Abstract
Tuberculosis (TB) is regarded as one of the highly infectious diseases which is caused by the species of Mycobacterium genus. Tuberculosis forms to be a major public health issue worldwide because it is anti-drug resistant; extensively drug-resistant (XDR) TB and multidrug-resistant (MDR) TB. Thus, there is an exigent need for the development of new anti-TB drugs. Various drugs are developed in the treatment of different ailments including chronic and TB related symptoms. The present study focuses on the evolution of drug resistance in Mycobacterium tuberculosis, the virulence of Mycobacterium tuberculosis, and preparation of model for evaluation of virulence caused by Mycobacterium tuberculosis. Efforts are also made to summarize the drug resistance mechanism in Mycobacterium tuberculosis including intrinsic and acquired drug resistance.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
16.1 Introduction
Tuberculosis (TB) is primarily caused by a single infectious agent, M. tuberculosis, which has remained the major cause of deaths worldwide (Glaziou et al. 2015). Even, the number of cases is subsequently increasing by the rate of 2% annually. In 1882, Robert Koch discovered Mycobacterium tuberculosis is responsible for TB, an airborne infection (Cambau and Drancourt 2014). M. tuberculosis is a pulmonary pathogen, but still it can exhibit dynamically from being asymptomatic to causing fatal disease (Smith 2003). Till date, a major pathogen of human TB is M. tuberculosis (Assam et al. 2013). Whereas, there are other causative agents of the same genus including M. bovis, M. microti, M. leprae, M. canetti, M. africanum, have also been found to cause of TB infection in humans (Banuls et al. 2015).
On the basis of public and clinical perspective, TB patients are categorized as LTBI (latent TB infection which is asymptomatic or non-transmissible state) or active TB (transmissible) (Lee 2016). Worldwide, about two billion people are suffering from LTBI. In the 17th report by WHO (World Health Organization), they cleared that there are 1.8 million death cases due to TB (Falzon et al. 2017). South Africa, India, China, and the Russian Federation are among the countries largely affected by TB (Jassal and Bishai 2010). Previously, primary drugs like para-aminosalicylic acid and streptomycin were thought to regulate the widespread disease. Furthermore, ethambutol, pyrazinamide, and rifampicin were also introduced (Murray et al. 2015). Due to this, nineteenth century was declared to be “Golden Age of TB Antibiotics.” During this time, these affordable drugs were able to control and decline the TB cases globally. In the 1980s, the reemergence of drug-resistant form of TB during the epidemic of AIDS (acquired immune deficiency syndrome) led to the spread of TB to all corners of the world (Lange et al. 2014). At present, MDR-TB (multidrug resistance-TB) is widespread, nearly 5,80,000 new cases were recorded in 2015. Globally, 84 countries have been reported to be infected by XDR-TB (extensively drug-resistant tuberculosis) (Prasad et al. 2017). Thus, dealing with TB is challenging and therefore it requires targeted diagnosis, screening of drug resistance, and direct evaluation of patient under treatment for 6 months minimum. Moreover, there is requirement for the discovery and effective formulation of novel TB drugs for effective treatment of TB (Chetty et al. 2017).
In this chapter, some major points about virulence, pathogenesis, and drug resistance mechanism of Mycobacterium tuberculosis are incorporated and also provide an insight on the update on new drugs effective against TB.
16.2 Mycobacterium tuberculosis Virulence
Generally, it is not simple to understand what makes M. tuberculosis virulent, in spite of the information gathered in the last 100 years (Jagielski et al. 2016). As it does not involve the traditional factors of virulence like those found in the major disease caused by E. coli O157:H7, Corynebacterium diphtheriae, Vibrio cholerae and Shigella dysenteriae (Forrellad et al. 2013). A very limited information is available which elucidates the mechanism opted by M. tuberculosis to spread disease and how its virulence can be assessed (Pym et al. 2002). On understanding the literature content, it can open a new option that can be used to determine the effect of alteration of the bacterium during disease progression. The two terms “morbidity” and “mortality” have been mostly used to report about M. tuberculosis (Connell et al. 2011). Mortality signifies the percentage of animals died due to infection by calculating the time taken to die after the onset of infection (Hawn et al. 2014). Microbial load (i.e., numbers of microbes presented inside the infested host after the onset infection) is another factor which is associated with virulence. This knowledge enables us to compare the fitness of diverse microbial stains to endure host response when the host is infected (Hoff et al. 2011).
In addition to this, mutant strains of M. tuberculosis exhibit lower bacterial load on assessing their growth curve of infected animals during the process (Ribeiro et al. 2014). Mutants are broadly divided into three broad categories, i.e., persistence genes (per) as they grow normally in early stage but on the on-set of cell-mediated immunity the number gets declined; severe growth in vivo (sgiv) as these mutant do not multiply themselves but either they persist at same cell number or gets rapidly cleared and growth in vivo (giv) as in this case mutant initially get multiplied but multiplication rate is relatively less as compared to wild-type (Glickman and Jacobs 2001). This classification of mutation aids in understanding the genetic mechanism of bacterial genes in regulating the different stages of infection (McGrath et al. 2013). To confirm the standard genetic nomenclature, M. tuberculosis showing the reduced growth in mice is categorized with similar terminology, i.e., per, sgiv, and giv (Smith 2003). Morbidity is a primary factor analyzed during histopathology studies and is the important factor to characterize the mutant class of M. tuberculosis virulence (Sakamoto 2012). For example, sigH mutant genes of M. tuberculosis showed normal growth and high survival rate in mice and macrophages but histopathology analysis of lungs of infected mouse showed reduced virulence in comparison to that of wild-type species of M. tuberculosis (Kaushal et al. 2002).
There is a need for a better understanding of pathogenesis related to TB in order to effectively measure the mortality and morbidity induced by M. tuberculosis (Abbara and Davidson 2011). The unregulated developmental stage of M. tuberculosis in human cells at common site relates to lung damage which ultimately led to death because of oxygen scarcity. This anoxia occurs due to the damaging of parenchymal cells of lung that are usually involved in oxygen uptake, impediment of bronchiolar passages because of granulomatous growths, and due to the release of blood in adjacent lung tissue because of the bursting of liquefied granulomas (Delogu et al. 2013). Another form of TB, also known as tuberculomas, effects the brain by forming enlarged brain granulomas, which may result because of inflammatory response or seizures (Rock et al. 2008). Moreover, inflammatory responses are also responsible for extrapulmonary manifestations in TB patients, especially in bones (Lee 2015).
Inflammation response plays a key role here as they aid in controlling the infection but it also damages tissues of the host (Sasindran and Torrelles 2011). Various proteases have been found to be involved in tissue damage, especially cathepsin D that is majorly involved with granulomas liquefaction (Ehlers and Schaible 2013). Moreover, uptake by M. tuberculosis leads to the apoptosis of macrophages and damaging of adjacent tissues. TNF (tumor necrosis factor), is the key cytokine which gets elicited during inflammatory reaction triggered by the cellular immune system to restrict the widespread of infection (Dutta and Karakousis 2014). Mice which were unable to synthesize or trigger the TNF- did not form granulomas to restrict bacterial dissemination. But, during the presence of a large number of these cytokines, it causes severe inflammation in the lung and early death of mice (Shaler et al. 2011). TNF- is now considered to be the determinant factor of TB meningitis in a rabbit model, as it allows us to directly linked with the severity of disease caused by various strains of both M. tuberculosis and M. bovis as well as with cytokine level in the fluid of cerebrospinal portion (Tsenova et al. 2005). On analyzing the cytokine response and virulence in infected mice, it revealed that there are other factors other than TNF- involved with TB progression (Domingo-Gonzalez et al. 2016). The clinical strain, M. tuberculosis CDC1551, was previously considered to be highly virulent but recent studies revealed that CDC1551 induces cytokines synthesis along with TNF- at a higher level in comparison to other strains of M. tuberculosis in mice. Also, it was less virulent than other strains stated on behalf of mortality rate and bacterial load (Manca et al. 1999). Even comparative study conducted on the rabbit model shows a similar result for the virulence of H37Rv and CDC1551. Another study evaluated the potential of two strains NHN5 and HN878 of M. tuberculosis to elicit the cytokine production and cause disease in the mouse model. For this, HN878 was found to be highly virulent in comparison to NHN5 (Manca et al. 2001).
