Abstract
The worldwide emergence of resistant bacteria to multiple antimicrobial drugs is one of the greatest hurdles to chemotherapy. Multidrug resistance (MDR) is the capability of pathogenic bacteria to survive lethal doses of antimicrobial drugs. One of the underlying mechanisms of survival under stressful conditions is the extrusion of drugs through membrane-embedded efflux proteins. These ubiquitous resistance elements, which confer resistance or cross resistance to multiple drugs, are considered MDR efflux transporters. Consequently, efflux pump inhibitors (EPIs) from various natural and synthetic sources have been developed to increase the therapeutic armamentarium for combating bacterial resistance and restoring the antibiotic activity. Owing to less toxicity issues than chemical-based EPIs, plant-based EPIs are gaining much importance, but none are yet undergoing clinical trials. In this review, we will introduce the concept of efflux pumps and their diversity and then provide a comprehensive understanding of efflux pump inhibitors from both plant and chemical sources, their mode of action and the recent advances in their development.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
In the present century, antimicrobial resistance (AMR) has become a global threat to chemotherapy of infection control as stated by data obtained from global surveillance (WHO 2014). Evolution of resistance leads to the emergence and spread of a large number of resistant bacteria globally that expresses sophisticated phenotypic or genotypic variants towards a variety of antimicrobial drugs. Antibiotic resistance has become a topic of interest in various science and political summits and has been referred to as a slow motion tsunami (Cox 2015). Bacterial resistance towards antibiotics develops as a result of mutations and/or gene acquisitions through genetic exchange mechanisms. Broadly, the mechanism of resistance is classified into three categories: first, production of drug hydrolysing or modifying enzymes; second, mutation in the transporters of antibiotics that hinders their entrance; and the third is by the impaired accessibility of an antibiotic to its target by using energy-dependent efflux pumps for its exclusion. In bacteria, active efflux pumps play an important role in both intrinsic and acquired multiple drug resistance as opposed to other various biochemical and molecular resistance mechanisms. Energy-dependent efflux pumps are ubiquitous and important to the drug resistance of all organisms from prokaryotes to mammals. In the case of humans, they are relevant contributors of anticancer drug resistance, and in bacteria, they contribute resistance to antibiotics (Amaral et al. 2012; Nikaido 2009). Efflux pumps were firstly described in the 1970s as the mechanism of resistance to P-glycoprotein in humans (Gottessman and Ling 2006) and resistance towards tetracycline in Escherichia coli (Levy S., 1992).
Considerable progress has been made in understanding the various types of efflux pumps prevalent in bacteria that are responsible for different types of drug resistance (Chitsaz and Brown 2017). Subsequently, researchers have found methods to inhibit efflux pump functions by using efflux pump inhibitors. The specific mechanisms involved may vary accordingly (Sjuts et al. 2016). Both synthetic and natural product-derived efflux pump inhibitors have been developed, and their clinical and therapeutic potential have been reported. In this chapter, we will provide a summary of recent updates on the diversity of efflux pumps and variance of both synthetic and natural efflux pump inhibitors and their significance in chemotherapy.
Drug efflux pumps are protein complexes embedded in the membranes of MDR bacteria. Their overexpression causes extrusion of structurally unrelated drugs and contributes to reduced susceptibly, thereby decreasing the concentration inside the cell to sub-toxic levels. This class of proteins are encoded by genes that are located on chromosomes or plasmids (Piddock et al. 2006; Nikaido and Pages 2012; Blanco et al. 2016). A generalized scheme of the five major families of bacterial efflux pumps is represented in Table 1.
2 Major Classes of Efflux Pumps
Efflux pump transporters in the prokaryotic kingdom are classified into five major superfamilies based on their composition, energy source, number of transmembrane spanning regions and substrate specificity: resistance-nodulation-division (RND) superfamily, the ATP (adenosine triphosphate)-binding cassette (ABC) superfamily, the major facilitator superfamily (MFS), the multi-drug toxic compound extrusion (MATE) superfamily and a small multidrug resistance (SMR) family (a member of a much larger metabolite or drug superfamily). These efflux pump transporters are found in both gram-positive and gram-negative bacteria except the RND efflux pumps that are predominantly distributed in gram-negative bacteria. The genes encoding this class of transporter proteins can be chromosomally encoded or plasmid encoded (Piddock 2006).
The RND superfamily have a tripartite composition that includes an inner membrane transporter, a periplasmic adapter protein and an outer membrane channel and are extensively associated with antibiotic resistance such as AcrB in Escherichia coli and MexB in Pseudomonas aeruginosa (Li and Nikiado 2009). With advancements in molecular biology and biochemistry, there has been a growing identification and characterization of MDR pumps in numerous problematic species of bacteria, particularly the pathogens belonging to ESKAPE group [Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumonia, Acinetobacter baumannii, P. aeruginosa, Enterobacter species], evoking their predominant role in clinical settings (Davin-Regli et al. 2016). Among gram-positive bacteria, the MFS family of efflux transporters is widely distributed like NorA efflux pumps in Staphylococcus aureus and PmrA from Streptococcus pneumonia (Jang 2016).
2.1 ATP-Binding Cassette (ABC) Family MDR Efflux Pumps
ABC transporters that are involved in drug extrusion are found in fungi and animal cells. There are only a few examples reported of ABC transporters in gram-negative bacteria, although MSbA transporters that export the biosynthetic intermediate of lipopolysaccharides carry out erythromycin extrusion when they are overexpressed in Lactococcus lactis. ABC pumps are composed of four domains in which two are hydrophobic and membrane-embedded and are responsible for substrate recognition and translocation. The other two domains are hydrophilic nucleotide-binding domains (NBDs) and serve as a site for ATP hydrolysis, which then generates the energy for the translocation of drugs (Crow et al. 2017). The most commonly studied ABC drug exporter in gram negatives is MacB of E. coli, which becomes functional when it comes into contact with the periplasmic MacA adapter and TolC of the outer membrane channel. Overexpression of MacA–TolC transporters increases the MIC of macrolide antibiotics and results in the emergence of resistance (Lu and Zgurskaya 2013). MacB also possesses moderate ATPase activity. ATP hydrolysis is triggered by the continuous presence of MacA, which becomes fully functional when it comes together with dimeric MacB and TolC trimer unit of the pump (Wilkens 2015).
2.2 SMR Transporters
SMR transporters are small, proton motive force-dependent drug transporters that consist of four transmembrane segments (TMSs). These proteins generally function as homodimers, but their arrangement is either in parallel or antiparallel and is a matter of controversy (Lloris-Garcera et al. 2013). Overexpression of these proteins by their corresponding plasmids causes reduced susceptibility to aminoglycoside antibiotics. EmrE, one of the transporters of the SMR superfamily, generates hyper-osmotolerant phenotypes in E. coli also harbouring AcrAB proteins. Most of the common substrates of EmrE pumps are mainly quaternary ammonium compounds like osmoprotectants of E. coli and betaine. The overexpression of such pumps increases the susceptibility of cells to alkalinity in the medium and hyperosmolarity conditions as well (Bay et al. 2008). In the case of S. aureus, the SMR proteins like QacC are mainly plasmid encoded, while in other bacteria, the genes for the related components are also present on chromosomes. QacC carries out the efflux of biocides such as ethidium bromide and also quaternary ammonium compounds (Furi et al. 2013; Wassenaar et al. 2015).
