Abstract
Microorganisms are highly versatile living beings that have survived nearly 4 billion years of evolutionary change. Microbes have adapted to environmental conditions that range from cold to hot and from anoxic to oxic. Their life forms range from free living to symbiotic. During the harsh environment, bacteria adopt various strategies to acquire multiple systems so as to thrive against adverse conditions. Among them, secreted proteins play a major role and facilitate most of the interactions of the cells with their surrounding environment as well as with neighbouring cells. Bacterial cells secrete the proteins, metabolites and various molecules to the extracellular environments to sustain nutrient depletion conditions, adhesion on substrate (biofilm formation), invasion of host or antagonism, etc. These microbial functions are managed through their secretion system, which are very sophisticated molecular machines that include export/secretion of required proteins or metabolic factors required for the conjugation and survival processes. Hence, the present chapter is aimed to describe the microbes, specially bacteria adapted to survival in extreme temperature. Further, the chapter highlights the secretion system to understand how these bacteria deal with the extreme environments.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Since the origin of life on earth, prokaryotic life is abundant on our planet and is evolving in all available niches wherever life is possible. On earth, various conditions, from favourable to extreme, are present, and living beings thrive in every possible way to overcome the swaths of unfavourability such as pH, pressure, temperature, salinity, energy, radiation and nutrients (Michels and Clark 1992; Subhashini et al. 2017). In any location of the earth, where liquid water is or was available for life, microorganisms have confirmed their presence. These statements demonstrate the adaptation capabilities of microbiota in the wide range of condition parameter. Hence, it is necessary to overview the machinery or system required in order to combat the conditions of maxima or extreme.
Among the prokaryotes, extremophilic bacteria are the best part of life to be used as an example for the understanding of mechanism and survival tactics. Extremophiles technically cover those bacteria which thrive under a variety of extreme conditions. Moreover, they represent vital objects of research in various fields and disciplines that reveal the ecological, agricultural, industrial and clinical treasures. Furthermore, extremophiles research has exposed the dark shadow of origin of life on the earth as well as on other terrestrial and celestial bodies. Extremophiles are completely opposite to extremotolerants because they are actually adapted creature of that condition with balanced metabolic, biochemical and cellular functionality which is harsh or extreme for normal or tolerant microflora. Merino et al. (2019) have reviewed the extremophiles and hypothesized them as the richest lifeforms on the earth. Moreover, Knoll (2015) speculated that mechanistic life cycle of extremophiles was the only way for survival on the earth during evolution. The environmental conditions and their physical as well as chemical parameters are generally used to categorize the microbiota in different groups. As an example, different temperature values categorize the thermophiles, i.e. bacteria capable of surviving at temperatures more than 60 °C are hyperthermophiles, while those below 15 C are known as psychrophiles. Similarly, the microbes surviving below pH 7 are acidophiles (pH <3) and above 7 are alkaliphiles (pH >9). Those capable of survival at high salt condition belong to halophiles. Similarly, bacteria in absence of oxygen or in low oxygen conditions are anaerobes or microanaerobes, those in low water are xerophiles, at high pressure are barophiles and in high radiations are radioresistant. Interestingly, some of extremophiles are poly-extremophiles which is due to their multi-resistance properties. Auernik et al. (2008) reviewed the thermoacidophiles, a very important poly-extremophilic microbiota and speculated that they majorly belong to the order Sulfolobales and Thermoplasmatales. By nature, they survive well in high temperature as well as in low pH. Similarly, high NaCl and pH tolerance was reported in some halophilic species (Green and Mecsas 2016).
Under the variety of extreme environments, the extremophiles have adopted various secretion mechanisms and a variety of secretomes to thrive normally. Studies have revealed that thermophiles have special features for degradation of complex extracellular carbohydrates (Vafiadi et al. 2009; Asada et al. 2009) and ability to secrete antimicrobial compounds (Esikova et al. 2002; Muhammad et al. 2009). Thermostable Taq polymerase enzyme of Thermus aquaticus is the most vibrant example of novel secretome of extremophiles. The secretome of microbiota and its released proteins in their surroundings or niche where it lives are completely mediated by bacterial cell wall and associated secretion system (Tjalsma et al. 2000). Moreover, cell-to-cell interaction, interaction with substrates, virulence factor and environmentally mediated stress are also governed by secretion system (Zhou et al. 2008). Hence, the following chapter is aimed to highlights the secretion strategies of various extremophile as well as the mechanisms of various types of bacterial secretion systems to understand the ecology of bacteria in extreme and bioprocessing environments.