Apoptosis is also one of the determinant factors, as infection of macrophages by M. tuberculosis depends on the TNF-. And, it was shown that the virulent strain of M. tuberculosis leads to less apoptosis (Behar et al. 2011). The result of above experiments highlights the complexity of the immune system as well as the effectors, but due to the inconsistency in the result, it is difficult to correlate the level of one or more cytokines like TNF- with the clinical model of the disease (Drain et al. 2018). It has become evident that the optimal balancing of these modulators of the immune system is very critical (Cooper 2009). In spite of the varied results, which makes the interpretation of data difficult but are valuable as they demonstrate that few species of Mycobacterium are highly virulent than other in clinical or animal models (Alvarez et al. 2009).
16.3 Model for Measuring Mycobacterium tuberculosis Virulence
Virulence of Mycobacterium tuberculosis is generally studied and measured on the animal or cell culture model. Therefore, different pathogenicity parameters are selected according to the model (Prozorov et al. 2014). The unique characteristic of M. tuberculosis to infect and survive in macrophages makes it the primary target; thus, cell lines and primary macrophages are used to check the effectiveness of M. tuberculosis as well as its mutants during the onset of infection (Pieters 2008). Thus, macrophages are chiefly targeted to assess the normal in vivo condition but difficult in propagating macrophages to a required number makes it incompetent for virulence experiment (Mehta et al. 2006). The immobilized cell lines like MH-S, THP, and J774 are most commonly used, whereas human macrophages from peripheral blood monocytes and murine bone-marrow derived macrophages are widely used macrophages to study the interaction among macrophages and M. tuberculosis (Majorov et al. 2003; Norris and Ernst 2018). Furthermore, besides the assessment of intracellular bacterial load, replication, and survival of M. tuberculosis in macrophage model, it can also be used to understand the mechanism of macrophage microbicide ability and how to counteract with it, like (a) generating resistance against reactive nitrogen/oxygen intermediates, (b) apoptosis inhibition, and (c) phagosome arresting (Bhat and Yaseen 2018).
Alternatively, the animal model aids in studying the diverse stages of TB infection. The most used animal models are rabbits, guinea pigs, and mice (Zhan et al. 2017). Most commonly used in vivo model is mice as it is genetically well-characterized; moreover, inbred strains and immunological reagents are also available (Singh and Gupta 2018). But species of mice are least susceptible to M. tuberculosis infection and their pathology is very different from humans (Kramnik and Beamer 2016). Similarly, guinea pigs are having high susceptible to infection of M. tuberculosis and show similar ailments like disease dissemination, lung necrosis, and lymphadenopathy (Clark et al. 2015). Also, rabbit model on infecting with M. bovis develops granulomas in the lung which resembles the histology of human TB, but because of their size, cost, and very less number of immunological reagents makes it the less tractable model in comparison to mice (Chen et al. 2017). Due to the high similarity of M. bovis and M. tuberculosis, cattle have become an attractive model to study the pathogenicity of TB (Aguilar León et al. 2009). Even, the TB pathology in bovine shows close similarity with humans, results in the formation of caveating lung granulomas and exhibiting similar latent phase after prolong infection (Waters et al. 2011). The benefit of conducting the experiment on the cattle model allows us to conduct field trials and also make it an attractive model for vaccination studies (Buddle et al. 2018). Non-human primate models are the one which shows all the clinical states of the disease that are found in human TB and have given the invaluable contribution in TB research. But the high cost and ethical issues restrict their usage in research (Scanga and Flynn 2014). The bacterial load is one of the most important parameters for measuring the virulence in animal models other than morbidity and mortality (Dormans et al. 2004). Lastly, zebrafish model has also been found to be effective in elucidating the initial stages of mycobacterial infection, especially during the granuloma formation and its function in regulating the infection (Van Leeuwen et al. 2015). In a study, when zebrafish was infected with M. marinum it showed great resemblance with different stages of human tuberculosis; in reality, host genes, virulence factors, and immune cell types are conserved in this interacting model. This model revealed that RD1 locus of bacteria was involved during granuloma formation, whereas ESX-1 system was found to be accountable for the death of infested macrophages (Meijer 2016).
16.4 Drug Resistance in Mycobacterium tuberculosis
The primary mechanism which drives the drug-resistance mechanism in M. tuberculosis is due to the mutation of compensatory genes which encodes for drug-activating enzymes or drug targets (Palomino and Martin 2014). These mutations generally occur due to the deletion, insertion of SNPs (single nucleotide polymorphism), and very rarely due to the deletion of nucleotide in high number (Nguyen 2016). Contrasting other bacteria, M. tuberculosis does not develop mutation due to horizontal transfer of genetic material. Therefore, two mechanisms were reported to generate drug resistance mechanism in M. tuberculosis: first one is transmission and second one is acquired drug resistance (Almeida Da Silva and Palomino 2011).
Various studies conducted to assess the progressive development in drug resistance via WGS revealed that M. tuberculosis initially acquainted the resistance to isoniazid, followed by developing resistance against ethambutol or rifampicin, then against pyrazinamide, and lastly developed resistance against second as well as third-line drugs. This assessment has provided worthy insight into the evolution of M. tuberculosis pathogenicity (Gygli et al. 2017). Furthermore, recent studies have stated mutation leading to the development of drug resistance differs with respect to the lineage to the recipient strain (Ford et al. 2013). Thus, we have summarized the existing anti-TB and new drugs, with the action mechanism of drug and genes linked with resistance development (Table 16.1).
16.4.1 Intrinsic Drug Resistance
M. tuberculosis has been considered to evolve as well as develop various molecular mechanisms to neutralize the cytotoxic of various chemicals such as antibiotics (Davies and Davies 2010). These intrinsic resistance mechanisms have aided the M. tuberculosis to develop resistance against anti-TB agents, which has not only reduced the number of available drugs against TB but have made the exploration of novel anti-TB agents more difficult (Hameed et al. 2018). There are various mechanisms that are responsible for growth intrinsic resistance in strains of M. tuberculosis and other pathogenic strains.
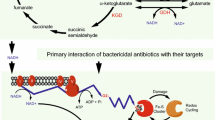
One of the mechanisms is cell wall permeability which regulates the entry and exists of the chemical from the cell membrane. The reduction in the permeability of drug via cell wall of mycobacteria serves as the active barrier and hindrance for antibiotic therapy (Sarathy et al. 2012). For example, a report revealed that β-lactams penetration through cell walls of mycobacteria species to be 100 times slower than the cell wall of E. coli. The function of cell wall permeability in antibiotic resistance in mycobacterial strains has well comprehended by studying the mutant defects during cell wall biosynthesis (Smith et al. 2012). Mycobacterial cell wall regulates the penetration of antibiotic, and there are other specialized resistance mechanisms which detoxify antibiotic molecules that were able to enter into the cytoplasmic region (Mukhopadhyay et al. 2012). The specialized mechanism involves alteration of the target, mimicking of the target, drug modification, drug degradation, and drug efflux (Fig. 16.1).
Target alteration strategy is generally applied by bacteria to avoid the antibiotic action by modifying the target structure of the antibiotics and is usually adopted by strains of M. tuberculosis species and other mycobacterial strains to decrease the chances of binding of lincosamides and macrolides to ribosomes of M. tuberculosis (Fair and Tor 2014). Recent studies revealed that Erm37 gene has the protecting roles in mycobacterial strains from lincosamides and macrolides. M. tuberculosis uses a similar mechanism to neutralize the activity of capreomycin and viomycin drugs used for treating multiple drug resistance TB (Buriánková et al. 2004; Fu and Shinnick 2007). The studies conducted on M. smegmatis and M. tuberculosis revealed the association of tlyA gene with viomycin and capreomycin resistance (Maus et al. 2005). Another specialized method of intrinsic drug resistance, mimicking of the target, is effective in neutralizing the effect of fluoroquinolones. Fluoroquinolones are anti-TB drug which has bactericidal effect as it inhibits the action mechanism during DNA replication, transcription, as well as repair (Von Groll et al. 2009). Generally, these drugs bind with DNA topoisomerase or gyrase enzyme resulting in the complexation of DNA which prevents resealing of DNA strands and finally leads to DNA degradation and cell cessation (Ginsburg et al. 2003). Thus, on mapping, the acquired fluoroquinolone resistance revealed that protein MfpA attributes for intrinsic resistance (Hegde et al. 2005). Another mechanism which mycobacterial species employs to directly deactivate the active drug is drug modification (D’Ambrosio et al. 2015). Aminoglycosides drugs have always held the main position, even in the history of TB therapy. The target function of these drugs remained the same, i.e., to inhibit the synthesis of protein (Xie et al. 2011). Studies revealed that acetyltransferase plays a key role in the survival of mycobacterium species in macrophages of the host (Kim et al. 2012). Lately, it was discovered that it aids in changes in innate immunity of the host in contradiction to infection of mycobacterial species. These modifications in the host signaling molecule have suppressed the immune response like apoptosis, autophagy, and inflammation of the host infected by M. tuberculosis (Zhai et al. 2019).