2.3 Multidrug and Toxin Extrusion (MATE) Family
MATE transporters were firstly identified in Vibrio parahaemolyticus as sodium or cationic antiporter, i.e. NorM with 12 TMS (Kuroda and Tsuchiya 2009). The MATE family of drug transporters act as a contributing factor for resistance and are in recent discovery. Most of the work has been done with these pumps, and well reports are obtained in gram-negative bacteria including the crystal structures of four proteins out of them (Lu and Zgurskaya 2013). Some of the significant work has been done to determine the function and structure of MATE transporters in gram-positive bacteria, such as S. aureus MATE pumps like MepA. Interestingly, it was reported that in methicillin-susceptible strains, the majority of the population overexpresses MepA, but the reason behind this remains uncertain (Schindler and Kaatz 2016). MATE proteins are dependent either on Na ions or the proton gradient and are regulated by the MepR regulatory protein. Dimers in single and dimeric form bind to MepR and MepA, respectively. MepR belongs to the MarR family of proteins and consists of winged helix-turn-helix that retards the expression of its own gene as well as mepA (Jang 2016). Additional MATE efflux pumps in gram positive have been described like PdrM of S. pneumoniae, FepA of Listeria monocytogenes and Cda of C. difficile (Hashimoto et al. 2013; Gurein et al. 2014). The substrate utilized by FepA is larger in number since they decrease susceptibility to several dyes, ciprofloxacin, norfloxacin and biocides. A regulatory protein that belongs to the TetR group regulates the expression of fepA, and mutation in FepR increases the overexpression of fepA and thus reduces the drug susceptibility.
2.4 Major Facilitator Superfamily (MFS) Transporters
There are 74 families of MFS transporters, grouped on the basis of their sequence homology. Most of them are embedded in an inner membrane in a free state, and these pumps transport the drugs from the cytosol only to the periplasm. Antimicrobial agents have the ability to cross the lipid bilayer through diffusion, and once they are pumped out, they are unable to come, but in this case, MFS pumped out drugs can easily come to the cytosol. Thus, this kind of transporters is not responsible for high level of resistance. It was found that in P. aeruginosa, these pumped out drugs are easily captured in the periplasm by MexAB–OprM RND pumps, which are constitutive expressed, synergistically enhancing the activity of singlet pumps in the development of resistance (Choudhury et al. 2015).
In case of E. coli, MFS forms a tripartite efflux system that includes an outer membrane protein channel, Tol-C and periplasmic adapter proteins. These efflux systems include EmrB and EmrY and carry out the extrusion of a variety of substrates and uncouplers (Nishino and Yamaguchi 2008). A central cavity which is surrounded by hydrophobic and hydrophilic side chains is found in most of the MFS pumps. It was also noted that the loops formed through connections between H4, H5, H10 and H11 are localized towards the cytosol and responsible for substrate recognition and binding (Yin et al. 2000). A dimer of EmrAB is formed when EmrA comes into association with EmrB. Among the singlet MFS transporters, MdfA pumps lead to MDR when they are overexpressed. In combination with the SMR-type transporter EmrE, these exporters carry out efflux of cationic agents like quaternary ammonium compounds (Tanabe et al. 2009).
NorA is an MDR pump predominantly found in S. aureus which is chromosomally encoded and also belongs to the MFS family of proteins. In Escherichia coli and S. aureus, expression of the plasmid-encoded norA gene leads to the resistance of hydrophilic quinolones like norfloxacin (Yu et al. 2002). MgrA is a global regulatory protein that controls expression of not only NorA pumps but also other pump proteins like TetG, NorB and NorC. Additionally, another regulatory protein involved in NorA expression is GntR which is able to interact with the promoters of NorC, NorB and NorA. In addition to interacting with these promoters, this regulatory protein reduces the transcription of NorG, leading to a fourfold decrease in fluoroquinolone susceptibility (Redgrave et al. 2014; van der Putten et al. 2018). A deletion of GntR does not affect NorA, NorB or NorC but does cause a significant increase in the transcription of the ABC family of transporters that play an important role in the susceptibility of S. aureus towards β-lactams (Foster 2017; Ranaweera et al. 2015).
2.5 Resistance-Nodulation-Cell Division (RND) Transporters
RND-type efflux pumps are predominant in gram-negative pathogens. These tripartite protein complexes span throughout the membrane and enable bacteria to pump antibiotics out of the cell. These protein assemblies are composed of an inner membrane protein (IMP), a periplasmic membrane fusion protein (MFP) and an outer membrane protein. The IMP exhibits selectivity towards drugs and carries out catalysis of the drug/proton antiport (Venter et al. 2015). The most widely studied RND-type efflux pumps are AcrAB–TolC in E. coli and MexAB–OprM in P. aeruginosa (Blair et al. 2014). In recent years, the elucidation of crystal structures of different efflux pumps that are complexed with their substrate or inhibitors has greatly increased our understanding of their specificities towards multiple substrates. The AcrAB pump protein of E. coli was the first crystal structure of an efflux pump to be resolved at 3.5 Å resolution (Murakami et al. 2002). Later the same research group identified the interaction of crystal structure with the substrates doxorubicin and minocycline. The crystal structure is comprised of a periplasmic headpiece, which constitutes a large portion and a transmembrane domain. The upper part represents the TolC docking domain, and the central region of the headpiece makes up the pore domain (Murakami et al. 2006).
The best characterized efflux pump is MexAb–AcrB produced by the opportunistic pathogen P. aeruginosa and confers resistance towards a broad spectrum of antimicrobials. The crystal structure of P. aeruginosa efflux pump components that closely resembles with E. coli tripartite AcrAB-TolC has also been reported and characterized (Sennhauser et al. 2009). There is structural similarity in the transmembrane domains of all RND-type transporters. The MexB docking domain in P. aeruginosa is composed of two sub domains, one of which forms a loop that inserts into the docking domain of an adjacent pump subunit. These findings clearly demonstrate that RND efflux pumps and outer membrane channels have low affinity and interact in transient ways (Yamaguchi et al. 2015; Daury et al. 2016).
3 Efflux Pump Inhibitors (EPIs)
Overexpression of multidrug efflux pumps is the principle cause of antibiotic resistance in pathogenic bacteria that have increased MIC towards variety of antibiotics. These efflux transporters have become major resistance determinants for the efficacy of both old and new antimicrobial drugs (Hernado-Amado et al. 2016). The structure of efflux transporters not only provides a deep insight into the mechanism of extrusion of drugs but also makes the discovery of efflux pump inhibitors possible (Lomovskaya and Bostian 2006). Broadly speaking, the agents that cause inhibition of efflux, either through disrupting the proton motive force, interacting with proteins or inhibiting the efflux of pump-associated genes, are called efflux pump inhibitors. Because of the increasing evidence of resistance emerging in pathogenic bacteria, alternative therapeutic strategies that counteract antibiotic efflux are needed. The strategies for inhibiting efflux pump include:
-
(i)
Structural alterations in the chemical design of existing antibiotics so that their binding affinity to the efflux pump protein sites is reduced.
-
(ii)
Interference with the energy source that is needed for the activity of efflux pumps.
-
(iii)
Downregulating the expression of genes required for the expression of active efflux pumps in bacterial cell envelopes.
-
(iv)
Interference with the functional subunit of efflux pump complexes.
-
(v)
Competitive or non-competitive inhibition of antibiotics towards substrate-binding sites in efflux pumps.
-
(vi)
Hindrance in the activity of membrane channels responsible for antibiotic extrusion by inserting designed molecular plugs.
EPIs act as therapeutic agents since they have the potential to restore the activity of conventional antibiotics. The combination of EPIs along with antibiotics is expected to reduce the intrinsic resistance of bacteria towards a varying range of antibiotics, decrease the frequency of emerging resistant mutant strains and facilitate the reversal of acquired resistance in strains that have multiple targeted mutations. Lomovskaya et al. (2001) provided the following criteria that a potent efflux pump inhibitor should satisfy:
-
(a)
It must potentiate the antibiotic activity against the resistant strain that has developed as a result of the drug efflux pump.
-
(b)
It must have the potential to enhance the accumulation and reduce the extrusion of a substrate that shows specificity towards the efflux pump.
-
(c)
It should not have outer membrane permeabilizing ability.
-
(d)
It should not decrease the MIC of antibiotics that are not the substrate of the efflux pump.
-
(e)
The proton gradient across the inner membrane must be unaffected by the EPI.