2 Secretion and Secretome of Some Extremophiles
Bacterial secretome are derived from cell surface by the specialized tools called secretion system. The secretions contain the numerous types of proteins that facilitated the microbes to interact with own community and with host, environment and substrate. Colonization of microbiota or adhesion on surfaces via biofilm is also facilitated by secretome. Moreover, breakdown of complex polymers into simpler monomers or their defence mechanism is also mediated by secretome. Overall, the physiology and survival as well as their application of microbiota are completely based on their secretome. Here, we have illustrated some extremophiles and their secretion to understand it clearly.
Acidophiles and Alkaliphiles
Acidophiles are microorganisms which grow generally at pH range from 0 to 5.5 and occur in all three life domains. Few well-known acidophiles include Cephalosporium sp., Ferroplasma acidarmanus, Acidithiobacillus ferrooxidans, Trichosporon cerebriae, etc. In natural condition, the acidophiles can be obtained from acidic mines, acidic soils and industrial areas (Rothschild and Mancinelli 2001). The acidophiles have unique ability to maintain an intracellular pH near neutral pHs result from high levels of positive cell surface charge, high internal buffer capacity and overexpression of H+ export proteins (Pick 1999). In recent years, the genome sequence of few acidophiles (e.g. Ferroplasma acidarmanus) unrevealed the physiological adaptation to acidic environments.
Alkaliphiles are microorganisms capable of growing optimally at a pH range of 8.5–11.5. These microorganisms represent archaea, prokaryotes and eukaryotes and are isolated from natural habitats such as alkaline soils, mines, hot springs, soda lakes, industrial areas, etc. The key point for alkaliphilic microbiota is to maintain the intracellular pH in their niche where they live. Krulwich (1998) have stated that alkaliphiles performed active import of H+ into the cells, cell wall polymers and use of complex membrane composition for survival. B. pseudofirmus strain OF4 is the best example to understand the molecular machinery of alkaliphiles. The bioenergetic study of cell surface of strain OF4 revealed that NADH dehydrogenase donates electrons in the cellular membrane via menaquinone (MQ) pool. Further, the electron of reduced MQ moves through the menaquinol:cytochrome c oxidoreductase and cytochrome c oxidase, that pumps the protons to deliver it outside the cell and resultantly the molecular O2 is reduced. Then, proton-motive force (PMF) catalysed the import of proton by electropotential charges and resultantly Na + goes lesser than proton. This inward movement maintained the low pH intracellularly in alkaliphiles (Preiss et al. 2015).
Halophiles
These are a versatile group of nature that optimally grown at or above 0.2 M salt concentration and are capable to adapt the rapid alteration of osmotic stress. The secretome of halophiles respond to stress and altered the secretion for homeostasis. For sustainability in saline environments, halophiles have evolved two types of mechanism. One is the cytoplasmic accumulation of inorganic ions that have balanced the osmotic pressure on environments and another is accumulation of specified organic osmolytes in high concentration, that acts as osmoprotectant for the cell, without any metabolic changes in the bacterial cells (Oren 2002). Halophilic bacterial proteomes are basically acidic in nature and possess the amino acids of hydrophilic nature (Weinisch et al. 2018). Expression of the ect gene, transcription of ectABC operon and biosynthesis of ectoine and hydroxyectoine also induce the salt stress (Ma et al. 2010).
Psychrophiles
These are a group of microorganism that thrive well at >15 °C temperature (Steven et al. 2006). However, the actual temperature limits for psychrophiles are not determined clearly. Although for reproduction and metabolism, temperatures of ~12 °C and ~20 °C have been reported, respectively (Rivkina et al. 2000). Moreover, cold niche negatively affects the physiology as well metabolism of psychrophiles, such as cellular integrity, viscosity, diffusion of solutes, fluidity of membrane, metabolome kinetics, etc. (Rodrigues and Tiedje 2008; Piette et al. 2011). Hence, psychrophiles have adapted specialized machinery to maintain the cellular function normally in cold environments and also evolved the polyextremophilic behaviour in adapted cell such as against the radiations, pH alterations, lower in nutrient (Tehei and Zaccai 2005). With the advent of ‘omics’ technologies, a boost in the molecular insight of psychrophile adaption to cold environment has been speculated. Till date, about 135 whole genomes are available on database and about 125 are ongoing. The analysis of published genomes has highlighted the presence of plasmids, fatty acid biosynthesis gene clusters and transposable elements that contribute to genome plasticity (Allen et al. 2009). These available transposables or mobile genes and genetic clusters directly mediate the cold adaptation mechanisms in psychrophilic microbiota. For example, the higher abundance and diversity in tRNA of the Alteromonas sp. SN2 and Psychromonas ingrahamii (Riley et al. 2008), though the G + C content is approximately similar across the genus (Rabus et al. 2004).