Another strategy used by M. tuberculosis to subvert the action of the anti-TB drug is to degrade them via hydrolases (Nguta et al. 2015). These mechanisms have been broadly studied in β-lactams drugs, which have no effect on M. tuberculosis as well as on other mycobacterial strain. This action mechanism of these drugs is to inhibit the synthesis of cell wall synthesis by binding on penicillin-binding proteins (PBPs) region which leads to apoptosis (Kohanski et al. 2010). On analyzing the M. tuberculosis genome, it revealed that genome contains four sites which encode for PBPs, where β-lactams bind within detectable concentrations. This clears the fact that the least target affinity is not acceptable for β-lactam resistance in mycobacterial strain (Li et al. 2018). Hydrolytic enzyme, β-lactamases are considered to be the determinant for β-lactams resistant, as this enzyme hydrolyzes the β-lactam ring. This was confirmed by conducting experiment on M. fallax (highly susceptible to β-lactams drug), result of permeability assay revealed that rate of penetration in cell walls of M. fallax by β-lactams was similar to other mycobacterial species and permits the accumulation of β-lactam drugs to lethal concentration (Wang et al. 2006). But on engineering the M. fallax with gene expressing β-lactamase from M. fortuitum showed the increase in resistance level similar with other species and revealed β-lactamases are the major cause of β-lactam susceptibility (Sauvage et al. 2006). BlaC is another β-lactamase which is effective against tuberculosis. Moreover, it has been found to have broad-substrate specificity because of flexible substrate binding nature. BlaI gene has been comprehended to regulate the function of BlaC in strains of M. tuberculosis. During β-lactams absence, there is the formation of homodimers of BlaI, which binds to the promoter region of BlaC by obstructing its transcription. But when this M. tuberculosis strain is subjected to β-lactams, it causes the dissociation of BlaI from DNA binding site and derepression of BlaC transcription, which results in the production of β-lactamase (Kurz and Bonomo 2012). Other than BlaC, M. tuberculosis also encodes other β-lactamases like BlaS, Rv3677c, and Rv0406c (Nampoothiri et al. 2008).
Lastly, the most commonly used method by microbes to avoid the action of drugs is to remove them from the cytoplasm via efflux mechanisms (Soto 2013). The trans-membrane proteins are the one which plays a key role in the mechanism. For example, there are 20 out of 36 genes which encode for membrane proteins in the genome of E. coli, which grant them the resistance to more than one drug (Niederweis et al. 2010). It is very improbable that now these transporter proteins have evolved themselves to act as specialized drug transporters (Feltcher et al. 2010). Various experiments revealed that mycobacteria contain 18 transporters which have conferred antibiotic resistance in them. Likewise, expression of EfpA and IniBAC is negatively regulated through Lsr2, which binds to AT-rich region of the sequence (Nguyen 2016). Significantly, the first-line drugs, isoniazid or ethambutol, have been found to have an inducible effect on Lsr2, which regulates the transcription of EfpA and IniBAC; thus, each transporter protein has evolved themselves to perform a specialized function in antibiotic resistant strain (Colangeli et al. 2007). Recent studies have also linked Lsr2 to changes in oxygen level involved in mycobacterial adaptation, thus providing us the connecting link between the pathogenesis and resistance of M. tuberculosis (Bartek et al. 2014). Another transporter protein that is effective in the efflux of anti-TB drugs like aminoglycosides, tetracycline, and spectinomycin is Tap. Some studies also confirmed the function of Tap in conferring the drug resistance to M. tuberculosis (Balganesh et al. 2012).
16.4.2 Acquired Resistance
The anti-TB drugs targeted binds to the target site with high affinity, as a result they obstruct the normal activity of the target molecule. But, modification in targeted site prevents the effective binding of the drug and generates resistant against the particular drug (Hoagland et al. 2016). In M. tuberculosis and other species, resistance occurs due to mutation (spontaneous) in the chromosomal genes encoding target molecules (Koch et al. 2018). Below, we have briefly discussed the point mutation allied with the resistance of strains of M. tuberculosis for first-line drugs like EMB, INH, PZA, and RIF as well as for second-line drugs like fluoroquinolones, bedaquiline, and macrolides.
16.5 Mutations Responsible for the Development of Acquired Resistance to First- and Second-Line TB Drugs
Recent studies have revealed that compensatory mutation in various genes like ahpC, inhA, kasA, katG, and ndh are all linked to INH resistance (Liu et al. 2018a). INH is one of the pro-drugs which uses peroxidase or catalase enzyme encoded by gene katG for its activation. A mutation in katG has been found to be linked with the reduced activity of catalase or peroxidase and is a common mechanism responsible for INH resistance (Cade et al. 2010). Another similar mechanism which confers low-level resistance towards INH occurs because of the mutation in inhA promoter (Bollela et al. 2016). Lately, Torres and his colleagues identified new mutation which comprehends for 98% of INH resistance induced by fabG1, katG mutation, or inhA promoter (Torres et al. 2015). Similar finding related to INH resistance and mutation is the discovery of harbinger mutation like katG S315T, which can serve as a valuable asset for reporting warning about the evolution of multidrug resistant. These results revealed the impact of these mutations on public health and have enabled to target treatment of the patients suffering from multiple-drug resistance TB (Pym et al. 2002), whereas 95% RIF resistant strains have reported about mutation in codons 507–533 of RNA polymerase beta-subunit gene (rpoB) (Van Deun et al. 2013).
Pyrazinamide (PZA) is an essential drug and considered to be short-term chemotherapy for TB as it is effective in reducing the treatment regimens (Chan et al. 2004). But now it has become ineffective due to the mutation in pncA gene, which reduces the activity of pyrazinamidase enzyme and becomes resistance against PZA (Ramirez-Busby and Valafar 2015). Various other studies also reported about the mutation in clpC1, rpsA, and panD which encodes for ATP-dependent ATPase, aspartate decarboxylase, and ribosomal protein S1, respectively, to be liable for PZA resistance (Zhang et al. 2017). Other first-line anti-TB drugs, such as EMB in combination with RIF, INH, and PZA, are used to treat TB and control the widespread of drug-resistant strains (Nasiri et al. 2016). On the contrary, various reports have documented showing that mutation in operon of embCAB especially in embB gene results in the development of resistance against EMB in M. tuberculosis making the treatment ineffective (Plinke et al. 2011). Although mutation of ubiA gene has also been established to be responsible for drug resistance in M. tuberculosis strains (Lingaraju et al. 2016), the other aminoglycosides drugs like amikacin and kanamycin are effective against TB. But, time strains of M. tuberculosis have developed the resistance to these aminoglycosides drug due to A1401G mutation in rrs gene which encodes for 16 s rRNA (Jugheli et al. 2009). In contrast to other bacteria, which contains multiple copies of genes, mycobacteria contain only one copy of this gene; hence, it defines that why mutation in this gene leads to aminoglycoside resistance (Garneau-Tsodikova and Labby 2016).
Capreomycin and viomycin are another set of drugs that have been used for the treatment of TB. But, due to similar function mechanism of these drugs with aminoglycosides drug they were found to be virulent against strains of M. tuberculosis having acquired resistance to kanamycin (Gualano et al. 2016). Fluoroquinolones, being the second-line anti-TB drugs, are used to treat infection caused by M. fortuitum, M. kansasii, and M. simiae (Ma et al. 2010). Generally, this drug targets the type II topoisomerases, DNA topoisomerase IV, and DNA gyrase enzyme which controls the functions like cell division, DNA replication, and supercoiling of DNA (Schluger 2013). The mutations in genes gyrA and gyrB are considered to be likely associated with fluoroquinolone resistance in mycobacterial strain. The substitution of codon 90 and 94 in gyrA gene is the mutation which is found to be involved with fluoroquinolone-resistant in M. tuberculosis (Maruri et al. 2012). Few studies also indicated that the efflux mechanism is also involved in fluoroquinolone resistance (Lu et al. 2014).
