Abstract
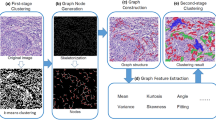
In order to apply the important topological information to solve a Cervical Histopathology Image Clustering (CHIC) problem, a Graph Based Unsupervised Learning (GBUL) approach is proposed in this paper. First, the GBUL method applies color features and k-means clustering for a first-stage “coarse” clustering. Then, a Skeletonization Based Node Generation (SBNG) approach is introduced to approximate the distribution of cervical cell nuclei. Thirdly, based on the SBNG nodes, a minimum spanning tree graph is constructed. Next, graph features and additional geometrical features are extracted based on the constructed graph. Finally, the k-means clustering is applied again for the second-stage clustering. In the experiment, a practical cervical histopathology image dataset with ten whole scanned images is tested, obtaining a promising CHIC result and showing a huge potential in the cancer risk prediction field.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Alpaydin, E.: Introduction to Machine Learning (2009)
Banu, P.N., Azar, A.T., Inbarani, H.H.: Fuzzy firefly clustering for tumour and cancer analysis. Int. J. Model. Ident. Control 27(2), 92–103 (2017)
Chaudhuriand, B., Rodenacker, K., Burger, G.: Characterization and featuring of histological section images. Patt. Recogn. Lett. 7(4), 245–252 (1988)
Cruz-Roa, A., Xu, J., Madabhushi, A.: A note on the stability and discriminability of graph-based features for classification problems in digital pathology. In: Proceedings of SPIE 9287, p. 928703 (2015)
Gonzalez, R., Woods, E., Eddins, S., et al.: Digital Image Processing Using MATLAB, vol. 624 (2004)
Miranda, G., Barrera, J., Soares, E., et al.: Structural analysis of histological images to aid diagnosis of cervical cancer. Proc. SIBGRAPI 2012, 316–323 (2012)
Otali, D., Fredenburgh, J., Oelschlager, D., et al.: A standard tissue as a control for histochemical and immunohistochemical staining. Biotech. Histochem. 91(5), 309–326 (2016)
Peng, Y., Park, M., Xu, M., et al.: Clustering nuclei using machine learning techniques. In: Proceedings of IEEE/ICME International Conference, pp. 52–57 (2010)
Ramos-Vara, J.: Principles and methods of immunohistochemistry. In: Gautier, J. (ed.) Drug Safety Evaluation, pp. 83–96 (2011)
Siegel, R., Miller, K., Jemal, A.: Cancer statistics. CA: A Cancer J. Clin. 67(1), 7–30 (2017)
Sornapudi, S.: Nuclei Segmentation of Histology Images Based on Deep Learning and Color Quantization and Analysis of Real World Pill Images (2017)
Sornapudi, S., Stanley, R., Stoecker, W., et al.: Deep learning nuclei detection in digitized histology images by superpixels. J. Pathol. Inf. 9 (2018)
Sudbø, J., Marcelpoil, R., Reith, A.: Caveats: numerical requirements in graph theory based quantitation of tissue architecture. Anal. Cell. Pathol. 21(2), 59–69 (2000)
Sukumarand, P., Gnanamurthy, R.: Computer aided detection of cervical cancer using pap smear images based on adaptive neuro fuzzy inference system classifier. J. Med. Imaging Health Inf. 6(2), 312–319 (2016)
Theodoridis, S., Pikrakis, A., Koutroumbas, K., et al.: Introduction to Pattern Recognition: A Matlab Approach. America (2010)
Torre, L., Bray, F., Siegel, R., et al.: Global cancer statistics. CA: A Cancer J. Clin. 65(2), 87–108 (2015)
Wang, X., Li, S., Li, J., Wang, J.: An adaptive and selective segmentation model based on local and global image information. Int. J. Model. Ident. Control 28(2), 114–124 (2017)
Wuand, Z., Leahy, R.: An optimal graph theoretic approach to data clustering: theory and its spplication to image segmentation. IEEE Trans. Patt. Anal. Mach. Intell. 15(11), 1101–1113 (1993)
Xiao, Y., Cao, Y., Yu, W., Tian, J.: Multi-level threshold selection based on artificial bee colony algorithm and maximum entropy for image segmentation. Int. J. Comput. Appl. Technol. 43(4), 343–350 (2012)
Acknowledgements
We thank the funds supported by the “National Natural Science Foundation of China” (No. 61806047), the “Fundamental Research Funds for the Central Universities” (No. N171903004), and the “Scientific Research Launched Fund of Liaoning Shihua University” (No. 2017XJJ-061). We also thank Zhijie Hu, due to his contribution is considered as important as the first author in this paper.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Li, C. et al. (2020). Cervical Histopathology Image Clustering Using Graph Based Unsupervised Learning. In: Wang, R., Chen, Z., Zhang, W., Zhu, Q. (eds) Proceedings of the 11th International Conference on Modelling, Identification and Control (ICMIC2019). Lecture Notes in Electrical Engineering, vol 582. Springer, Singapore. https://doi.org/10.1007/978-981-15-0474-7_14
Download citation
DOI: https://doi.org/10.1007/978-981-15-0474-7_14
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-0473-0
Online ISBN: 978-981-15-0474-7
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)