Genomic data and bioinformatic interpretations of psychrophiles revealed the significance of genes encoded for cold-shock proteins (CSPs). CSPs are very prominent, small, upregulated nucleic acid binders of proteins. CSPs have been responsible for the regulation of various microbial molecular phenomena such as membrane fluidity, genetic transcription and regulation of expression, protein folding and strong upregulated response to cold shock or exposure (Rodrigues and Tiedje 2008; Margesin and Miteva 2011). This cold shock response of CSP genes induces the RNA helicase enzymes to destabilize the DNA/RNA secondary structures, chaperons, HSPs and mediator genes of sugar metabolism and cellular biogenesis (Chen et al. 2012). Cold environments induce the cellular freezing via formation of cytoplasmic ice crystal in the native microbiota. Resultantly, osmotic balance and cell fluidity gets damaged. Psychrophiles fight against these stresses by producing the antifreeze protein (AFP) that binds the ice crystal of cytoplasm and prevents the crystallization by thermal hysteresis. Exopolysaccharide (EPS) production is another tactic of psychrophiles for cryoprotection. Bordat et al. (2005) have found the excess production of EPSs in Antarctic marine microbes and winter ocean ice. Cornelis and Wolf-Watz (1997) have reviewed that EPSs regulate the cellular surface maintenance of bacteria in cold water and appropriate management of nutrients. Moreover, EPSs facilitate the adherence of cells to surfaces via biofilm formation and synergism with other bacterial communities.
3 Process of Bacterial Secretome
Bacterial secretome is defined as the set of proteins that contains extracellular matrix, vesicle (outer and inner) and membrane secretory proteins of cell (Makridakis and Vlahou 2010). This term was coined by Tjalsma et al. (2000) and described the secretion system of bacteria as well as their secretome. Secretion system and secretome belong to the cellular complexity, virulence, transports, receptors, toxin, etc. and mediated the cellular communication, signalling with in situ environments, biofilm formation, biodegradation, nutrient mineralization, host defence, etc. (Walsh 2000; Wooldridge 2009; Dalbey and Kuhn 2012). Interestingly, a bacterial genome has been occupied 10–30% by the gene that encoded the bacterial secretion system (Kudva et al. 2013). Now, the development of advanced spectrometry such as 2-D electrophoresis, MS/MS and NanoLC/MS has increased our knowledge in secretome of microbiota (Berman et al. 2013; Anwar et al. 2019). Secretome can also facilitate the pathogens to directly or indirectly infect the host cellular surfaces and to survive within it (Aizawa 2001). In non-pathogenic and saprophytic organisms, secretome facilitates the adaptation to its habitat by means of the shape of the bacterial cell envelope (Wattiau et al. 1996).
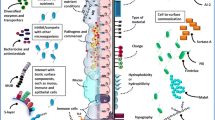
Recent development within the molecular analysis of the protein secretion pathways of bacteria has revealed the 12 types of secretion apparatus as Sec, Tat, T1SS, T2SS, T3SS, T5SS, T6SS, T7SS, SecA2, sortase and injectosome. Sec, Tat and T4SS are common in bacteria, while T1SS, T2SS, T3SS, T4SS, T5SS, T6SS has described in gram-negative bacteria and T7SS, SecA2, sortase and injectosome are described in gram-positive bacteria (Green and Mecsas 2016). T1SS-T4SS has been illustrated in the Fig. 6.1. Examples and classes of bacterial protein secretion system are displayed in Table 6.1.
Gram-positive bacterial cell wall is arranged in the following manner: a single plasma membrane, the cytoplasmic membrane, observed by way of a thick cellular wall layer, while in gram-negative microorganism, it is a double-membrane system, a cytoplasmic membrane and an outer membrane which sandwich the peptidoglycan and periplasmic area among them. The protein secretion system is found across the inner membrane in both gram-positive and gram-negative organisms and normally follows the Sec-based pathway (regularly referred to as the overall secretory pathway (GSP), despite the fact that other pathways have recently been recognized, the Tat and signal recognition particle (SRP) pathways) (Herskovits et al. 2000, Mori and Ito 2001; Driessen 2001). Different types of secretion systems have been described in detail in the following points.
3.1 Type I Secretion System
This secretion system (T1ss) is described widely in gram-negative cells. It governs the secretion of proteins (varied in size) and allowed a single-step transport functions from the cytoplasm to the extracellular medium. The secretion of scavenging molecules, outer membrane vesicles proteins, proteases, lipases and different kinds of virulence factors is mediated by T1ss system. T1ss is similar to the ATP-binding cassette (ABC) transporters family and is involved in the transfer of small molecules such as antibiotics and toxins out of the cell cytoplasm (Symmons et al. 2009). The acquisition of iron by HasAp, a heme-binding protein of P. aeruginosa, is the best example for T1ss protein secretion (Letoffe et al. 1998).