Another bacteriostatic drug, linezolid which inhibits the synthesis of protein by forming complexes on the 50S ribosomal subunit, is now clinically used for treating drug-resistant TB (Chetty et al. 2017). But, mutation in rplC and rrl gene has been discovered in linezolid resistant strain (Zhang et al. 2016). Recently approved diarylquinoline drug, bedaquiline, was also assessed for its resistance in mycobacterial strains. To our surprise, mutations in the atpE gene were found to be accountable for drug resistance (Andries et al. 2014). This was the brief discussion about the mutations responsible for the growth of acquired resistance in relation to TB drugs of the first or second line. And, we tried to highlight how these acquired mutations are making the situation difficult to regulate drug-resistance TB.
16.6 Mechanism of Drug Resistance
In 1948, the phenomenon of drug resistance was recorded while the first trial of TB therapy was being conducted (Gillespie 2002). As each novel anti-TB drug was discovered and brought into clinical trials, the prevalence of resistant strains was encountered within a decade (Rawal and Butani 2016). Genetic mutation is the key reason for the drug resistance in M. tuberculosis as there is no evidence or report for resistance development due to the acquisition of new DNA (Parida et al. 2015). Allelic exchange experiment has established the interconnection between drug resistance and mutation, which occurs due to a mutation in a subset of genes. There are two primary mechanisms involved in drug resistance: a) modification of the targeted molecules and b) due to defect in the enzyme function which changes its activity (Caminero et al. 2010).
Limitation in both genotypic and phenotypic drug-susceptibility test hampers the basic understanding of resistance mechanisms. Generally, the phenotypic test shows the dichotomous result, i.e., strain of M. tuberculosis is either resistant or susceptible to a specific set of drugs like ethambutol, rifampicin, and isoniazid (Ocheretina et al. 2014). Besides this, the genotypic test fails to detect the mutation present in the phenotypic resistant strain. Conclusively, identifying the mutation in phenotypic resistant strain with the help of genome or gene sequencing does not ensure to check the mutation responsible for the resistance (Yakrus et al. 2014). Hence, the phenotypic mutation could be any mutation from contemporary, intermediator, or causal mutation (Motiwala et al. 2010). This prompts them to design the diagnostic assay based on the causal mutation to identify the drug-resistant strains. That is why it is difficult to determine the mutation and categorized according to its type (Desjardins et al. 2016).
Till now, various groups have started to sequencing the whole genome of clinical isolates to find the novel mutation linked with resistance and long-term goal to develop a diagnostic test which could detect the resistant strain and can replace the culture-dependent drug susceptibility test (Iketleng et al. 2018). This approach has shown the feasibility in preliminary studies but lack of precision and high cost prevented its usage. Still, culture-based approach remained the reliable option for clinical care (Nahid et al. 2012).
16.7 Evolution of Drug Resistance in Mycobacterium tuberculosis
Bacterial epistasis and fitness are two main factors which influence the progress of drug resistance of M. tuberculosis strains (Al-Saeedi and Al-Hajoj 2017). Epistasis signifies genetic interaction of a certain set of genes, in which the phenotypic effect of the first mutation solely depends on the second mutations (Wong 2017). As observed, resistance strains carry the same resistant mutation which varies in their capacity during transmission from one to another patient, providing the evidence that genetic background of the strain can aid in determining the course of evolution to develop drug resistance (Trauner et al. 2014). On the contrary, bacterial fitness is the function of growth rate, transmissibility, and virulence. Thus, mutation results in the reduction of the bacterial number in contrast to wild-strain are considered to carry the “fitness cost” (Schulz et al. 2010). If one needs to immediately estimate the relative fitness of bacteria, he can determine it by measuring the growth rate of bacteria present in the culture (Ayabina et al. 2016). As evolution is a continuous process, various studies have provided the evidence in support that fitness of resistant mutant cannot be fixed (O’Neill et al. 2012). Another example of epistasis is the acquisition of compensatory mutation, which also plays vital role in the formation of drug-resistant strains imposing a great risk on human health (Müller et al. 2013). As of now, we do not have the adequate information to predict the epistasis interactions using bioinformatic tools; thus, we have to rely on the conventional approaches to gather knowledge about the genetics behind the development of drug resistance (Ngo and Teo 2019).
Recently, various research groups used WGS (whole genome sequencing) to gather information about molecular epidemiology, mutation frequency, and phylogeny to compare drug-resistant and drug-susceptible in M. tuberculosis strains. This approach also helps us to address the key contributor involved in the evolution of strains of M. tuberculosis (Ilina et al. 2013). Generally, there are various genes involved in transcriptional control, cell wall homeostasis, lipid metabolism, and purine metabolism during anti-TB therapy (Fonseca et al. 2015). Henceforth, these genes can assist us in understanding the drug-resistant mechanism. For example, ponA1 gene, whose actual function is unknown but is discovered to involve in the evolution of drug resistance in species of M. tuberculosis (Smith et al. 2012). Evidence from other studies prompted us to investigate the role of these genes and how these genes can be further used for diagnosis purpose. With time number of these genes is growing exponentially and we are also getting the supporting evidence to prove their role as epistasis from adaptation to resistance (Daya et al. 2015). rpoC, which act as a mediator from adaptation to RIF (rifampicin) resistance development and Rv3806c, which mediated the EMB (ethambutol) resistance are the examples of the genes identified in recent studies (Somoskovi et al. 2001).
Recently, RIF resistance is found to be induced due to the mutation of RNA polymerase by rpoB enzyme (Kumar and Jena 2014). Hence, mutation in multidrug-resistant strains of M. tuberculosis is nearly ubiquitous and is mostly found to be associated with compensatory mutations in rpoA, rpoB, and rpoC genes (RNA polymerase genes). Instead of this, the compensatory mutation also restores the baseline profile of cells (De Vos et al. 2013). Especially, mutants rpoB were found to improve the lipid profile and alter the expression of various proteins involved in lipid metabolism, specifically phthiocerol dimycocerosates (PDIMs). Therefore, lipid metabolism involving PDIMs has positive influence during the progress evolution of drug resistance (Lahiri et al. 2016). In addition to this, in vitro studies revealed that resistant mutant embB M306v contains a synonymous mutation in Rv3792 and non-synonymous mutation in Rv 3806c, which are a major contributor for developing EMB resistance (Safi et al. 2013). Thus, it is evident that epistasis does not depend on one drug. Therefore, different studies are being conducted to assess the interaction among the disparate drugs and mutations, and their eminent role in the growth and development of drug resistance. This proves the fact that positive epistasis can trigger multidrug resistance (Trauner et al. 2014).
Recently, it has been accorded that continuous exposure of drug imposes some constraint on the evolution of TB, which increase the chances of compensatory mutation in already resistant strains (Liu et al. 2018b). Once these strains get mutated, the strains possess the ability to transmit the mutated gene to the next generation alone (Banuls et al. 2015). Furthermore, continuous drug exposure is known to start accumulating the mutant which results in an increased level of resistant towards the particular drug. This is one of the factors, which influences isoniazid (INH) resistance in multidrug-resistant strains and also found to be contributing for high resistance against fluoroquinolones (FQ) (Dookie et al. 2018).
16.8 Conclusion
Due to the development of resistance to first- and second-line drugs, TB has remained the biggest concern worldwide. WHO has also issued some recommendations and guidelines for the proper care of TB patient in the public or private sector and are ensuring that precise diagnosis is being used and found effective in treating tuberculosis infection. With the failure of second-line drug like fluoroquinolones, which was effective in reducing the duration of chemotherapy and has limited drug treatment options for multiple drug-resistant tuberculosis. Therefore, the development of a new drug is urgently required along with that there is a need for exploring alternative treatments like host-directed therapy, personalized medicine, and more. Though we are still investigating basic biology as well as the pathogenesis of M. tuberculosis and exploring the different therapeutic options and new various anti-TB drugs. But there is a need to scale up various approaches, tools, and health care service in reliance with the government as it provides us to regulate the chaos induced by tuberculosis and the growing issue of drug-resistant tuberculosis.