The use of a combination of EPI and antibacterial agents have the potential to enhance the activity of antibiotics against efflux pumps and lead to a reduction in mutant frequency. For example, in the case of the food-borne pathogen Campylobacter jejuni, the combination of PAβN with antimicrobials caused a 2000-fold reduction in the minimum inhibitory concentration of the substrate used against CmeABC efflux of C. jejuni and a 1000-fold decrease in the development of erythromycin-resistant mutants (McCrackin et al. 2016).
Several EPIs are used in combination with a photosensitizer dye in the presence of light and exhibit broad-spectrum antibacterial activity. The photoactivable dye generates reactive oxygen species and singlet oxygen, which kill microbial cells. Tegos et al. (2008) used a cationic phenodizidium dye, Toluidine Blue (TBO), under red light in combination with a variety of known EPIs that inhibit the NorA and MexAB–OprM efflux pumps of S. aureus and P. aeruginosa, respectively. Quinolone derivatives such as alkylaminoquinolone, alkoxyquinolone, chloroquinolone and pyrridoquinolone are also used as EPIs and exhibit structural similarity with the quinolone group of antibiotics (Pages and Amaral 2009). These derivatives inhibit antibacterial extrusion and increase antibiotic susceptibility in clinical strains of Klebsiella pneumonia and Enterobacter aerogenes (Cheveleir et al. 2004).
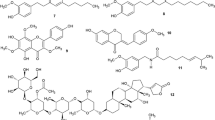
The efflux transporters between different gram-negative bacteria exhibit structural similarity so that the EPI used against pumps such as the AcrA–AcrB–TolC of E. coli can also be effectively used against other gram-negative pathogens. At the present time, two classes of EPIs have been extensively characterized such as pyridopyrimidines and peptidomimetics that exhibit broad-spectrum activity. A third class of EPIs have also been established known as quinolone derivatives (Sun et al. 2014; Aygul 2015). The chemical structure of some synthetic and natural efflux pump inhibitors are shown in Fig. 1.
4 Synthetic EPIs for Modulating the Reversal of MDR Phenotypes
L-Phenyl alanine-L arginine β Napthylamide (PAβN) [MC-207,110] was the first compound developed as an EPI for gram-negative bacteria and is characterized as a dipeptide amide compound, MC-207,110. It inhibits all four efflux systems of Pseudomonas aeruginosa, MexAB–OprM, MexCD–OprJ, MexEF–OprN and MexXY–OprM. Overexpression of the AcrAB–TolC efflux pumps of E. coli, S. typhimurium and S. pneumoniae is also reduced by combining PAβN with fluoroquinolones (Kourtesi et al. 2013). However, the use of PAβN is still limited because of issues associated with its bioavailability and toxicity (Bhardwaj and Mohanty 2012). A derivative of PAβN, MC-04,124, has been developed to target the overexpressed pumps of Pseudomonas aeruginosa with less toxicity and greater stability in biological fluids (Pages and Amaral 2009). Additionally, a number of pyranopyridine EPIs have been developed like MBX2319 that exhibits potent activity against the members of Enterobacteriaceae, but it has reduced activity in P. aeruginosa strains (Oppeman et al. 2014). The efficacy of this compound has not been tested in animal models and is the stages of lead optimization.
Quinoline compounds also show enhanced activity against the AcrAB–TolC efflux pumps of Enterobacter aerogenes. These compounds effectively increase the intracellular accumulation of chloramphenicol by inhibiting drug extrusion.
More recently, a molecular modelling and docking study conducted to determine promising EPI activities of aminoguanidine hydrazones found that they act as competitive inhibitors of the NorA efflux pump of S. aureus against norfloxacin. By binding with NorA, they restored the activity of norfloxacin and ultimately reduced resistance emergence (Dantas et al. 2018).
5 EPIs from Natural Sources
EPIs have been screened from both microbial and plant sources like in study two compounds, Ea-371α and EA-371δ, as effective efflux pump inhibitors through the microbial fermentation process have been identified (Lee et al. 2001). These two compounds have the potential to suppress the MexAB–OprM pumps of P. aeruginosa.
In developing countries, medicinal plants have had a long history to treat various health-related issues. Natural phytochemical compounds are routinely used as antibacterial agents, and their strength is their ability to synergize with antibacterial drugs, which can make antibiotics that are not used in infection control due to resistance, relevant again (Lee et al. 2011). These compounds cripple the resistance mechanisms of bacteria making, and due to the complexity of plant EPIs, bacteria may take decades to develop resistance to them (Wink 2012). Researchers have classified the phytocompounds that potentiate the activity of antibiotics and exhibit activity against multidrug-resistant pathogens as modifying, modulating or reversal agents (Fankam et al. 2017).
A well-known example of an EPI from a natural source is the plant alkaloid reserpine, obtained from the roots of the poisonous devil’s pepper (Rauwolfia vomitoria Afz), which inhibits the Bacillus subtilis efflux pump Bmr (Ahmed et al. 1993). Phytochemistry has played a significant role in the search for EPIs against the NorA efflux pumps of S. aureus, and it has been reported that plant-derived EPIs containing varied chemical constituents, such as porphyrin phaeophorbide, acylated glycosides, flavones and isoflavones, have the ability to inhibit NorA pumps (Holler et al. 2012). Moreover, in case of gram-negative bacteria, it was found that a synergistic combination of cefixime and aqueous extracts of Terminalia chebula and gallactotannin, isolated from Terminalia chebula, enhances the antibiotic potential and competes with multidrug-resistant efflux pumps of E. coli as well (Bag and Chattopadhyay 2014).
Some plants produce a variety of antimicrobials along with compounds that inhibit the extrusion of antibiotics from the bacterial cell. This was first exemplified by the discovery of the antimicrobial compound berberine from berberis, a medicinal plant, which also synthesizes 5′-methoxyhydnocarpin (5′MHC), a S. aureus NorA efflux pump inhibitor, which reduces the MIC of berberine (Stermitz et al. 2000). Unfortunately, its use is limited because of toxicity issues. There are some EPIs of plant origin that have been patented, such as a tetrandrine-based EPI that reduces the resistance of E. coli to fluoroquinolones and also a natural compound called geraniol that is effective against Enterobacter aerogenes (Berti et al. 2010).
Maisuria et al. (2015) extracted maple syrup with methanol and demonstrated EPI activity against P. aeruginosa ATCC15692, Proteus mirabilis and E. coli ATCC 700928. The active compound catechol was isolated from maple syrup and found to inhibit ethidium bromide efflux but not to a larger extent than maple syrup extract itself. Lysergol which is a clavine alkaloid obtained from Ipomoea muricata (L) Jack (Convolvulaceae) exhibited efflux pump inhibitory potential against both sensitive and resistant strains of E. coli. It was reported that the active compound lysergol and its derivative 17-O-3″,4″,5″ trimethoxy benzyl lysergol have promising EPI activity compared to that of reserpine in an EtBr accumulation assay (one of the assays for the screening of efflux overexpressing strains), and it was shown that the YojI efflux pump of E. coli is the target of this compound (Maurya et al., 2013). In another case, the essential oil obtained from Salvia fruticosa Mill (Lamiaceae) by means of hydrodistillation is found to be active against the TetK pumps of tetracycline-resistant S. epidermis. It was reported that this oil caused a 63% reduction in the overexpression of Tet proteins (Chavanova et al. 2015). Furthermore, a known fatty acid, linoleic acid, was obtained from the herb Portulaca oleracea L (Portulacaceae) and found to inhibit EtBr efflux at a concentration of 64 mg/L compared to reserpine against MRSA strains. This unsaturated fatty acid restored the activity of erythromycin in overexpressing ABC efflux pumps of MRSA (Chan et al. 2015). The overexpression of the NorA efflux pump can be inhibited by Olympian A obtained from Hypericum Oylmicum L.cf. uniflorum. This patented acylphloroglucinol compound is active against S. aureus 1199B strains with decreased cytotoxicity at about 8.9 μM in cancer cell lines (Shiu et al. 2013). Previously, it was observed that carnosic acid from Rosmarinus officinalis can block NorA-induced efflux of EtBr in MDR S. aureus 1199B strains (Smith et al. 2007). In a study of triterpenes from plant sources, active compound such as karavilagenin C and balsaminagenin B from the medicinal plant Momordica balsamina L (Cucurbitaceae) was obtained and was checked for the EPI activity against MRSA COLoxa (MRSA that are highly resistant to oxacillin) strains and E. faecalis ATCC 29212, and it was found that balsaminagenin B was most active at a concentration of 30 μM in E. faecalis cells (Ramalhete et al. 2016). Another triterpenes isolated from the same plant were Karavilagenin C and balsaminol F, found effective at a concentration of 3 μM in NorA pump overexpressed MRSA COLoxa cells (Ramalhete et al. 2011a, b).
