Abstract
This paper proposes a graph based unsupervised learning (GBUL) clustering approach that uses features of graphs to identify different tissue structures in images, aiding histopathologists to quickly identify lesion areas and improve the accuracy of diagnosis. The first-stage rough clustering was performed by applying color features and k-means based on graph theory in Hematoxylin-eosin (H&E) stained cervical histopathology images. By applying a skeletonization based node generation (SBNG) approach, the generated nodes are approximated as a distribution of cervical cell nuclei, a minimum spanning tree (MST) is constructed from the generated nodes and their geometric features are extracted, followed by clustering by applying the k-means algorithm again. The important topological information hidden in histopathology images is applied to solve the cervical histopathology image clustering (CHIC) problem, and the function of mouse manual annotation to assist histopathologists is provided. CHIC based on MST can more accurately distinguish the tissue structure of high, medium and low density adherent cells. Combined with suspicious lesion points added manually by the physician, it can semi-automatically predict the cancer risk of tissues, rapidly identify lesion areas and improve the accuracy and efficiency of diagnosis.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Cervical cancer is one of the most common malignant tumors of the female reproductive tract, ranking second in incidence among female malignant tumors, occurring mainly in developing countries, with a trend of younger incidence in recent years [1,2,3]. With the increasing incidence of cancer and the increasing workload of histopathology departments, the disadvantages of the traditional histopathological diagnosis mode, which is subjective and cannot be accurately quantified in the era of “precision medicine”, have become increasingly apparent. How to apply Artificial Intelligence (AI) in cervical histopathology images to make rapid and accurate diagnosis is an urgent problem [4].
Squamous cell carcinoma is the most common histopathological tissue type of cervical cancer [5]. When the squamous epithelium becomes cancerous, the cell dysplasia and the nuclear-cytoplasmic ratio increase, and the distribution and topology in the tissue will also change [6]. Therefore, this study contributes a GBUL clustering method by studying the topology of cervical squamous cell carcinoma, which can rapidly identify lesion regions and thus assist in the histopathological diagnosis of cervical cancer [7].
2 Materials and Methods
2.1 Histopathology Image Source
The H&E stained cervical histopathology images of 20 cases used in this experiment were provided by the Department of Pathology, Liaoning Cancer Hospital, and each slice was imaged by an Olympus digital scanning microscope.
2.2 Research Method
Data Settings.
The acquired images were 20 H&E stained cervical histopathology images with a magnification of 40 times, the size of each image was between 200 M and 2024 M, and the number of pixels was between 30000 × 30000 and 60000 × 60000. Each scanned image was cropped into multiple sub-images, with sub-image sizes between 1 M and 2 M and pixel values of 976 × 881. Ten more representative images among the multiple sub-images were selected by the histopathologists as the main study subjects, and the clustering results were examined and evaluated.
Lab Environment.
The programming software used in this paper is MATLAB, and the operating system is Windows10.
Main Work.
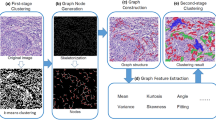
Firstly, the color features of the histopathology image are extracted, and the image is processed for the first time by k-means, and then the node is generated by the method of SBNG, which is approximated to the distribution of cervical cell nuclei. Based on the generated nodes, a MST is constructed and geometric features are extracted, the k-means algorithm is applied again for clustering, and the function of manual correction is provided to the histopathologist. The specific research content is shown in Fig. 1.
Schematic diagram of the research process. (a) Original cervical histopathology images. (b) Extracting color features and using k-means to make the first clustering result. (c) is the result of skeletonization based on (b). (d) Nodes are generated on the basis of skeletonization, and the distribution of nuclei can be represented by the generated skeleton nodes. (e) Using the MST image constructed from the generated nodes, the adjacent relationship and spatial arrangement between the nodes is extracted using the MST. (f), (g), (h), (i) Calculate statistical values as features of the graph, describing the topological spatial structure of the image. (j), (k) utilize graph features, again using k-means clustering. (l), (m) Manually annotate new regions where nuclei form and group them into known clusters.
2.3 k-Means Algorithm
The k-means algorithm is a clustering algorithm that divides different clusters according to the similarity of different data objects. It initializes k different clusters, calculates the distance between the remaining points and the initially selected points, and assigns each sample to the cluster with the closest distance, and updates the cluster center until it is stable to generate k categories [8]. Rahmadwati et al. used k-means clustering and color segmentation algorithm to classify cervical cancer cells [9].