References
Abbara A, Davidson RN (2011) Etiology and management of genitourinary tuberculosis. Nat Rev Urol 8(12):678–688
Aguilar León D, Zumárraga MJ, Jiménez Oropeza R, Gioffré AK, Bernardelli A, Orozco Estévez H, Cataldi AA, Hernández Pando R (2009) Mycobacterium bovis with different genotypes and from different hosts induce dissimilar immunopathological lesions in a mouse model of tuberculosis. Clin Exp Immunol 157(1):139–147
Almeida Da Silva PE, Palomino JC (2011) Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J Antimicrob Chemother 66(7):1417–1430
Al-Saeedi M, Al-Hajoj S (2017) Diversity and evolution of drug resistance mechanisms in Mycobacterium tuberculosis. Infect Drug Resist 10:333–342
Alvarez AH, Estrada-Chávez C, Flores-Valdez MA (2009) Molecular findings and approaches spotlighting Mycobacterium bovis persistence in cattle. Vet Res 40(3):1–6
Andries K, Villellas C, Coeck N, Thys K, Gevers T, Vranckx L, Lounis N, de Jong BC, Koul A (2014) Acquired resistance of Mycobacterium tuberculosis to bedaquiline. PLoS One 9(7):e102135
Assam JP, Beng VP, Cho-Ngwa F, Toukam M, Ngoh AA, Kitavi M, Nzuki I, Nyonka JN, Tata E, Tedom JC, Skilton RA (2013) Mycobacterium tuberculosis is the causative agent of tuberculosis in the southern ecological zones of Cameroon, as shown by genetic analysis. BMC Infect Dis 13(1):431
Ayabina D, Hendon-Dunn C, Bacon J, Colijn C (2016) Diverse drug-resistant subpopulations of Mycobacterium tuberculosis are sustained in continuous culture. J R Soc Interface 13(124):20160745
Balganesh M, Dinesh N, Sharma S, Kuruppath S, Nair AV, Sharma U (2012) Efflux pumps of Mycobacterium tuberculosis play a significant role in antituberculosis activity of potential drug candidates. Antimicrob Agents Chemother 56(5):2643–2651
Banuls AL, Sanou A, Van Anh NT, Godreuil S (2015) Mycobacterium tuberculosis: ecology and evolution of a human bacterium. J Med Microbiol 64(11):1261–1269
Bartek IL, Woolhiser LK, Baughn AD, Basaraba RJ, Jacobs WR, Lenaerts AJ, Voskuil MI (2014) Mycobacterium tuberculosis Lsr2 is a global transcriptional regulator required for adaptation to changing oxygen levels and virulence. MBio 5(3):e01106–e01114
Behar SM, Martin CJ, Booty MG, Nishimura T, Zhao X, Gan HX, Divangahi M, Remold HG (2011) Apoptosis is an innate defense function of macrophages against Mycobacterium tuberculosis. Mucosal Immunol 4(3):279–287
Bhat KH, Yaseen I (2018) Mycobacterium tuberculosis: macrophage takeover and modulation of innate effector responses. In: Ribón W (ed) Mycobacterium-research and development. IntechOpen, London. https://doi.org/10.5772/intechopen.75003
Bollela VR, Namburete EI, Feliciano CS, Macheque D, Harrison LH, Caminero JA (2016) Detection of katG and inhA mutations to guide isoniazid and ethionamide use for drug-resistant tuberculosis. Int J Tuberc Lung Dis 20(8):1099–1104
Buddle BM, Vordermeier HM, Chambers MA, de Klerk-Lorist LM (2018) Efficacy and safety of BCG vaccine for control of tuberculosis in domestic livestock and wildlife. Front Vet Sci 5:259
Buriánková K, Doucet-Populaire F, Dorson O, Gondran A, Ghnassia JC, Weiser J, Pernodet JL (2004) Molecular basis of intrinsic macrolide resistance in the Mycobacterium tuberculosis complex. Antimicrob Agents Chemother 48(1):143–150
Cade CE, Dlouhy AC, Medzihradszky KF, Salas-Castillo SP, Ghiladi RA (2010) Isoniazid-resistance conferring mutations in Mycobacterium tuberculosis KatG: Catalase, peroxidase, and INH-NADH adduct formation activities. Protein Sci 19(3):458–474
Cambau E, Drancourt M (2014) Steps towards the discovery of Mycobacterium tuberculosis by Robert Koch, 1882. Clin Microbiol Infect 20(3):196–201
Caminero JA, Sotgiu G, Zumla A, Migliori GB (2010) Best drug treatment for multidrug-resistant and extensively drug-resistant tuberculosis. Lancet Infect Dis 10(9):621–629
Chan ED, Laurel V, Strand MJ, Chan JF, Huynh ML, Goble M, Iseman MD (2004) Treatment and outcome analysis of 205 patients with multidrug-resistant tuberculosis. Am J Respir Crit Care Med 169(10):1103–1109
Chen H, Liu X, Ma X, Wang Q, Yang G, Niu H, Li S, He B, He S, Dannenberg AM Jr, Zhu B (2017) A new rabbit-skin model to evaluate protective efficacy of tuberculosis vaccines. Front Microbiol 8:842
Chetty S, Ramesh M, Singh-Pillay A, Soliman ME (2017) Recent advancements in the development of anti-tuberculosis drugs. Bioorg Medicinal Chem Lett 27(3):370–386
Clark S, Hall Y, Williams A (2015) Animal models of tuberculosis: guinea pigs. Cold Spring Harb Perspect Med 5(5):a018572
Colangeli R, Helb D, Vilchèze C, Hazbón MH, Lee CG, Safi H, Sayers B, Sardone I, Jones MB, Fleischmann RD, Peterson SN (2007) Transcriptional regulation of multi-drug tolerance and antibiotic-induced responses by the histone-like protein Lsr2 in M. tuberculosis. PLoS Pathog 3(6):e87
Connell DW, Berry M, Cooke G, Kon OM (2011) Update on tuberculosis: TB in the early 21st century. Eur Respir Rev 20(120):71–84
Cooper AM (2009) Cell-mediated immune responses in tuberculosis. Annu Rev Immunol 27:393–422
D’Ambrosio L, Centis R, Sotgiu G, Pontali E, Spanevello A, Migliori GB (2015) New anti-tuberculosis drugs and regimens: 2015 update. ERJ Open Res 1(1):00010–02015
Davies J, Davies D (2010) Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74(3):417–433
Daya M, Van der Merwe L, Van Helden PD, Möller M, Hoal EG (2015) Investigating the role of gene-gene interactions in TB susceptibility. PLoS One 10(4):e0123970
De Vos M, Müller B, Borrell S, Black PA, Van Helden PD, Warren RM, Gagneux S, Victor TC (2013) Putative compensatory mutations in the rpoC gene of rifampin-resistant Mycobacterium tuberculosis are associated with ongoing transmission. Antimicrob Agents Chemother 57(2):827–832
Delogu G, Sali M, Fadda G (2013) The biology of Mycobacterium tuberculosis infection. Mediterr J Hematol Infect Dis 5(1):e2013070
Desjardins CA, Cohen KA, Munsamy V, Abeel T, Maharaj K, Walker BJ, Shea TP, Almeida DV, Manson AL, Salazar A, Padayatchi N (2016) Genomic and functional analyses of Mycobacterium tuberculosis strains implicate ald in D-cycloserine resistance. Nat Genet 48(5):544–551
Domingo-Gonzalez R, Prince O, Cooper A, Khader SA (2016) Cytokines and Chemokines in Mycobacterium tuberculosis Infection. Microbiol Spectr 4(5). https://doi.org/10.1128/microbiolspec.TBTB2-0018-2016
Dookie N, Rambaran S, Padayatchi N, Mahomed S, Naidoo K (2018) Evolution of drug resistance in Mycobacterium tuberculosis: a review on the molecular determinants of resistance and implications for personalized care. J Antimicrob Chemother 73(5):1138–1151
Dormans J, Burger M, Aguilar D, Hernandez-Pando R, Kremer K, Roholl P, Arend SM, Van Soolingen D (2004) Correlation of virulence, lung pathology, bacterial load and delayed type hypersensitivity responses after infection with different Mycobacterium tuberculosis genotypes in a BALB/c mouse model. Clin Exp Immunol 137(3):460–468
Drain PK, Bajema KL, Dowdy D, Dheda K, Naidoo K, Schumacher SG, Ma S, Meermeier E, Lewinsohn DM, Sherman DR (2018) Incipient and subclinical tuberculosis: a clinical review of early stages and progression of infection. Clin Microbiol Rev 31(4):e00021–e00018