The secretion signal of secreted protein is basically located at the C-terminal, and it is not cleavable at the time of functioning. The overall machinery is designed by three kinds of proteins. Among it, ABC transporter and membrane fusion protein were found in the inner membrane of bacteria cell and an outer membrane channel forming porins. All of these proteins are localized in the bacterial cell envelope and in the presence of a substrate; these proteins form a ‘tunnel channel’ which is transverse to the periplasmic space. Morgan et al. (2017) has described the T1ss-mediated translocation of unfolded polypeptides and proved the significance of T1ss system for protein secretion.
3.2 Type II Secretion System
Secreted proteins have a major function in the pathogenesis of bacterial infections, along with vital sicknesses of humans, animals and plants. Mostly, type II SS (T2ss) is mostly described in the gram-negative bacterium. It facilitates the translocation of proteins from the periplasm to extracellular environments via outer membrane. It is common in pathogenic bacteria as well as in non-pathogenic bacteria. For example, human pathogens V. cholerae, E. coli, P. aeruginosa, Klebsiella sp., L. pneumophila, Yersinia enterocolitica, etc. mediated the pathogenesis via T2ss (Korotkov et al. 2012). Similar, T2ss machinery was also studied in A. hydrophila, an amphibian animal pathogens, by Jiang and Howard (1992).
The T2ss is a classical multi-protein system that contains 12 ‘centre’ proteins, and it was denoted by Anacker et al. (1987) as T2S C, D, E, F, G, H, I, J, K, L, M and O. In recent years, with the development of molecular biology, T2ss structure has been defined well. Presently, it has been divided into four sub-complexes: (i) an OM ‘secretin’, which is a pentadecamer of the T2S D protein that offers a pore via the membrane; (ii) an IM platform composed of T2S C, F, L and M, with T2S C imparting a connection to the OM secretin; (iii) a cytoplasmic ATPase, which is a hexamer of T2S E that is recruited to the IM platform; and (iv) a periplasm-spanning pseudopilus that is a helical filament of the most important pseudopilin T2S G capped with the minor pseudopilins T2S H, I, J and K. Finally, T2S O, an IM peptidase, cleaves and methylates the pseudopilins for the incorporation it into the pseudopilus (Bachovchin et al. 1990).
3.3 Type III Bacterial Secretion System
Many bacterial pathogens along with salmonella use tiny molecular syringes so that it can inject proteins into host cellular cytoplasm. This type of transportation is done by type III secretion device. Type III secretion system (T3ss) presents as microscopic needle complexes and comes from inner to the outer membrane of the bacterium. The needle projected is far away from the cell (Aepfelbacher 2004). T3ss device basically contains two rings that can help throughout the secretion across the cellular membrane including the peptidoglycan of bacterial cell. Moreover, the ring present in inner membrane is larger in size, and the ring protein is also identified in several bacteria. The outer membrane ring is composed of the secretin protein family. It is a needle-like structure that associates with the outer membrane ring and projects from the bacterial surface. T3ss is also reported for the involvement in T2ss secretion and also present in the type IV bacterial pili complex. End of the needle system bought the secretion machinery to touch on substrate for initiate the secretion. Salmonella sp. uses type III secretion supply a cocktail of at least 13 kinds of protein toxin known as effector protein without delay into host cells. Inside the bacterium, sheperon protein binds to effector protein for the handing of over loosely folded from the needle. ATpase triggers the release of effector protein and helps them to start out their journey through narrow channel inside the needle (Frankel et al. 2001). Direct transport to cytoplasm of the host cellular is right concept because it gets rid of the dilution that arises in the cellular within the gut. These effector proteins interfere with the signal transduction cascade to host reaction. The effector protein intrudes with sign transduction cascades to host response. Once the salmonella enters in the host cell, it invades by the vacuoles via the ‘phagosome’, a normal course of activities (Hapfelmeier et al. 2005). The lysosomes then loose with phagosomes and introduce its digestive content to kill the invader.
3.4 Type IV Bacterial Secretion System
Bacterial cells mediate the translocation of DNA (in addition to proteins) into target cells by this unique secretion system type IVss which is found in both gram-negative and gram-positive bacteria and also in some archaea (Alvarez-Martinez and Christie 2009). Conjugation of plasmid DNA has been mediated only by type IVss, and due to these systems, it contributes into the spread of plasmid-borne antibiotic resistance genes, and hence it is the most omnipresent secretion systems in nature. Moreover, TIVss is also involved in bacterial pathogenesis characteristics of few organisms. In Helicobacter pylori and Bordetella pertussis, it is mediated through the secretion of transforming proteins toxins.