2.4 Graph Theory
Graph theory takes graphs as the research object to study the properties of graphs. The algorithm of graph theory provides a simple and systematic model, which can remove specific edges and divide the graph into several subgraphs for image segmentation, which plays an important role in the computer field [10]. Normalized Cut, MST, and Dominate Set based method are commonly used graph theory methods [11,12,13].
2.5 CHIC Method Based on MST
Color Histogram Feature Extraction.
Color histogram is to extract features by counting the proportion of each color of the selected whole image in the image [14].
Preprocessing.
Image preprocessing using k-means image segmentation method. When k-means performs image segmentation, each pixel in the image in the RGB color space is treated as a three-dimensional vector. In k-means clustering, both the number of clusters k and the maximum number of iterations N have a certain influence on the image segmentation results. In this paper, the number of clusters is k = 2, the maximum number of iterations is 50, and the pixels of different clusters are represented by different colors. The results are shown in Fig. 2.
The image segmentation result with cluster number k = 2 is selected and transformed into a binary image with simple morphological processing for a series of operations such as subsequent skeletonization extraction. The segmentation effect of the binary image after morphological processing is more obvious, filling the tiny holes that appear in the cells during the segmentation. The effect is shown in Fig. 3.
MST Construction.
Connected Area Filter. According to the density of epithelial cells, it is artificially divided into three categories: high density, medium density, and low density, and grouped according to the area. Let L, M, and N all be matrices that mark four connected regions on the processed binary image. Let the number of pixels in the marked area be S(i). (1) Eliminate the connected regions of S(i) > 500 in L as low density regions. (2) Eliminate the connected regions of S(i) < 500|| S(i) > 1700 in M as medium density regions. (3) Eliminate the connected regions of S(i) < 1700 in N as high density regions.
Skeletonization.
The binary image is skeletonized, and the refinement operation is performed on the basis that the skeleton structure is not broken, as shown in Fig. 4.
Skeletonization of the three connected region matrices mentioned earlier. As shown in Fig. 5. The cytoskeleton extracted by the L, M, N connected region matrix in turn:
Node Generation.
The extracted skeletonized image is attached to the histopathology binary image. From Fig. 6, it can be observed that its bifurcation point is similar to the position of the nucleus, which can be used to replace the position of the nucleus and extracted.
In the extracted skeleton, look for a point in its eight neighborhoods that is also in the skeleton. When there are more than three points in its eight neighborhoods that are also in the skeleton, this point must be a bifurcation point, which is node. Figure 7 is the result generated by the node, and the red mark is the location of the node.
In this study, after finding the node, the node position was calculated, some redundant nodes were deleted, the interference of adjacent bifurcation points was excluded, and the one-to-one correspondence between a node and a nucleus was ensured.
Build a MST.
Since most of the cells in the low density area were screened for a small amount of cell adhesion, the MST was constructed only for the large areas in the medium density and high density areas. The black line is the MST constructed in the medium density area. The colored lines are the MST constructed in the high density area, and Fig. 8 is the visualization result of the MST construction.
Graph Feature Extraction.
According to the graph generated by the MST, various statistical values are calculated, and these statistical values are used as shape and geometric features to describe different tissues. Shape features extracted in this study include mean, variance, skewness, and kurtosis of edge lengths, as well as the angle of each figure. Geometric features include the perimeter of the tissue, and the independent arrangement of nodes within each tissue.
k-means Clustering.
Cells in the low density region were extracted in the first clustering. Subsequent processing of medium and high density regions again uses k-means clustering. According to the graphic features and set features extracted by the MST, k = 2 is selected for the number of clusters in this study, and the high density and medium density are clustered respectively. The results are shown in Fig. 9.
2.6 Manual Annotation and Results Evaluation
This study provides physicians with the ability to manually label cell nuclei to mark dense areas or suspicious lesions. The yellow area shown in Fig. 10 is the result displayed by the manually marked area. In this study, the mean, variance, skewness and kurtosis were calculated from the constructed MST, presented in a visual statistical plot and compared across categories to evaluate the clustering results. In addition, silhouette plots were used in the study to further evaluate the effectiveness of the clustering, with a high mean value indicating a better overall performance.
3 Result
3.1 Research Results and Analysis
The visualization of the statistical values calculated from the constructed MST is shown in Fig. 11 and Fig. 12. The histopathology images of cervical cancer show more distinct information on the organization of the nuclei in the high density areas, which are less numerous than the medium density nuclei.
As can be seen in Fig. 11 and Fig. 12, the mean of lengths and angles of high density tissue are relatively stable and not significantly different, but there are significant differences in variance, skewness and kurtosis. The average value of the length and angle of the medium density region shows a certain fluctuation, among which the variance, skewness and kurtosis are more obvious. Therefore, high density areas and medium density areas are divided into two or three categories using k-means.