Dutta NK, Karakousis PC (2014) Latent tuberculosis infection: myths, models, and molecular mechanisms. Microbiol Mol Biol Rev 78(3):343–371
Ehlers S, Schaible UE (2013) The granuloma in tuberculosis: dynamics of a host–pathogen collusion. Front Immunol 3:411
Fair RJ, Tor Y (2014) Antibiotics and Bacterial Resistance in the 21st Century. Perspect Medicin Chem 6:25–64
Falzon D, Schünemann HJ, Harausz E, González-Angulo L, Lienhardt C, Jaramillo E, Weyer K (2017) World Health Organization treatment guidelines for drug-resistant tuberculosis, 2016 update. Eur Respiratory Soc 49(3):1602308
Feltcher ME, Sullivan JT, Braunstein M (2010) Protein export systems of Mycobacterium tuberculosis: novel targets for drug development? Future Microbiol 5(10):1581–1597
Fonseca JD, Knight GM, McHugh TD (2015) The complex evolution of antibiotic resistance in Mycobacterium tuberculosis. Int J Infect Dis 32:94–100
Ford CB, Shah RR, Maeda MK, Gagneux S, Murray MB, Cohen T, Johnston JC, Gardy J, Lipsitch M, Fortune SM (2013) Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nature Genet 45(7):784–790
Forrellad MA, Klepp LI, Gioffré A, Sabio y Garcia J, Morbidoni HR, Santangelo MD, Cataldi AA, Bigi F (2013) Virulence factors of the Mycobacterium tuberculosis complex. Virulence 4(1):3–66
Fu LM, Shinnick TM (2007) Genome-wide exploration of the drug action of capreomycin on Mycobacterium tuberculosis using Affymetrix oligonucleotide GeneChips. J Infect 54(3):277–284
Fujiwara M, Kawasaki M, Hariguchi N, Liu Y, Matsumoto M (2018) Mechanisms of resistance to delamanid, a drug for Mycobacterium tuberculosis. Tuberculosis 108:186–194
Garneau-Tsodikova S, Labby KJ (2016) Mechanisms of resistance to aminoglycoside antibiotics: overview and perspectives. Med Chem Comm 7(1):11–27
Gillespie SH (2002) Evolution of drug resistance in Mycobacterium tuberculosis: clinical and molecular perspective. Antimicrob Agents Chemother 46(2):267–274
Ginsburg AS, Grosset JH, Bishai WR (2003) Fluoroquinolones, tuberculosis, and resistance. Lancet Infect Dis 3(7):432–442
Glaziou P, Sismanidis C, Floyd K, Raviglione M (2015) Global epidemiology of tuberculosis. Cold Spring HarbPerspect Med 5(2):a017798
Glickman MS, Jacobs WR (2001) Microbial pathogenesis of Mycobacterium tuberculosis: dawn of a discipline. Cell 104(4):477–485
Gualano G, Capone S, Matteelli A, Palmieri F (2016) New Antituberculosis Drugs: From Clinical Trial to Programmatic Use. Infect Dis Rep 8(2):6569
Gygli SM, Borrell S, Trauner A, Gagneux S (2017) Antimicrobial resistance in Mycobacterium tuberculosis: mechanistic and evolutionary perspectives. FEMS Microbiol Rev 41(3):354–373
Hameed HM, Islam MM, Chhotaray C, Wang C, Liu Y, Tan Y, Li X, Tan S, Delorme V, Yew WW, Liu J (2018) Molecular targets related drug resistance mechanisms in MDR-, XDR-, and TDR-Mycobacterium tuberculosis strains. Front Cell Infect Microbiol 8:114
Hawn TR, Day TA, Scriba TJ, Hatherill M, Hanekom WA, Evans TG, Churchyard GJ, Kublin JG, Bekker LG, Self SG (2014) Tuberculosis vaccines and prevention of infection. Microbiol Mol Biol Rev 78(4):650–671
Hegde SS, Vetting MW, Roderick SL, Mitchenall LA, Maxwell A, Takiff HE, Blanchard JS (2005) A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science 308(5727):1480–1483
Hoagland DT, Liu J, Lee RB, Lee RE (2016) New agents for the treatment of drug-resistant Mycobacterium tuberculosis. Adv Drug Deliv Rev 102:55–72
Hoff DR, Ryan GJ, Driver ER, Ssemakulu CC, De Groote MA, Basaraba RJ, Lenaerts AJ (2011) Location of intra-and extracellular M. tuberculosis populations in lungs of mice and guinea pigs during disease progression and after drug treatment. PLoS One 6(3):e17550
Iketleng T, Lessells R, Dlamini MT, Mogashoa T, Mupfumi L, Moyo S, Gaseitsiwe S (2018) Mycobacterium tuberculosis Next-generation whole genome sequencing: opportunities and challenges. Tuberc Res Treat 2018:1298542
Ilina EN, Shitikov EA, Ikryannikova LN, Alekseev DG, Kamashev DE, Malakhova MV, Parfenova TV, Afanas’ev MV, Ischenko DS, Bazaleev NA, Smirnova TG (2013) Comparative genomic analysis of Mycobacterium tuberculosis drug resistant strains from Russia. PLoS One 8(2):e56577
Jagielski T, Minias A, Van Ingen J, Rastogi N, Brzostek A, Żaczek A, Dziadek J (2016) Methodological and clinical aspects of the molecular epidemiology of Mycobacterium tuberculosis and other mycobacteria. Clin Microbiol Rev 29(2):239–290
Jassal MS, Bishai WR (2010) Epidemiology and challenges to the elimination of global tuberculosis. Clin Infect Dis 50:S156–S164
Jugheli L, Bzekalava N, de Rijk P, Fissette K, Portaels F, Rigouts L (2009) High level of cross-resistance between kanamycin, amikacin, and capreomycin among Mycobacterium tuberculosis isolates from Georgia and a close relation with mutations in the rrs gene. Antimicrob Agents Chemother 53(12):5064–5068
Kaushal D, Schroeder BG, Tyagi S, Yoshimatsu T, Scott C, Ko C, Carpenter L, Mehrotra J, Manabe YC, Fleischmann RD, Bishai WR (2002) Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative σ factor, SigH. Proc Natl Acad Sci U S A 99(12):8330–8335
Kim KH, An DR, Song J, Yoon JY, Kim HS, Yoon HJ, Im HN, Kim J, Lee SJ, Kim KH, Lee HM (2012) Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7. Proc Natl Acad Sci U S A 109(20):7729–7734
Koch A, Cox H, Mizrahi V (2018) Drug-resistant tuberculosis: challenges and opportunities for diagnosis and treatment. Curr Opin Pharmacol 42:7–15
Kohanski MA, Dwyer DJ, Collins JJ (2010) How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8(6):423–435
Kramnik I, Beamer G (2016) Mouse models of human TB pathology: roles in the analysis of necrosis and the development of host-directed therapies. Semin Immuno Pathol 38(2):221–237
Kumar S, Jena L (2014) Understanding rifampicin resistance in tuberculosis through a computational approach. Genomics Inform 12(4):276–282
Kurz SG, Bonomo RA (2012) Reappraising the use of β-lactams to treat tuberculosis. Expert Rev Anti-Infect Ther 10(9):999–1006
Lahiri N, Shah RR, Layre E, Young D, Ford C, Murray MB, Fortune SM, Moody DB (2016) Rifampin resistance mutations are associated with broad chemical remodeling of Mycobacterium tuberculosis. J Biol Chem 291(27):14248–14256
Lange C, Abubakar I, Alffenaar JW, Bothamley G, Caminero JA, Carvalho AC, Chang KC, Codecasa L, Correia A, Crudu V, Davies P (2014) Management of patients with multidrug-resistant/extensively drug-resistant tuberculosis in Europe: a TBNET consensus statement. The Eur Respir J 44(1):23–63
Lee JY (2015) Diagnosis and treatment of extrapulmonary tuberculosis. Tuberc Respir Dis 78(2):47–55
Lee SH (2016) Tuberculosis infection and latent tuberculosis. Tuberc Respir Dis 79(4):201–206
Li F, Wan L, Xiao T, Liu H, Jiang Y, Zhao X, Wang R, Wan K (2018) In vitro activity of β-lactams in combination with β-lactamase inhibitors against Mycobacterium tuberculosis clinical isolates. Biomed Res Int 2018:3579832