In others, effector proteins are required to support an intracellular lifestyle in bacteria such as Legionella pneumophila. Christie et al. (2014) has reported the detailed mechanism and structure of the bacterial type IVss and revealed that it is encoded by the Ti plasmid of Agrobacterium tumefaciens, together with the TIVss encoded by the pKM101 and R388 conjugative plasmids of E. coli. A total of 12 proteins were notified for the arrangement of IVss which is VirB1, VirB2, VirB3, VirB4, VirB5, VirB6, VirB7, VirB8, VirB9, VirB10, VirB11 and VirD4. The protein categorization in different domains of TIVss has been described in Fig. 6.2.
3.5 Type V Bacterial Secretion System
Type V secretion systems (TVss) is a single-membrane-spanning secretion system, which is also known as the autotransporter system. The TVss is mainly for virulence factors but also biofilm formation as well as cell-cell adhesion (Leo et al. 2012). TVss transfers an unfolded autotransporter polypeptide through the IM to the periplasm via SecYEG translocon. The autotransporters are known as ‘passenger’ domain, which is composed of a secreted domain and that is either semi-unfolded or fully unfolded in the periplasm. For the OM (transmembrane domain), it is also called ‘translocator’ or ‘β-domain’. On the basis proteins’ number involved in the secretion process, Green and Mecsas (2016) have separated the TVss into three classes which is autotransporter secretion, two-partner secretion and chaperone-usher secretion.
In autotransporter secretion, it is allowed to secrete themselves by self-contained components (Leyton et al. 2012). These components are described as a translocator domain, a linker domain and a protease domain which were responsible for the C-terminus that forms the outer membrane channel, a passenger domain that contains the functional part of the autotransporter protein and cleaves off the passenger domain once it passes through the channel, respectively. Besides the common autotransporter secretion, few rely on the two-partner secretion, a different polypeptide for transport outside of the cells. In this process, a pair of proteins, on for carrying the carries the β-barrel domain and other, serves as the secreted protein, had participates in the secretion process (Henderson et al. 2004).
The chaperone-usher secretion categories are secreted with the usher protein and chaperone. Usher protein forms the β-barrel, a periplasmic protein channel in the outer membrane that is for the facilitation of folding the secreted protein before delivering to the channel (Waksman and Hultgren 2009). This category is commonly used to assemble pilins on the surface of gram-negative bacteria like P pilus of uropathogenic E. coli.
3.6 Type VI Secretion Systems
The term TVIss is a pivotal secretion system activated during pathogenesis and bacterial competition (Ho et al. 2014), coined firstly by Pukatzki et al. (2006) in the human pathogen V. cholerae. However, the actual coded gene (the imp genes) for the TVIss was reported by Bladergroen et al. (2003) in the symbiotic phytobacterium R. leguminosarum and reported broadly among Proteobacteria (Boyer et al. 2009). TVIss is a cell envelope-spanning machine that is applied to inject toxic effectors into eukaryotic and prokaryotic cells (Ho et al. 2013). Evidences revealed that the TVIss complex is somewhat equivalent to a contractile phage tail that is divided into a tail sheath, an inner tube and a base plate which is anchored to the cell envelope by the membrane complex. The tail sheath extends deep from bacterial outer membrane to cell cytoplasm (Basler et al. 2012). TVIss is composed by several accessory components and 13 essential conserve core components (Zheng and Leung 2007). Generally, all were encoded within the same gene cluster (Taylor et al. 2016). This macromachinery has a membrane and a tail complex. Both are the main complex of TVIss and comprises IM proteins (T4SS homolog) and a tail complex that contains components that are evolutionarily related to contractile bacteriophage tails, respectively (Ma et al. 2009; Leiman et al. 2009).
3.7 Type VII Secretion System
Type VII secretion system (TVIIss) is a specialized protein secretion machinery of gram-positive bacterium, described first in M. tuberculosis and M. bovis. This secretion system is commonly found in phyla of Actinobacteria and Firmicutes. TVIIss is a ~1.5 MDa protein complex that shares a conserved IM and cytosolic apparatus. Central channel of IM is predicted to be composed by membrane proteins EccB, EccC, EccD and EccE. Among them, EccC possesses an ATPase cytoplasmic domain (Solomonson et al. 2013) and EccD have hydrophobic domains.