The organizational structures of the clustering results were extracted separately, as shown in Fig. 13, (a) is the clustering result of the cell structure in the medium density area, (b) is the clustering result of the cell structure in the high density area.
In the final image display, the clustering results of the high density area (red), medium density area (blue) and low density area (green) are marked with different colors and shades, and the darker the area is, the higher the density of the area represents the topology of its organization The more complex the structure is, as shown in Fig. 14(a).
In the visualization image of the clustering results, there are still a small number of highly suspicious areas that have not been identified. When viewing the histopathology image, doctors can manually mark the nuclei through the panel, as shown in Fig. 14 (b). The manually annotated regions are then subjected to graph structure feature extraction and classified into closer clusters.
The silhouette plot of the clustering results before and after the doctor's labeling is different. As shown in Fig. 15, the effect of re-clustering after labeling is not very stable, which may be related to the structure of the doctor's manual labeling.
4 Conclusion
This paper presents a GBUL method for CHIC. The method focuses on the density of cell distribution in histopathology images, aiming to solve the problems of cancer risk prediction and rapid detection of lesion points, and proposes a SBNG approach to approximate the distribution of cell nuclei in the images, which is more effective in achieving a semi-automatic annotation of cell nuclei structure.
References
Hui, Z., Dongyan, W., Ming, L., et al.: FIGO 2018 Women’s Cancer Report – Interpretation of cervical cancer guidelines. Chinese J. Pract. Gynecol. Obstet. 35(01), 99–107 (2019)
Siegel, R., Miller, K., Jemal, A.: Cancer Statistics. CA: A Cancer J. Clin. 67(1), 7–30 (2017)
Jiaomei, G.: Research on New Technology of Cervical Cancer Screening. Zhengzhou University, Zhengzhou (2013)
Rahaman, M.M., Li, C., Yao, Y., et al.: Identification of COVID-19 samples from chest X-Ray images using deep learning: a comparison of transfer learning approaches. J. Xray Sci. Technol. 28(5), 821–839 (2020)
Li, C., et al.: A review for cervical histopathology image analysis using machine vision approaches. Artif. Intell. Rev. 53(7), 4821–4862 (2020). https://doi.org/10.1007/s10462-020-09808-7
Sukumarand, P., Gnanamurthy, R.: Computer aided detection of cervical cancer using pap smear images based on adaptive neuro fuzzy inference system classifier. J. Med. Imaging Health Inform. 6(2), 312–319 (2016)
Li, C., Xue, D., Zhou, X., et al.: Transfer learning based classification of cervical cancer immunohistochemistry images. In: Proceedings of the Third International Symposium on Image Computing and Digital Medicine, pp. 102–106 (2019)
Lee, H.B., Macqueen, J.B.: A k-means cluster analysis computer program with cross-tabulations and next-nearest-neighbor analysis. Educ. Psychol. Measur. 40(1), 133–138 (1980)
Rahmadwati, R., Naghdy, G., Todd, C.: Computer aided decision support system for cervical cancer classification. In: SPIE Optical Engineering + Applications. International Society for Optics and Photonics, pp. 105–121 (2012)
Xing, C., Jun, L.: Research review of segmentation algorithm based on graph theory. Comput. Dig. Eng. 44(10), 1–12 (2016)
Shi, J., Malik, J.: Normalized cut and image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 22(8), 26–33 (2000)
Felzenszwalb, P.F., Huttenlocher, D.P.: Efficiet graph-based image segmentation. Int. J. Comput. Vision 59(2), 167–181 (2004)
Pavan, M., Pelillo, M.: A new graph-theoretic approach to clusterin and segmentation. In: Proceedings of 2003 IEEE Computer Society Conference on CVPR, vol. 1, pp. 145–152 (2003)
Yanhua, G., Aihong, Z., Lingyun, F.: Color feature extraction based on color histogram. Fujian Comput. 5, 96–97 (2017)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Yang, X., Shang, L., He, R., Li, X., Li, C. (2023). Clustering of Cervical Histopathology Images Based on Minimum Spanning Tree. In: You, P., Li, H., Chen, Z. (eds) Proceedings of International Conference on Image, Vision and Intelligent Systems 2022 (ICIVIS 2022). ICIVIS 2022. Lecture Notes in Electrical Engineering, vol 1019. Springer, Singapore. https://doi.org/10.1007/978-981-99-0923-0_10
Download citation
DOI: https://doi.org/10.1007/978-981-99-0923-0_10
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-99-0922-3
Online ISBN: 978-981-99-0923-0
eBook Packages: EngineeringEngineering (R0)