It can be emphasized that natural compounds are effective at inhibiting the SMR, MFS and ABC transporter superfamilies found in gram-positive bacteria; however, the transporters in gram negatives are highly complex because of the presence of an outer membrane permeability barrier. Furthermore, there are certain compounds that inhibit human transporters. Thus, EPIs should be designed in such a way that can specifically target the efflux pumps of bacterial origin (Amaral et al. 2014). Recently, in a screening of a small molecule library containing 8000 molecules, a compound (IIR08027) was found to potently inhibit the proton-driven efflux pump AbeS of Acinetobacter baumannii. Although the molecule does not have growth inhibitory action, it restored the activity of ciprofloxacin against Acinetobacter baumannii-resistant strains (Bhattacharyya et al. 2017). In Table 2, we list some of the newly synthesized EPIs from natural sources. Previously identified EPIs are discussed elsewhere (Tegos et al. 2011; Abreu et al. 2012 and Kourtesi et al. 2013
).
6 Mechanisms of Action of the Major Classes of EPIs
Bacterial efflux pumps are novel target for a variety of antimicrobial compounds for combating MDR resistance. EPIs are used as adjuvant to enhance the efficacy of antibiotics. Efflux pump inhibitors have been exploited from both natural and synthetic sources, including gram-positive and gram-negative bacteria. RND pumps in gram negatives play a predominant role in virulence and pathogenicity. EPIs that are targeted towards RND pumps are useful as adjunctive therapies in combination with antibiotics to reduce spread and emergence, decrease virulence and inhibit biofilm formation in Klebsiella pneumoniae and E. coli (Kvist et al. 2008; El-Banna et al. 2016). A variety of EPIs have been developed, but recent reports suggest that only three of the synthetic EPIs are optimized and in preclinical trials (Mahmood et al. 2016). Owing to the structural complexity of the RND-type efflux pumps, the mechanism of action of EPIs complicates their development and discovery. Generally, efflux pump inhibitors are designed to target pump components as depicted in Fig. 2.
Possible EPI targets of RND-type efflux pumps. (Source Venter et al. 2015)
6.1 1-(1-Naphthylmethyl)-Piperazine (NMP) and Analogs of Arylpiperazine
NMP was discovered as an EPI against E. coli by Bohnert and Kern (2005). This research group screened a library of N-heterocyclic compounds and found that phenylpiperazines act as effective potentiators of levofloxacin in AcrAB and AcrEF-resistant strains of E. coli.
Schuster et al. (2014) were the first research group to determine the mode of action of NMP-resistant mutants. They screened a library of NMR-resistant mutants that were able to grow on linezolid, and focusing their study on the binding and extrusion domains of the AcrB pumps in these mutants. Subsequently, they found a mutation in the amino acid residue, Phe610, near the substrate-binding pocket that plays an important role in the extrusion process. Mutated amino acid residues were also present across the G-loop that separates the distal and proximal binding sites (Eicher et al. 2012). A predicted study of MD simulations also suggested Nmp bound to these locations across the G-loop and allowed for the extrusion of linezolid in resistant mutants (Vargiu et al. 2014). This clearly demonstrates that NMP interferes with the G-loop and is responsible for substrate extrusion, inhibiting the action of AcrB. Similar to the mode of action of PAβN, NMP is able to cause conformational changes that lead to reduction in the width of the substrate-binding domain as predicted by MD simulations (Vargiu and Nikaido 2012; Bohnert et al. 2016) (Table 3).
6.2 Pyranopyridines (MBX2319)
MBX2319 is a novel inhibitor of the AcrAB efflux pumps of E. coli, reported by Opperman et al. (2014). The efflux proteins of other members of Enterobacteriales like K. pneumoniae, Shigella flexneri and E. cloacae are also inhibited by this compound. MBX2319 possesses no antibacterial activity (MIC≥100 μg/ml) but has profound effects on the MIC of antibiotics like beta-lactams, fluoroquinolones, chloramphenicol, linezolid and erythromycin, acting in the range of 3.1–12.5 μg/m as substrate for AcrB pumps. The underlying mechanism of this efflux transporter inhibition involves conformational changes in binding sites due to the functional rotation of AcrB. Once MBX2319 binds these sites, it hinders their rearrangement, and as a result, the translocation of proteins from the periplasm to the cytoplasm occurs. Additionally, a distal binding site, which is also the site for doxorubicin and minocycline, becomes shrunk when the compound binds to them (Nakashima et al. 2013). The results obtained through MD simulation closely resemble those from crystallographic methods and generate a molecular hypothesis that supports the mode of action of this EPI on the basis of biochemical and genetic experiments. The compound is effective against the RND pumps of gram negatives since it does not possess any antibacterial activity nor has any additional targets like outer/inner membranes (Opperman et al. 2014; Nguyen et al. 2015).
6.3 D13–9001 and Pyridopyrimidinone Analogs
D13–9001 was discovered by Yoshida et al. (2007) and possesses in vivo activity and good solubility against MexAB–OprM overexpressing P. aeruginosa. The three-dimensional structure of this compound was elucidated by Nakashima et al. (2013). The hydrophobic tert-butyl thiazolyl amonocarboxyl pyridopyrimide moeity unit of D13–9001 comes into contact with a depression in the hydrophobic trap of the substrate-binding site. This narrow depression is lined with hydrophobic residues and branches off from the substrate translocation channels. The substrate-binding regions are surrounded by aceto amino-ethylene amino acetate moieties that are the sites for minocycline and doxorubicin. The crystal structures suggest that the binding sites are similar for both AcrB and MexB, as the compound strongly bound to the hydrophobic trap of the MexB binding domain and inhibited conformational changes required for pump activity (Nakashima et al. 2013). The silver lining of this study is the fact that these data lead to an understanding of the MOA of EPIs as well as provide information concerning the unknown binding sites of RND pumps.
7 Conclusion
Efflux-mediated antibiotic resistance can be abolished by screening or designing EPIs against multidrug-resistant organisms. EPIs have the promising potential to restore the activity of antibiotics by decreasing their MIC. Significant advancement has been made in understanding the physiology, mechanisms of action and regulation of MDR pumps. Efflux pump crystal structures bound to their substrate/specific inhibitors increase our knowledge of MDR pumps as well as aiding in the development of EPI. However, to date, there are still no efflux pump inhibitors that are used in treating human/animal infections caused by bacteria. However, EPIs have been developed from synthetic and natural sources and some of them are in preclinical trials. Meanwhile, numerous researchers have suggested that the secondary metabolites of plants display greater activity as EPIs against gram-positive than gram-negative bacteria. So, there is need to develop effective EPIs against gram negatives. A silver lining is the fact that in the near future, standardized methods and techniques for discovering EPIs from natural sources will likely be established. Future endeavours involve the search for EPIs that exhibit low cytotoxicity, improved solubility and a broad spectrum of activity against clinically significant different classes of efflux proteins.
References
Abreu, A. C., McBain, A. J., & Simoes, M. (2012). Plants as sources of new antimicrobials and resistance-modifying agents. Natural Product Reports, 29(9), 1007–1021.