Generally, TVIIss is divided into two types of component: (a) FtsK/SpoIIIE protein and (b) EsxA-/EsxB-related protein (Pallen 2002; Sysoeva et al. 2014). The atomic structures of EsxA–EsxB, EsxG–EsxH and PE25–PPE41 have been resolved, but their functional attributes remain uncleared (Houben et al. 2014). Hence, the bioinformatic predictions and homology modelling studies revealed that each type of VIIss machinery secretes numerous substrates and establish the several components based complex and multi-protein machinery.
4 Conclusion
Microbes utilize the various methods to survive in the extreme conditions and establish immune system to thwart against it. For that, bacterial secretome and secretion system, across the phospholipid membranes, is foremost and essential component in promoting bacterial virulence, attaching to the substrate, formation of biofilm, to scavenging resources in extreme niche. Moreover, this chapter is illustrating the bacterial secretion system and secretome. It is offering the detailed molecular and functional details of secretion system which is important for the survival of microbiota in the extreme conditions.
References
Aepfelbacher M (2004) Modulation of Rho GTPases by type III secretion system translocated effectors of Yersinia. Rev Physiol Biochem Pharmacol 152:65–77
Aizawa SI (2001) Bacterial flagella and type III secretion systems. FEMS Microbiol Lett 202:157–116
Allen MA, Lauro FM, Williams TJ, Burg D, Siddiqui KS et al (2009) The genome sequence of the psychrophilic archaeon, Methanococcoides burtonii: the role of genome evolution in cold adaptation. ISME J 3:1012–1035
Alvarez-Martinez CE, Christie PJ (2009) Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev 73:775–808
Anacker RL, Mann RE, Gonzales C (1987) Reactivity of monoclonal antibodies to Rickettsia rickettsii with spotted fever and typhus group rickettsiae. J Clin Microbiol 25:167–171
Anwar MN, Li ZF, Gong Y, Singh RP, Li YZ (2019) Omics studies revealed the factors involved in the formation of Colony boundary in Myxococcus xanthus. Cell 8(6):530. https://doi.org/10.3390/cells8060530
Asada Y, Endo S, Inoue Y, Mamiya H, Hara A et al (2009) Biochemical and structural characterization of a short-chain dehydrogenase/reductase of Thermus thermophilus HB8 A hyperthermostable aldose-1-dehydrogenase with broad substrate specificity. Chem Biol Interact 178:117–126
Auernik KS, Cooper CR, Kelly RM (2008) Life in hot acid: pathway analyses in extremely thermoacidophilic archaea. Curr Opin Biotechnol 19(5):445–453
Bachovchin WW, Plaut AG, Flentke GR, Lynch M, Kettner CA (1990) Inhibition of IgA1 proteinases from Neisseria gonorrhoeae and Haemophilus influenzae by peptide prolyl boronic acids. J Biol Chem 265:3738–3743
Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ (2012) Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186
Berman HM, Coimbatore NB, Di CL, Dutta S, Ghosh S et al (2013) Trendspotting in the protein data bank. FEBS Lett 587:1036–1045
Bladergroen MR, Badelt K, Spaink HP (2003) Infection-blocking genes of a symbiotic strain that are involved in temperature-dependent protein secretion. Mol Plant-Microbe Interact 16(1):53–64
Bordat P, Lerbret A, Demaret J-P, Affouard F, Descamps M (2005) Comparative study of trehalose, sucrose and maltose by molecular modelling. Europhys Lett 65:41–47
Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I (2009) Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104
Chen Z, Yu H, Li L, Hu S, Dong X (2012) The genome and transcriptome of a newly described psychrophilic archaeon, Methanolobus psychrophilus R15, reveal its cold adaptive characteristics. Environ Microbiol Rep 4:633–641
Christie PJ, Whitaker N, González-Rivera C (2014) Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta (BBA) Mol Cell Res 1843(8):1578–1591
Cornelis GR, Wolf-Watz H (1997) The Yersinia Yopvirulon: a bacterial system for subverting eukaryotic cells. Mol Microbiol 23(5):861–867
Dalbey RE, Kuhn A (2012) Protein traffic in Gram-negative bacteria – how exported and secreted proteins find their way. FEMS Microbiol Rev 36(6):1023–1045
Driessen AJ (2001) SecB, a molecular chaperone with two faces. Trends Microbiol 9:193–196
Esikova TZ, Temirov YV, Sokolov SL, Alakhov YB (2002) Secondary antimicrobial metabolites produced by thermophilic Bacillus spp. strains VK2 and VK21. Appl Biochem Microbiol 38:226–231