Lingaraju S, Rigouts L, Gupta A, Lee J, Umubyeyi AN, Davidow AL, German S, Cho E, Cho SN, Kim CT, Alland D (2016) Geographic differences in the contribution of ubiA mutations to high-level ethambutol resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 60(7):4101–4105
Liu L, Jiang F, Chen L, Zhao B, Dong J, Sun L, Zhu Y, Liu B, Zhou Y, Yang J, Zhao Y (2018a) The impact of combined gene mutations in inhA and ahpC genes on high levels of isoniazid resistance amongst katG non-315 in multidrug-resistant tuberculosis isolates from China. Emerg Microbes Infect 7(1):1–10
Liu Q, Zuo T, Xu P, Jiang Q, Wu J, Gan M, Yang C, Prakash R, Zhu G, Takiff HE, Gao Q (2018b) Have compensatory mutations facilitated the current epidemic of multidrug-resistant tuberculosis? Emerg Microbes Infect 7(1):1–8
Lu J, Liu M, Wang Y, Pang Y, Zhao Z (2014) Mechanisms of fluoroquinolone monoresistance in Mycobacterium tuberculosis. FEMS Microbiol Lett 353(1):40–48
Ma Z, Lienhardt C, McIlleron H, Nunn AJ, Wang X (2010) Global tuberculosis drug development pipeline: the need and the reality. Lancet 375(9731):2100–2109
Majorov KB, Lyadova IV, Kondratieva TK, Eruslanov EB, Rubakova EI, Orlova MO, Mischenko VV, Apt AS (2003) Different innate ability of I/St and A/Sn mice to combat virulent Mycobacterium tuberculosis: phenotypes expressed in lung and extrapulmonary macrophages. Infect Immun 71(2):697–707
Manca C, Tsenova L, Barry CE, Bergtold A, Freeman S, Haslett PA, Musser JM, Freedman VH, Kaplan G (1999) Mycobacterium tuberculosis CDC1551 induces a more vigorous host response in vivo and in vitro, but is not more virulent than other clinical isolates. J Immunol 162(11):6740–6746
Manca C, Tsenova L, Bergtold A, Freeman S, Tovey M, Musser JM, Barry CE, Freedman VH, Kaplan G (2001) Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-α/β. Proc Natl Acad Sci U S A 98(10):5752–5757
Maruri F, Sterling TR, Kaiga AW, Blackman A, van der Heijden YF, Mayer C, Cambau E, Aubry A (2012) A systematic review of gyrase mutations associated with fluoroquinolone-resistant Mycobacterium tuberculosis and a proposed gyrase numbering system. J Antimicrob Chemother 67(4):819–831
Maus CE, Plikaytis BB, Shinnick TM (2005) Mutation of tlyA confers capreomycin resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 49(2):571–577
McGrath M, Gey van Pittius NC, Van Helden PD, Warren RM, Warner DF (2013) Mutation rate and the emergence of drug resistance in Mycobacterium tuberculosis. J Antimicrob Chemother 69(2):292–302
Mehta PK, Pandey AK, Subbian S, El-Etr SH, Cirillo SL, Samrakandi MM, Cirillo JD (2006) Identification of Mycobacterium marinum macrophage infection mutants. Microb Pathog 40(4):139–151
Meijer AH (2016) Protection and pathology in TB: learning from the zebrafish model. Semin Immuno pathol 38(2):261–273
Motiwala AS, Dai Y, Jones-López EC, Hwang SH, Lee JS, Cho SN, Via LE, Barry CE III, Alland D (2010) Mutations in extensively drug-resistant Mycobacterium tuberculosis that do not code for known drug-resistance mechanisms. J Infect Dis 201(6):881–888
Mukhopadhyay S, Nair S, Ghosh S (2012) Pathogenesis in tuberculosis: transcriptomic approaches to unraveling virulence mechanisms and finding new drug targets. FEMS Microbiol Rev 36(2):463–485
Müller B, Borrell S, Rose G, Gagneux S (2013) The heterogeneous evolution of multidrug-resistant Mycobacterium tuberculosis. Trends Genet 29(3):160–169
Murray JF, Schraufnagel DE, Hopewell PC (2015) Treatment of tuberculosis. A historical perspective. Ann Am Thorac Soc 12(12):1749–1759
Nahid P, Kim PS, Evans CA, Alland D, Barer M, Diefenbach J, Ellner J, Hafner R, Hamilton CD, Iademarco MF, Ireton G (2012) Clinical Research and Development of Tuberculosis Diagnostics: Moving From Silos to Synergy. J Infect Dis 205:S159–S168
Nampoothiri KM, Rubex R, Patel AK, Narayanan SS, Krishna S, Das SM, Pandey A (2008) Molecular cloning, overexpression and biochemical characterization of hypothetical β-lactamases of Mycobacterium tuberculosis H37Rv. J Appl Microbiol 105(1):59–67
Nasiri MJ, Imani Fooladi AA, Dabiri H, Pormohammad A, Salimi Chirani A, Dadashi M, Houri H, Heidary M, Feizabadi MM (2016) Primary ethambutol resistance among Iranian pulmonary tuberculosis patients: a systematic review. Ther Adv Infect Dis 3(5):133–138
Ngo TM, Teo YY (2019) Genomic prediction of tuberculosis drug-resistance: benchmarking existing databases and prediction algorithms. BMC Bioinformatics 20(1):68
Nguta JM, Appiah-Opong R, Nyarko AK, Yeboah-Manu D, Addo PG (2015) Current perspectives in drug discovery against tuberculosis from natural products. Int J Mycobacteriol 4(3):165–183
Nguyen L (2016) Antibiotic resistance mechanisms in M. tuberculosis: an update. Arch Toxicol 90(7):1585–1604
Niederweis M, Danilchanka O, Huff J, Hoffmann C, Engelhardt H (2010) Mycobacterial outer membranes: in search of proteins. Trends Microbiol 18(3):109–116
Norris BA, Ernst JD (2018) Mononuclear cell dynamics in M. tuberculosis infection provide opportunities for therapeutic intervention. PLoS Pathog 14(10):e1007154
O’Neill MB, Mortimer TD, Pepperell CS (2012) Diversity of Mycobacterium tuberculosis across evolutionary scales. PLoS Pathog 11(11):e1005257
Ocheretina O, Escuyer VE, Mabou MM, Royal-Mardi G, Collins S, Vilbrun SC, Pape JW, Fitzgerald DW (2014) Correlation between genotypic and phenotypic testing for resistance to rifampin in Mycobacterium tuberculosis clinical isolates in Haiti: investigation of cases with discrepant susceptibility results. PLoS One 9(3):e90569
Palomino J, Martin A (2014) Drug resistance mechanisms in Mycobacterium tuberculosis. Antibiotics 3(3):317–340
Parida SK, Axelsson-Robertson R, Rao MV, Singh N, Master I, Lutckii A, Keshavjee S, Andersson J, Zumla A, Maeurer M (2015) Totally drug-resistant tuberculosis and adjunct therapies. J Intern Med 277(4):388–405
Pieters J (2008) Mycobacterium tuberculosis and the macrophage: maintaining a balance. Cell Host Microbe 3(6):399–407
Plinke C, Walter K, Aly S, Ehlers S, Niemann S (2011) Mycobacterium tuberculosis embB codon 306 mutations confer moderately increased resistance to ethambutol in vitro and in vivo. Antimicrob Agents Chemother 55(6):2891–2896
Prasad R, Singh A, Balasubramanian V, Gupta N (2017) Extensively drug-resistant tuberculosis in India: Current evidence on diagnosis & management. Indian J Med Res 145(3):271–293
Prozorov AA, Fedorova IA, Bekker OB, Danilenko VN (2014) The virulence factors of Mycobacterium tuberculosis: genetic control, new conceptions. Russ J Genet 50(8):775–797
Pym AS, Saint-Joanis B, Cole ST (2002) Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect Immun 70(9):4955–4960
Ramirez-Busby SM, Valafar F (2015) Systematic review of mutations in pyrazinamidase associated with pyrazinamide resistance in Mycobacterium tuberculosis clinical isolates. Antimicrob Agents Chemother 59(9):5267–5277
Rawal T, Butani S (2016) Combating tuberculosis infection: a forbidding challenge. Indian J Pharm Sci 78(1):8–16