Aghayan, S. S., Mogadam, H. K., Fazli, M., et al. (2017). The effects of berberine and palmatine on efflux pumps inhibition with different gene patterns in Pseudomonas Aeruginosa isolated from burn infections. AJMB9, 9(1), 2.
Ahmed, M. A., Borsch, C. M., Neyfakh, A. A., & Schuldiner, S. (1993, May 25). Mutants of the Bacillus subtilis multidrug transporter Bmr with altered sensitivity to the antihypertensive alkaloid reserpine. Journal of Biological Chemistry, 268(15), 11086–11089.
Amaral, L., Spengler, G., Martins, A., Armada, A., Handzlik, J., Kiec-Kononowicz, K., & Molnar, J. (2012). Inhibitors of bacterial efflux pumps that also inhibit efflux pumps of cancer cells. Anticancer Research, 32(7), 2947–2957.
Amaral, L., Martins, A., Spengler, G., & Molnar, J. (2014). Efflux pumps of gram-negative bacteria: What they do, how they do it, with what and how to deal with them. Frontiers in Pharmacology, 4, 168.
Anes, J., McCusker, M. P., Fanning, S., & Martins, M. (2015). The ins and outs of RND efflux pumps in Escherichia coli. Frontiers in Microbiology, 6, 587.
Aparna, V., Dineshkumar, K., Mohanalakshmi, N., Velmurugan, D., & Hopper, W. (2014). Identification of natural compound inhibitors for multidrug efflux pumps of Escherichia coli and Pseudomonas aeruginosa using in silico high-throughput virtual screening and in vitro validation. PLoS One, 9(7), 101840.
Avrain, L., Mertens, P., & Van Bambeke, F. (2013). RND efflux pumps in P. aeruginosa: An underestimated resistance mechanism. Antibiotic Susceptibility, 26321, 26–28.
Aygül, A. (2015). The importance of efflux systems in antibiotic resistance and efflux pump inhibitors in the management of resistance. Mikrobiyoloji Bülteni, 49(2), 278–291.
Bag, A., & Chattopadhyay, R. R. (2014). Efflux-pump inhibitory activity of a gallotannin from Terminalia chebula fruit against multidrug-resistant uropathogenic Escherichia coli. Natural Product Research, 28(16), 1280–1283.
Barreto, H. M., Coelho, K. M., Ferreira, J. H., dos Santos, B. H., de Abreu, A. P., Coutinho, H. D., da Silva, R. A., de Sousa TO, Citó, A. M. D. G., & Lopes, J. A. (2016). Enhancement of the antibiotic activity of aminoglycosides by extracts from Anadenanthera colubrine (Vell.) Brenan var. cebil against multi-drug resistant bacteria. Natural Product Research, 30(11), 1289–1292.
Bay, D. C., Rommens, K. L., & Turner, R. J. (2008). Small multidrug resistance proteins: A multidrug transporter family that continues to grow. Biochimica et Biophysica Acta, 1778(9), 1814–1838.
Beheshti, M., Talebi, M., Ardebili, A., Bahador, A., & Lari, A. R. (2014). Detection of AdeABC efflux pump genes in tetracycline-resistant Acinetobacter baumannii isolates from burn and ventilator-associated pneumonia patients. Journal of Pharmacy & Bioallied Sciences, 6(4), 229.
Berti, L., Lorenzi, V., Casanova, J., Muselli, A., Pagès, J.M. and Bolla, J.M. (2010) Geraniol as bacterial efflux pump inhibitor. European Patent No. EP 2 184 061 A1, 12 Jun 2010.
Bhardwaj, A. K., & Mohanty, P. (2012). Bacterial efflux pumps involved in multidrug resistance and their inhibitors: Rejuvinating the antimicrobial chemotherapy. Recent Patents on Anti-Infective Drug Discovery, 7(1), 73–89.
Bhattacharyya, T., Sharma, A., Akhter, J., & Pathania, R. (2017). The small molecule IITR08027 restores the antibacterial activity of fluoroquinolones against multidrug-resistant Acinetobacter baumannii by efflux inhibition. International Journal of Antimicrobial Agents, 50(2), 219–226.
Bialek-Davenet, S., Lavigne, J. P., Guyot, K., Mayer, N., Tournebize, R., Brisse, S., Leflon-Guibout, V., & Nicolas-Chanoine, M. H. (2014). Differential contribution of AcrAB and OqxAB efflux pumps to multidrug resistance and virulence in Klebsiella pneumoniae. The Journal of Antimicrobial Chemotherapy, 70(1), 81–88.
Blair, J. M., Richmond, G. E., & Piddock, L. J. (2014). Multidrug efflux pumps in gram-negative bacteria and their role in antibiotic resistance. Future Microbiology, 9(10), 1165–1177.
Blanco, P., Hernando-Amado, S., Reales-Calderon, J. A., Corona, F., Lira, F., Alcalde-Rico, M., Bernardini, A., Sanchez, M. B., & Martinez, J. L. (2016). Bacterial multidrug efflux pumps: Much more than antibiotic resistance determinants. Microorganism, 4(1), 14.
Bohnert, J. A., & Kern, W. V. (2005). Selected arylpiperazines are capable of reversing multidrug resistance in Escherichia coli overexpressing RND efflux pumps. Antimicrobial Agents and Chemotherapy, 49(2), 849–852.
Bohnert, J. A., Schuster, S., Kern, W. V., Karcz, T., Olejarz, A., Kaczor, A., Handzlik, J., & Kieć-Kononowicz, K. (2016). Novel piperazine arylideneimidazolones inhibit the AcrAB-TolC pump in Escherichia coli and simultaneously act as fluorescent membrane probes in a combined real-time influx and efflux assay. Antimicrobial Agents and Chemotherapy, 60(4), 1974–1983.
Boncoeur, E., Durmort, C., Bernay, B., Ebel, C., Di Guilmi, A. M., Croizé, J., Vernet, T., & Jault, J. M. (2012). PatA and PatB form a functional heterodimeric ABC multidrug efflux transporter responsible for the resistance of Streptococcus pneumoniae to fluoroquinolones. Biochemistry, 51(39), 7755–7765.
Bremner, J. B. (2007). Some approaches to new antibacterial agents. Pure and Applied Chemistry, 79(12), 2143–2153.
Chan, B. C., Han, X. Q., Lui, S. L., Wong, C. W., Wang, T. B., Cheung, D. W., Cheng, W., Ip, M., Han, S. Q., Yang, X. S., & Jolivalt, C. (2015). Combating against methicillin-resistant Staphylococcus aureus–two fatty acids from Purslane (Portulaca oleracea L.) exhibit synergistic effects with erythromycin. The Journal of Pharmacy and Pharmacology, 67(1), 107–116.
Chevalier, J., Bredin, J., Mahamoud, A., Malléa, M., Barbe, J., & Pagès, J. M. (2004). Inhibitors of antibiotic efflux in resistant Enterobacter aerogenes and Klebsiella pneumoniae strains. Antimicrobial Agents and Chemotherapy, 48(3), 1043–1046.
Chitsaz, M., & Brown, M. H. (2017). The role played by drug efflux pumps in bacterial multidrug resistance. Essays in Biochemistry, 61(1), 127–139.
Choudhury, D., Talukdar, A. D., Choudhury, M. D., Maurya, A. P., Paul, D., Chanda, D., Chakravorty, A., & Bhattacharjee, A. (2015). Transcriptional analysis of MexAB-OprM efflux pumps system of Pseudomonas aeruginosa and its role in carbapenem resistance in a tertiary referral hospital in India. PLoS One, 10(7), 0133842.
Chovanová, R., Mezovská, J., Vaverková, Š., & Mikulášová, M. (2015). The inhibition the Tet (K) efflux pump of tetracycline resistant Staphylococcus epidermidis by essential oils from three Salvia species. Letters in Applied Microbiology, 61(1), 58–62.
Cox, D. (2015). Antibiotic resistance: The race to stop the silent tsunami facing modern medicine. The Guardian.
Coyne, S., Courvalin, P., & Périchon, B. (2011). Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrobial Agents and Chemotherapy, 55(3), 947–953.