Frankel GA, Phillips L, Trabulsi S, Knutton GD et al (2001) Intimin and the host cell-is it bound to end in Tir(s)? Trends Microbiol 9:214–218
Green ER, Mecsas J (2016) Bacterial secretion systems: an overview. Microbiol Spectr 4(1). https://doi.org/10.1128/microbiolspec.VMBF-0012-2015
Hapfelmeier S, Stecher B, Barthel M, Kremer M, Muller AJ (2005) The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar Typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J Imunol 174:1675–1685
Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala’Aldeen D (2004) Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev 68(4):692–744
Herskovits AA, Bochkareva ES, Bibi E (2000) New prospects in studying the bacterial signal recognition particle pathway. Mol Microbiol 38:927–939
Ho B, Basler M, Mekalanos J (2013) Type 6 secretion system–mediated immunity to type 4 secretion system–mediated gene transfer. Science 342:250–253
Ho BT, Dong TG, Mekalanos JJ (2014) A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15:9–21
Houben EN, Korotkov KV, Bitter W (2014) Take five- type VII secretion systems of Mycobacteria. Biochim Biophys Acta 1843:1707–1716
Jiang B, Howard SP (1992) The Aeromonas hydrophila exeE gene, required both for protein secretion and normal outer membrane biogenesis, is a member of a general secretion pathway. Mol Microbiol 6(10):1351–1361
Knoll AH (2015) Life on a young planet: the first three billion years of evolution on earth. Princeton University Press, Princeton. https://doi.org/10.1515/9781400866045
Korotkov KV, Sandkvist M, Hol WG (2012) The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol 10(5):336–351. https://doi.org/10.1038/nrmicro2762. Published 2012 April 2
Krulwich TA (1998) Alkaliphilic prokaryotes, the prokaryotes, pp 283–308
Kudva R, Denks K, Kuhn P, Vogt A, Müller M et al (2013) Protein translocation across the inner membrane of Gram-negative bacteria: the Sec and Tat dependent protein transport pathways. Res Microbiol 164:505–534. https://doi.org/10.1016/j.resmic.2013.03.016
Leiman PG et al (2009) Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A 106:4154–4159
Leo JC, Grin I, Linke D (2012) Type V secretion: mechanism(s) of autotransport through the bacterial outer membrane. Philos Trans R Soc B 367:1088–1101
Letoffe S, Redeker V, Wandersman C (1998) Isolation and characterization of an extracellular haem-binding protein from Pseudomonas aeruginosa that shares function and sequence similarities with the Serratia marcescens HasA haemophore. Mol Microbiol 28:1223–1234
Leyton DL, Rossiter AE, Henderson IR (2012) From self sufficiency to dependence: mechanisms and factors important for autotransporter biogenesis. Nat Rev Microbiol 10:213–225. https://doi.org/10.1038/nrmicro2733
Ma LS, Lin JS, Lai EM (2009) An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its walker a motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J Bacteriol 19:4316–4329
Ma Y, Galinski EA, Grant WD, Oren A, Ventosa A (2010) Halophiles 2010: life in saline environments. Appl Environ Microbiol 76(21):6971–6981. https://doi.org/10.1128/AEM.01868-10
Makridakis M, Vlahou A (2010) Secretome proteomics for discovery of cancer biomarkers. J Proteome 73:2291–2305
Margesin R, Miteva V (2011) Diversity and ecology of psychrophilic microorganisms. Res Microbiol 162:346–361
Merino N, Aronson HS, Bojanova DP, Feyhl-Buska J, Wong ML et al (2019) Living at the extremes: extremophiles and the limits of life in a planetary context. Front Microbiol 10:780
Michels PC, Clark DS (1992) Pressure dependence of enzyme catalysis. In: Adams MWW, Kelly R (eds) Biocatalysis at extreme environments. American Chemical Society Books, Washington, DC, pp 108–121
Morgan JLW, Justin FA, Jochen Z (2017) Structure of a Type-1 secretion system ABC transporter. Structure 25(3):522–529
Mori H, Ito K (2001) The Sec protein-translocation pathway. Trends Microbiol 9:494–500
Muhammad SA, Ahmad S, Hameed A (2009) Antibiotic production by thermophilic bacillus specie Sat-4. Pak J Pharm Sci 22:339–345
Oren A (2002) Diversity of halophilic microorganisms: environments, phylogeny, physiology, and applications. J Ind Microbiol Biotechnol 28:58–63
Pallen MJ (2002) The ESAT-6/WXG100 superfamily and a new Gram-positive secretion system? Trends Microbiol 10:209–212
Pick U (1999) Dunaliella acidophila: a most extreme acidophilic alga. In: Enigmatic microorganisms and life in extremophiles, vol 2. Springer, Dordrecht, pp 141–148