Rengarajan J, Sassetti CM, Naroditskaya V, Sloutsky A, Bloom BR, Rubin EJ (2004) The folate pathway is a target for resistance to the drug para-aminosalicylic acid (PAS) in mycobacteria. Mol Microbiol 53(1):275–282
Ribeiro SC, Gomes LL, Amaral EP, Andrade MR, Almeida FM, Rezende AL, Lanes VR, Carvalho EC, Suffys PN, Mokrousov I, Lasunskaia EB (2014) Mycobacterium tuberculosis strains of the modern sublineage of the Beijing family are more likely to display increased virulence than strains of the ancient sublineage. J Clin Microbiol 52(7):2615–2624
Rock RB, Olin M, Baker CA, Molitor TW, Peterson PK (2008) Central nervous system tuberculosis: pathogenesis and clinical aspects. Clin Microbiol Rev 21(2):243–261
Safi H, Lingaraju S, Amin A, Kim S, Jones M, Holmes M, McNeil M, Peterson SN, Chatterjee D, Fleischmann R, Alland D (2013) Evolution of high-level ethambutol-resistant tuberculosis through interacting mutations in decaprenylphosphoryl-β-D-arabinose biosynthetic and utilization pathway genes. Nat Genet 45(10):1190–1197
Sakamoto K (2012) The pathology of Mycobacterium tuberculosis infection. Vet Pathol 49(3):423–439
Sarathy J, Dartois V, Lee E (2012) The role of transport mechanisms in Mycobacterium tuberculosis drug resistance and tolerance. Pharmaceuticals 5(11):1210–1235
Sasindran SJ, Torrelles JB (2011) Mycobacterium tuberculosis Infection and Inflammation: what is Beneficial for the Host and for the Bacterium? Front Microbiol 2:2
Sauvage E, Fonzé E, Quinting B, Galleni M, Frere JM, Charlier P (2006) Crystal structure of the Mycobacterium fortuitum class A β-lactamase: structural basis for broad substrate specificity. Antimicrob Agents Chemother 50(7):2516–2521
Scanga CA, Flynn JL (2014) Modeling tuberculosis in nonhuman primates. Cold Spring HarbPerspect Med 4(12):a018564
Schluger NW (2013) Fluoroquinolones in the Treatment of Tuberculosis: Which Is Best? Am J Respir Crit Care Med 188(7):768–769
Schulz zurWiesch P, Engelstädter J, Bonhoeffer S (2010) Compensation of fitness costs and reversibility of antibiotic resistance mutations. Antimicrob Agents Chemother 54(5):2085–2095
Shaler CR, Kugathasan K, McCormick S, Damjanovic D, Horvath C, Small CL, Jeyanathan M, Chen X, Yang PC, Xing Z (2011) Pulmonary mycobacterial granuloma: increased IL-10 production contributes to establishing a symbiotic host–microbe microenvironment. Am J Pathol 178(4):1622–1634
Singh AK, Gupta UD (2018) Animal models of tuberculosis: lesson learnt. Indian J Med Res 147(5):456–463
Smith I (2003) Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin Microbiol Rev 16(3):463–496
Smith T, Wolff KA, Nguyen L (2012) Molecular biology of drug resistance in Mycobacterium tuberculosis. In: Pieters J, McKinney J (eds) Pathogenesis of Mycobacterium tuberculosis and its Interaction with the Host Organism. Springer, Berlin, pp 53–80
Somoskovi A, Parsons LM, Salfinger M (2001) The molecular basis of resistance to isoniazid, rifampin, and pyrazinamide in Mycobacterium tuberculosis. Respir Res 2(3):164
Soto SM (2013) Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 4(3):223–229
Spies FS, Ribeiro AW, Ramos DF, Ribeiro MO, Martin A, Palomino JC, Rossetti ML, da Silva PE, Zaha A (2011) Streptomycin resistance and lineage-specific polymorphisms in Mycobacterium tuberculosis gidB gene. J Clin Microbiol 49(7):2625–2630
Torres JN, Paul LV, Rodwell TC, Victor TC, Amallraja AM, Elghraoui A, Goodmanson AP, Ramirez-Busby SM, Chawla A, Zadorozhny V, Streicher EM (2015) Novel katG mutations causing isoniazid resistance in clinical M. tuberculosis isolates. Emerg Microbes Infect 4(1):1–9
Trauner A, Borrell S, Reither K, Gagneux S (2014) Evolution of drug resistance in tuberculosis: recent progress and implications for diagnosis and therapy. Drugs 74(10):1063–1072
Tsenova L, Ellison E, Harbacheuski R, Moreira AL, Kurepina N, Reed MB, Mathema B, Barry CE III, Kaplan G (2005) Virulence of selected Mycobacterium tuberculosis clinical isolates in the rabbit model of meningitis is dependent on phenolic glycolipid produced by the bacilli. J Infect Dis 192(1):98–106
Van Deun A, Aung KJ, Bola V, Lebeke R, Hossain MA, de Rijk WB, Rigouts L, Gumusboga A, Torrea G, de Jong BC (2013) Rifampin drug resistance tests for tuberculosis: challenging the gold standard. J Clin Microbiol 51(8):2633–2640
Van Leeuwen LM, Van der Sar AM, Bitter W (2015) Animal models of tuberculosis: zebrafish. Cold Spring HarbPerspect Med 5(3):a018580
Von Groll A, Martin A, Jureen P, Hoffner S, Vandamme P, Portaels F, Palomino JC, da Silva PA (2009) Fluoroquinolone resistance in Mycobacterium tuberculosis and mutations in gyrA and gyrB. Antimicrob Agents Chemother 53(10):4498–4500
Wang F, Cassidy C, Sacchettini JC (2006) Crystal structure and activity studies of the Mycobacterium tuberculosis β-lactamase reveal its critical role in resistance to β-lactam antibiotics. Antimicrob Agents Chemother 50(8):2762–2771
Waters WR, Palmer MV, Thacker TC, Davis WC, Sreevatsan S, Coussens P, Meade KG, Hope JC, Estes DM (2011) Tuberculosis immunity: opportunities from studies with cattle. Clin Dev Immunol 2011:768542
Wong A (2017) Epistasis and the evolution of antimicrobial resistance. Front Microbiol 8:246
Xie J, Talaska AE, Schacht J (2011) New developments in aminoglycoside therapy and ototoxicity. Hear Res 281(1–2):28–37
Yakrus MA, Driscoll J, Lentz AJ, Sikes D, Hartline D, Metchock B, Starks AM (2014) Concordance between molecular and phenotypic testing of Mycobacterium tuberculosis complex isolates for resistance to rifampin and isoniazid in the United States. J Clin Microbiol 52(6):1932–1937
Zhai W, Wu F, Zhang Y, Fu Y, Liu Z (2019) The Immune Escape Mechanisms of Mycobacterium tuberculosis. Int J Mol Sci 20(2):340
Zhan L, Tang J, Sun M, Qin C (2017) Animal models for tuberculosis in translational and precision medicine. Front Microbiol 8:717
Zhang S, Chen J, Cui P, Shi W, Zhang W, Zhang Y (2015) Identification of novel mutations associated with clofazimine resistance in Mycobacterium tuberculosis. J Antimicrob Chemother 70(9):2507–2510
Zhang S, Chen J, Cui P, Shi W, Shi X, Niu H, Chan D, Yew WW, Zhang W, Zhang Y (2016) Mycobacterium tuberculosis mutations associated with reduced susceptibility to linezolid. Antimicrob Agents Chemother 60(4):2542–2544
Zhang S, Chen J, Shi W, Cui P, Zhang J, Cho S, Zhang W, Zhang Y (2017) Mutation in clpC1 encoding an ATP-dependent ATPase involved in protein degradation is associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerg Microbes Infect 6(1):1–2
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Kumar, V. et al. (2020). Current Trends in Mycobacterium tuberculosis Pathogenesis and Drug Resistance. In: Siddhardha, B., Dyavaiah, M., Syed, A. (eds) Model Organisms for Microbial Pathogenesis, Biofilm Formation and Antimicrobial Drug Discovery. Springer, Singapore. https://doi.org/10.1007/978-981-15-1695-5_16
Download citation
DOI: https://doi.org/10.1007/978-981-15-1695-5_16
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-1694-8
Online ISBN: 978-981-15-1695-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)