Crow, A., Greene, N. P., Kaplan, E., & Koronakis, V. (2017). Structure and mechanotransmission mechanism of the MacB ABC transporter superfamily. Proceedings of the National Academy of Sciences, 114(47), 12572–12577.
Dantas, N., de Aquino, T. M., de Araújo-Júnior, J. X., da Silva-Júnior, E., Gomes, E., Gomes, A. A. S., Siqueira-Júnior, J. P., & Junior, F. J. B. M. (2018). Aminoguanidine hydrazones (AGH’s) as modulators of norfloxacin resistance in Staphylococcus aureus that overexpress NorA efflux pump. Chemico-Biological Interactions, 280, 8–14.
Daury, L., Orange, F., Taveau, J. C., Verchère, A., Monlezun, L., Gounou, C., Marreddy, R. K., Picard, M., Broutin, I., Pos, K. M., & Lambert, O. (2016). Tripartite assembly of RND multidrug efflux pumps. Nature Communications, 12(7), 1073.
Davin-Regli, A., Masi, M., Bialek, S., Nicolas-Chanoine, M. H., & Pagès, J. M. (2016). Antimicrobial drug efflux pumps in Enterobacter and Klebsiella. In L. Xianzhi, C. A. Elkins, & H. I. Zgurskaya (Eds.), Efflux-mediated antimicrobial resistance in Bacteria (Ist ed.). Cham: Springer International Publishing.
Dhanarani, S., Congeevaram, S., Piruthiviraj, P., Park, J. H., & Kaliannan, T. (2017). Inhibitory effects of reserpine against efflux pump activity of antibiotic resistance bacteria. Chemical Biology Letters, 4(2), 69–72.
Dwivedi, G. R., Maurya, A., Yadav, D. K., Khan, F., Darokar, M. P., & Srivastava, S. K. (2015). Drug resistance reversal potential of Ursolic acid derivatives against Nalidixic acid-and multidrug-resistant Escherichia coli. Chemical Biology & Drug Design, 86(3), 272–283.
Eicher, T., Cha, H. J., Seeger, M. A., Brandstätter, L., El-Delik, J., Bohnert, J. A., Kern, W. V., Verrey, F., Grütter, M. G., Diederichs, K., & Pos, K. M. (2012). Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proceedings of the National Academy of Sciences of the United States of America, 109, 5687–5692.
El-Banna, T. E., Sonbol, F. I., El-Aziz, A. A., & Al-Fakharany, O. M. (2016). Modulation of antibiotic efficacy against Klebsiella pneumoniae by antihistaminic drugs. Journal of Medical Microbiology & Diagnosis, 5(225), 2161–0703.
Fankam, A. G., Kuiate, J. R., & Kuete, V. (2017). Antibacterial and antibiotic resistance modulatory activities of leaves and bark extracts of Recinodindron heudelotii (Euphorbiaceae) against multidrug-resistant gram-negative bacteria. BMC Complementary and Alternative Medicine, 17(1), 168.
Ferreira, S., Silva, F., Queiroz, J. A., Oleastro, M., & Domingues, F. C. (2014). Resveratrol against Arcobacter butzleri and Arcobacter cryaerophilus: Activity and effect on cellular functions. Int J Food Microbiol, 180, 62–68.
Foster, T. J. (2017). Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiology Reviews, 1;41(3), 430–449.
Furi, L., Ciusa, M. L., Knight, D., Di Lorenzo, V., Tocci, N., Cirasola, D., Aragones, L., Coelho, J. R., Freitas, A. T., Marchi, E., & Moce, L. (2013). Evaluation of reduced susceptibility to quaternary ammonium compounds and bisbiguanides in clinical isolates and laboratory-generated mutants of Staphylococcus aureus. Antimicrobial Agents and Chemotherapy, 57(8), 3488–3497.
Garvey, M. I., & Piddock, L. J. (2008). The efflux pump inhibitor reserpine selects multidrug-resistant Streptococcus pneumoniae strains that overexpress the ABC transporters PatA and PatB. Antimicrobial Agents and Chemotherapy, 52(5), 1677–1685.
Goli HR, Nahaei MR, Rezaee MA, Hasani A, Kafil HS, Aghazadeh M, Nikbakht M and Khalili Y (2017) Role of MexAB-OprM and MexXY-OprM efflux pumps and class 1 integrons in resistance to antibiotics in burn and intensive care unit isolates of Pseudomonas aeruginosa. Journal Infect Public Health 11(3):364-372.
Gottesman, M. M., & Ling, V. (2006). The molecular basis of multidrug resistance in cancer: The early years of P-glycoprotein research. FEBS Letters, 580(4), 998–1009.
Guérin, F., Galimand, M., Tuambilangana, F., Courvalin, P., & Cattoir, V. (2014). Overexpression of the novel MATE fluoroquinolone efflux pump FepA in Listeria monocytogenes is driven by inactivation of its local repressor FepR. PLoS One, 9(9), 106340.
Hashimoto, K., Ogawa, W., Nishioka, T., Tsuchiya, T., & Kuroda, T. (2013). Functionally cloned pdrM from Streptococcus pneumoniae encodes a Na+ coupled multidrug efflux pump. PLoS One, 8(3), e59525.
Hernando-Amado, S., Blanco, P., Alcalde-Rico, M., Corona, F., Reales-Calderón, J. A., Sánchez, M. B., & Martínez, J. L. (2016). Multidrug efflux pumps as main players in intrinsic and acquired resistance to antimicrobials. Drug Resistance Updates, 28, 13–27.
Holler, J. G., Christensen, S. B., Slotved, H. C., Rasmussen, H. B., Gúzman, A., Olsen, C. E., Petersen, B., & Mølgaard, P. (2012). Novel inhibitory activity of the Staphylococcus aureus NorA efflux pump by a kaempferol rhamnoside isolated from Persea lingue Nees. The Journal of Antimicrobial Chemotherapy, 67(5), 1138–1144.
Hou, P. F., Chen, X. Y., Yan, G. F., Wang, Y. P., & Ying, C. M. (2012). Study of the correlation of imipenem resistance with efflux pumps AdeABC, AdeIJK, AdeDE and AbeM in clinical isolates of Acinetobacter baumannii. Chemotherapy, 58(2), 152–158.
Hürlimann, L. M., Corradi, V., Hohl, M., Bloemberg, G. V., Tieleman, D. P., & Seeger, M. A. (2016). The heterodimeric ABC transporter EfrCD mediates multidrug efflux in Enterococcus faecalis. Antimicrobial Agents and Chemotherapy, 60(9), 5400–5411.
Jang, S. (2016). Multidrug efflux pumps in Staphylococcus aureus and their clinical implications. Journal of Microbiology, 54(1), 1–8.
Kenana, J., Langat, B., Kalicki, C., Inthavong, E., & Kanna, A. (2017). The structure of EmrE and its role in antibiotic resistance. The FASEB Journal, 31(1 Supplement), 777–723.
Kourtesi, C., Ball, A. R., Huang, Y. Y., Jachak, S. M., Vera, D. M. A., Khondkar, P., Gibbons, S., Hamblin, M. R., & Tegos, G. P. (2013). Suppl 1: Microbial efflux systems and inhibitors: Approaches to drug discovery and the challenge of clinical implementation. Open Microbiologica Journal, 7, 34.
Kovač, J., Šimunović, K., Wu, Z., Klančnik, A., Bucar, F., Zhang, Q., & Možina, S. S. (2015). Antibiotic resistance modulation and modes of action of (−)-α-pinene in campylobacter jejuni. PLoS One, 10(4), e0122871.
Krishnamoorthy, S., Shah, B. P., Lee, H. H., & Martinez, L. R. (2016). Microbicides alter the expression and function of RND-type efflux pump AdeABC in biofilm-associated cells of Acinetobacter baumannii clinical isolates. Antimicrobial Agents and Chemotherapy, 60(1), 57–63.