Piette A, D’Amico S, Mazzucchelli G, Danchin A, Leprince P et al (2011) Life in the cold: a proteomic study of cold-repressed proteins in the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Appl Environ Microbiol 77:3881–3883
Preiss L, Hicks DB, Suzuki S, Meier T, Krulwich TA (2015) Alkaliphilic bacteria with impact on industrial applications, concepts of early life forms, and bioenergetics of ATP synthesis. Front Bioeng Biotechnol 3:75. https://doi.org/10.3389/fbioe.2015.00075
Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D et al (2006) Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A 103:1528–1533
Rabus R, Ruepp A, Frickey T, Rattei T, Fartmann B et al (2004) The genome of Desulfotalea psychrophila, a sulfate-reducing bacterium from permanently cold Arctic sediments. Environ Microbiol 6:887–902
Riley M, Staley JT, Danchin A, Wang TZ, Brettin TS et al (2008) Genomics of an extreme psychrophile, Psychromonas ingrahamii. BMC Genomics 9:210
Rivkina EM, Friedmann EI, McKay CP, Gilichinsky D (2000) Metabolic activity of permafrost bacteria below the freezing point. Appl Environ Microbiol 66(8):3230–3233
Rodrigues DF, Tiedje JM (2008) Coping with our cold planet. Appl Environ Microbiol 74:1677–1686
Rothschild LJ, Mancinelli RL (2001) Life in extreme environment. Nature 409:1092–1101
Solomonson M, Wasney GA, Watanabe N, Gruninger RJ et al (2013) Structure of the mycosin-1 protease from the mycobacterial ESX-1 protein type VII secretion system. J Biol Chem 288:17782–17790
Steven B, Leveille R, Pollard WH, Whyte LG (2006) Microbial ecology and biodiversity in permafrost. Extremophiles 10:259–267
Subhashini DV, Singh RP, Manchanda G (2017) OMICS approaches: tools to unravel microbial systems. Directorate of Knowledge Management in Agriculture, Indian Council of Agricultural Research. ISBN: 9788171641703. https://books.google.co.in/books?id=vSaLtAEACAAJ
Symmons MF, Bokma E, Koronakis E, Hughes C, Koronakis V (2009) The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc Natl Acad Sci U S A 106(17):7173–7178
Sysoeva TA, Zepeda-Rivera MA, Huppert LA, Burton BM (2014) Dimer recognition and secretion by the ESX secretion system in Bacillus subtilis. Proc Natl Acad Sci U S A 111:7653–7658
Taylor NM, Prokhorov NS, Guerrero-Ferreira RC, Shneider MM, Browning C et al (2016) Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 533:346–352
Tehei M, Zaccai G (2005) Adaptation to extreme environments: macro- molecular dynamics in complex systems. Biochim Biophys Acta 1724:404–410
Tjalsma H, Bolhuis A, Jongbloed JDH, Bron S, van Dijl JM (2000) Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol Mol Biol Rev 64:515–547
Vafiadi C, Topakas E, Biely P, Christakopoulos P (2009) Purification, characterization and mass spectrometric sequencing of a thermophilic glucuronoyl esterase from Sporotrichum thermophile. FEMS Microbiol Lett 296:178–184
Waksman G, Hultgren SJ (2009) Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat Rev Microbiol 7(11):765–774
Walsh C (2000) Molecular mechanisms that confer antibacterial drug resistance. Nature 406(6797):775–781
Wattiau P, Woestyn S, Cornelis GR (1996) Customized secretion chaperones in pathogenic bacteria. Mol Microbiol 20:255–262
Weinisch L, Kühner S, Roth R, Grimm M, Roth T et al (2018) Identification of osmoadaptive strategies in the halophile, heterotrophic ciliate Schmidingerothrix salinarum. PLoS Biol 16(1):e2003892
Wooldridge K (2009) Bacterial secreted proteins: secretory mechanisms and role in pathogenesis. Academic, Norfolk
Zheng J, Leung KY (2007) Dissection of a type VI secretion system in Edwardsiella tarda. Mol Microbiol 66:1192–1206
Zhou M, Boekhorst J, Francke C, Siezen RJ (2008) LocateP: Genome-scale subcellular-location predictor for bacterial proteins. BMC Bioinformatics 9:173
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Mishra, M.S., Singh, R.K., Chauhan, S., Gupta, P. (2020). Secretome of Microbiota in Extreme Conditions. In: Singh, R., Manchanda, G., Maurya, I., Wei, Y. (eds) Microbial Versatility in Varied Environments. Springer, Singapore. https://doi.org/10.1007/978-981-15-3028-9_6
Download citation
DOI: https://doi.org/10.1007/978-981-15-3028-9_6
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-3027-2
Online ISBN: 978-981-15-3028-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)