plot that illustrates the diagnostic capability of a binary classifier system. The ROC curve is created by plotting the True Positive Rate against the False Positive Rate.
References
Campos, H.d.S., Lemos, A.C.M., et al.: A asma e a dpoc na visão do pneumologista (2009)
Mannino, D.M., Thorn, D., Swensen, A., Holguin, F.: Prevalence and outcomes of diabetes, hypertension and cardiovascular disease in COPD. Eur. Respir. J. 32(4), 962–969 (2008)
Larrey-Ruiz, J., Morales-Sánchez, J., Bastida-Jumilla, M.C., Menchón-Lara, R.M., Verdú-Monedero, R., Sancho-Gómez, J.L.: Automatic image-based segmentation of the heart from CT scans. EURASIP J. Image Video Process. 2014(1), 52 (2014). https://doi.org/10.1186/1687-5281-2014-52
Bishop, C.M.: Pattern Recognition and Machine Learning. Springer, Boston (2006). https://doi.org/10.1007/978-1-4615-7566-5
Mitchell, T.M.: The discipline of machine learning, vol. 9. Carnegie Mellon University, School of Computer Science, Machine Learning (2006)
Witten, I.H., Frank, E., Hall, M.A., Pal, C.J.: Data Mining: Practical Machine Learning Tools and Techniques. Morgan Kaufmann, Burlington (2016)
Niknejad, A., Petrovic, D.: Introduction to computational intelligence techniques and areas of their applications in medicine. In: Medical Applications of Artificial Intelligence, vol. 51 (2013)
Spathis, D., Vlamos, P.: Diagnosing asthma and chronic obstructive pulmonary disease with machine learning. Health Inf. J. (2017). https://journals.sagepub.com/doi/10.1177/1460458217723169#_i7, 1460458217723169
Metting, E.I., et al.: Development of a diagnostic decision tree for obstructive pulmonary diseases based on real-life data. ERJ Open Res. 2(1), 00077–2015 (2016)
Mohktar, M.S., et al.: Predicting the risk of exacerbation in patients with chronic obstructive pulmonary disease using home telehealth measurement data. Artif. Intell. Med. 63(1), 51–59 (2015)
Prosperi, M.C., Marinho, S., Simpson, A., Custovic, A., Buchan, I.E.: Predicting phenotypes of asthma and eczema with machine learning. BMC Med. Genomics 7(1), S7 (2014). https://doi.org/10.1186/1755-8794-7-S1-S7
Prasad, B.D.C.N., Prasad, P.E.S.N.K., Sagar, Y.: A comparative study of machine learning algorithms as expert systems in medical diagnosis (asthma). In: Meghanathan, N., Kaushik, B.K., Nagamalai, D. (eds.) CCSIT 2011. CCIS, vol. 131, pp. 570–576. Springer, Heidelberg (2011). https://doi.org/10.1007/978-3-642-17857-3_56
Dexheimer, J.W., Brown, L.E., Leegon, J., Dominik, A., et al.: Comparing decision support methodologies for identifying asthma exacerbations. In: Medinfo 2007: Proceedings of the 12th World Congress on Health (Medical) Informatics; Building Sustainable Health Systems, p. 880, IOS Press (2007)
Leidy, N.K., et al.: Insight into best variables for COPD case identification: a random forests analysis. Chronic Obstr. Pulm. Dis. J. COPD Found. 3(1), 406 (2016)
Kohavi , R., et al.: A study of cross-validation and bootstrap for accuracy estimation and model selection. In: Ijcai, vol. 14, pp. 1137–1145, Montreal, Canada (1995)
Pedregosa, F., et al.: Scikit-learn: Machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Rahman, U., Bhattacharyya, P., Saha, S. (2020). Obstructive Pulmonary Disease Prediction Through Heart Structure Analysis. In: Thampi, S., et al. Advances in Signal Processing and Intelligent Recognition Systems. SIRS 2019. Communications in Computer and Information Science, vol 1209. Springer, Singapore. https://doi.org/10.1007/978-981-15-4828-4_10
Download citation
DOI: https://doi.org/10.1007/978-981-15-4828-4_10
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-4827-7
Online ISBN: 978-981-15-4828-4
eBook Packages: Computer ScienceComputer Science (R0)