Kumar, S., Floyd, J. T., He, G., & Varela, M. F. (2013). Bacterial antimicrobial efflux pumps of the MFS and MATE transporter families: A review. Recent Res Dev Antimicrob Agents Chemother, 7, 1–21.
Kumar, R., & Pooja Patial, S. J. (2016). A review on efflux pump inhibitors of gram-positive and gram-negative bacteria from plant sources. International Journal of Current Microbiology and Applied Sciences, 5, 837–855.
Kuroda, T., & Tsuchiya, T. (2009). Multidrug efflux transporters in the MATE family. Biochimica et Biophysica Acta. Proteins and Proteomics, 1794(5), 763–768.
Kvist, M., Hancock, V., & Klemm, P. (2008). Inactivation of efflux pumps abolishes bacterial biofilm formation. Applied and Environmental Microbiology, 74(23), 7376–7382.
Lee, M. D., Galazzo, J. L., Staley, A. L., Lee, J. C., Warren, M. S., Fuernkranz, H., Chamberland, S., Lomovskaya, O., & Miller, G. H. (2001). Microbial fermentation-derived inhibitors of efflux-pump-mediated drug resistance. Farmaco, 56(1–2), 81–85.
Lee, Y. S., Jang, K., & Cha, J. D. (2011). Synergistic antibacterial effect between silibinin and antibiotics in oral bacteria. Journal of Biomedicine & Biotechnology, 2012, 7.
Lekshmi, M., Ammini, P., Adjei, J., Sanford, L. M., Shrestha, U., Kumar, S., & Varela, M. F. (2017). Modulation of antimicrobial efflux pumps of the major facilitator superfamily in Staphylococcus aureus. Review, 4(1), 1–18.
Li, X. Z., & Nikaido, H. (2009). Efflux-mediated drug resistance in bacteria. Drugs, 69(12), 1555–1623.
Lloris-Garcerá, P., Slusky, J. S., Seppälä, S., Prieß, M., Schäfer, L. V., & von Heijne, G. (2013). In vivo Trp scanning of the small multidrug resistance protein EmrE confirms 3D structure models. Journal of Molecular Biology, 425(22), 4642–4651.
Lomovskaya, O., & Bostian, K. A. (2006). Practical applications and feasibility of efflux pump inhibitors in the clinic—A vision for applied use. Biochemical Pharmacology, 71(7), 910–918.
Lomovskaya, O., Warren, M. S., Lee, A., Galazzo, J., Fronko, R., Lee, M., Blais, J., Cho, D., Chamberland, S., Renau, T., & Leger, R. (2001). Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: Novel agents for combination therapy. Antimicrobial Agents and Chemotherapy, 45(1), 105–116.
Lu, S., & Zgurskaya, H. I. (2013). MacA, a periplasmic membrane fusion protein of the macrolide transporter MacAB-TolC, binds lipopolysaccharide core specifically and with high affinity. Journal of Bacteriology, 195(21), 4865–4872.
Mahmood, H. Y., Jamshidi, S., Sutton, J. M., & Rahman, K. M. (2016). Current advances in developing inhibitors of bacterial multidrug efflux pumps. Current Medicinal Chemistry, 23(10), 1062–1081.
Maisuria, V. B., Hosseinidoust, Z., & Tufenkji, N. (2015). Polyphenolic extract from maple syrup potentiates antibiotic susceptibility and reduces biofilm formation of pathogenic bacteria. Applied and Environmental Microbiology, 81(11), 3782–3792.
Marchi, E., Furi, L., Arioli, S., Morrissey, I., Di Lorenzo, V., Mora, D., Giovannetti, L., Oggioni, M. R., & Viti, C. (2015). Novel insight into antimicrobial resistance and sensitivity phenotypes associated to qac and norA genotypes in Staphylococcus aureus. Microbiological Research, 170, 184–194.
Maurya, A., Dwivedi, G. R., Darokar, M. P., & Srivastava, S. K. (2013). Antibacterial and synergy of Clavine alkaloid Lysergol and its derivatives against Nalidixic acid-resistant Escherichia coli. Chemical Biology and Drug Design, 81(4), 484–490.
McCrackin, M. A., Helke, K. L., Galloway, A. M., Poole, A. Z., Salgado, C. D., & Marriott, B. P. (2016). Effect of antimicrobial use in agricultural animals on drug-resistant foodborne campylobacteriosis in humans: A systematic literature review. Critical Reviews in Food Science and Nutrition, 56(13), 2115–2132.
Murakami, S., Nakashima, R., Yamashita, E., & Yamaguchi, A. (2002a). Crystal structure of bacterial multidrug efflux transporter AcrB. Nature, 419(6907), 587.
Murakami, S., Nakashima, R., Yamashita, E., Matsumoto, T., & Yamaguchi, A. (2006). Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature, 443(7108), 173.
Nakashima, R., Sakurai, K., Yamasaki, S., Hayashi, K., Nagata, C., Hoshino, K., Onodera, Y., Nishino, K., & Yamaguchi, A. (2013). Structural basis for the inhibition of bacterial multidrug exporters. Nature, 500(7460), 102.
Nguefack, J., Dongmo, J. L., Dakole, C. D., Leth, V., Vismer, H. F., Torp, J., Guemdjom, E. F. N., Mbeffo, M., Tamgue, O., Fotio, D., & Zollo, P. A. (2009). Food preservative potential of essential oils and fractions from Cymbopogon citratus, Ocimum gratissimum and Thymus vulgaris against mycotoxigenic fungi. International Journal of Food Microbiology, 131(2–3), 151–156.
Nguyen, S. T., Kwasny, S. M., Ding, X., Cardinale, S. C., McCarthy, C. T., Kim, H. S., Nikaido, H., Peet, N. P., Williams, J. D., Bowlin, T. L., & Opperman, T. J. (2015). Structure–activity relationships of a novel pyranopyridine series of Gram-negative bacterial efflux pump inhibitors. Bioorganic & Medicinal Chemistry, 23(9), 2024–2034.
Nikaido, H. (2009). Multidrug resistance in bacteria. Annual Review of Biochemistry, 78, 119–146.
Nikaido, H., & Pagès, J. M. (2012). Broad-specificity efflux pumps and their role in multidrug resistance of gram-negative bacteria. FEMS Microbiology Reviews, 36(2), 340–363.
Nishino, K., & Yamaguchi, A. (2008). Virulence and drug resistance roles of multidrug efflux pumps in Escherichia coli and Salmonella. Bioscience and Microflora, 27(3), 75–85.
Opperman, T. J., Kwasny, S. M., Kim, H. S., Nguyen, S. T., Houseweart, C., D’Souza, S., Walker, G. C., Peet, N. P., Nikaido, H., & Bowlin, T. L. (2014). Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrobial Agents and Chemotherapy, 58(2), 722–733.
Pagès, J. M., & Amaral, L. (2009). Mechanisms of drug efflux and strategies to combat them: Challenging the efflux pump of gram-negative bacteria. Biochimica et Biophysica Acta. Proteins and Proteomics, 1794(5), 826–833.
Pérez, A., Poza, M., Fernández, A., Fernández Mdel, C., Mallo, S., Merino, M., Rumbo-Feal, S., Cabral, M. P., & Bou, G. (2012). Involvement of the AcrAB-TolC efflux pump in the resistance, fitness, and virulence of Enterobacter cloacae. Antimicrobial Agents and Chemotherapy, 56(4), 2084–2090.
Perez, F., Rudin, S. D., Marshall, S. H., Coakley, P., Chen, L., Kreiswirth, B. N., Rather, P. N., Hujer, A. M., Toltzis, P., Van Duin, D., & Paterson, D. L. (2013). OqxAB, a quinolone and olaquindox efflux pump, is widely distributed among multidrug-resistant Klebsiella pneumoniae isolates of human origin. Antimicrobial Agents and Chemotherapy, 57(9), 4602–4603.
Piddock, L. J. (2006). Multidrug-resistance efflux pumps – not just for resistance. Nature Reviews. Microbiology, 4(8), 629.
Piddock, L. J., Garvey, M. I., Rahman, M. M., & Gibbons, S. (2010). Natural and synthetic compounds such as trimethoprim behave as inhibitors of efflux in gram-negative bacteria. The Journal of Antimicrobial Chemotherapy, 65(6), 1215–1223.
Radchenko, M., Symersky, J., Nie, R., & Lu, M. (2015). Structural basis for the blockade of MATE multidrug efflux pumps. Nature Communications, 6, 7995.
Ramalhete, C., Spengler, G., Martins, A., Martins, M., Viveiros, M., Mulhovo, S., Ferreira, M. J. U., & Amaral, L. (2011a). Inhibition of efflux pumps in methicillin-resistant Staphylococcus aureus and Enterococcus faecalis resistant strains by triterpenoids from Momordica balsamina. International Journal of Antimicrobial Agents, 37(1), 70–74.
Ramalhete, C., Spengler, G., Martins, A., Martins, M., Viveiros, M., Mulhovo, S., Ferreira, M. J., & Amaral, L. (2011b). Inhibition of efflux pumps in methicillin-resistant Staphylococcus aureus and Enterococcus faecalis resistant strains by triterpenoids from Momordica balsamina. International Journal of Antimicrobial Agents, 37(1), 70–74.
Ramalhete, C., Mulhovo, S., Molnar, J., & Ferreira, M. J. (2016). Triterpenoids from Momordica balsamina: Reversal of ABCB1-mediated multidrug resistance. Bioorganic & Medicinal Chemistry, 1;24(21), 5061–5067.
Ranaweera, I., Shrestha, U., Ranjana, K. C., Kakarla, P., Willmon, T. M., Hernandez, A. J., Mukherjee, M. M., Barr, S. R., & Varela, M. F. (2015). Structural comparison of bacterial multidrug efflux pumps of the major facilitator superfamily. Trends in Cell & Molecular Biology, 10, 131.
Reddy, V. S., Shlykov, M. A., Castillo, R., Sun, E. I., & Saier, M. H. (2012). The major facilitator superfamily (MFS) revisited. The FEBS Journal, 279(11), 2022–2035.
Redgrave, L. S., Sutton, S. B., Webber, M. A., & Piddock, L. J. (2014). Fluoroquinolone resistance: Mechanisms, impact on bacteria, and role in evolutionary success. Trends in Microbiology, 22(8), 438–445.
Schindler, B. D., & Kaatz, G. W. (2016). Multidrug efflux pumps of gram-positive bacteria. Drug Resistance Updates, 27, 1–13.
Schuster, S., Kohler, S., Buck, A., Dambacher, C., König, A., Bohnert, J. A., & Kern, W. V. (2014). Random mutagenesis of the multidrug transporter AcrB from Escherichia coli for identification of putative target residues of efflux pump inhibitors. Antimicrobial Agents and Chemotherapy, 58(11), 6870–6878.
Sennhauser, G., Bukowska, M. A., Briand, C., & Grütter, M. G. (2009). Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. Journal of Molecular Biology, 389(1), 134–145.
Shiu, W. K., Malkinson, J. P., Rahman, M. M., Curry, J., Stapleton, P., Gunaratnam, M., Neidle, S., Mushtaq, S., Warner, M., Livermore, D. M., & Evangelopoulos, D. (2013). A new plant-derived antibacterial is an inhibitor of efflux pumps in Staphylococcus aureus. International Journal of Antimicrobial Agents, 42(6), 513–518.
Singh, K. V., Malathum, K., & Murray, B. E. (2001). Disruption of an Enterococcus faecium species-specific gene, a homologue of acquired macrolide resistance genes of staphylococci, is associated with an increase in macrolide susceptibility. Antimicrobial Agents and Chemotherapy, 45(1), 263–266.
Singh, M., Yau, Y. C., Wang, S., Waters, V., & Kumar, A. (2017). MexXY efflux pump overexpression and aminoglycoside resistance in cystic fibrosis isolates of Pseudomonas aeruginosa from chronic infections. Canadian Journal of Microbiology, 63(12), 929–938.
Sjuts, H., Vargiu, A. V., Kwasny, S. M., Nguyen, S. T., Kim, H. S., Ding, X., Ornik, A. R., Ruggerone, P., Bowlin, T. L., Nikaido, H., & Pos, K. M. (2016). Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives. Proceedings of the National Academy of Sciences of the United States of America, 29;113(13), 3509–3514.
Smith, E. C., Williamson, E. M., Wareham, N., Kaatz, G. W., & Gibbons, S. (2007). Antibacterials and modulators of bacterial resistance from the immature cones of Chamaecyparis lawsoniana. Phytochemistry, 68(2), 210–217.
Stermitz, F. R., Tawara-Matsuda, J., Lorenz, P., Mueller, P., Zenewicz, L., & Lewis, K. (2000). 5′-Methoxyhydnocarpin-D and Pheophorbide A: Berberis species components that Potentiate Berberine growth inhibition of resistant Staphylococcus aureus. Journal of natural products, 63(8), 1146–1149.
Su, C. C., Yin, L., Kumar, N., Dai, L., Radhakrishnan, A., Bolla, J. R., Lei, H. T., Chou, T. H., Delmar, J. A., Rajashankar, K. R., & Zhang, Q. (2017). Structures and transport dynamics of a Campylobacter jejuni multidrug efflux pump. Nature Communications, 8(1), 171.
Sun, J., Deng, Z., & Yan, A. (2014). Bacterial multidrug efflux pumps: Mechanisms, physiology and pharmacological exploitations. Biochemical and Biophysical Research Communications, 453(2), 254–267.
Tanabe, M., Szakonyi, G., Brown, K. A., Henderson, P. J., Nield, J., & Byrne, B. (2009). The multidrug resistance efflux complex, EmrAB from Escherichia coli forms a dimer in vitro. Biochemical and Biophysical Research Communications, 380(2), 338–342.
Tegos, G., Masago, K., Aziz, F., Higginbotham, A., Stermitz, F. R., & Hamblin, M. R. (2008). Inhibitors of bacterial multidrug efflux pumps potentiate antimicrobial photoinactivation. Antimicrobial Agents and Chemotherapy, 52(9), 3202–3209.
Tegos, G. P., Haynes, M., Jacob Strouse, J., Khan, M. M. T., Bologa, C. G., Oprea, T. I., & Sklar, L. A. (2011). Microbial efflux pump inhibition: Tactics and strategies. Current Pharmaceutical Design, 17(13), 1291–1302.
Tocci, N., Iannelli, F., Bidossi, A., Ciusa, M. L., Decorosi, F., Viti, C., Pozzi, G., Ricci, S., & Oggioni, M. R. (2013). Functional analysis of pneumococcal drug efflux pumps associates the MATE DinF transporter with quinolone susceptibility. Antimicrobial Agents and Chemotherapy, 57(1), 248–253.
van der Putten, B. C., Pasquini, G., Remondini, D., Janes, V. A., Matamoros, S., & Schultsz, C. (2018 Jan). Quantifying the contribution of four resistance mechanisms to ciprofloxacin minimum inhibitory concentration in Escherichia coli: A systematic review. bioRxiv, 1, 372086.
Vargiu, A. V., Ruggerone, P., Opperman, T. J., Nguyen, S. T., & Nikaido, H. (2014). Molecular mechanism of MBX2319 inhibition of Escherichia coli AcrB multidrug efflux pump and comparison with other inhibitors. Antimicrob Agents Chemother, 58(10), 6224–6234.
Vargiu, A. V., & Nikaido, H. (2012). Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc Nat Acad Sci, 109(50), 20637–20642.
Venter, H., Mowla, R., Ohene-Agyei, T., & Ma, S. (2015). RND-type drug efflux pumps from gram-negative bacteria: Molecular mechanism and inhibition. Frontiers in Microbiology, 6, 377.
Wassenaar, T., Ussery, D., Nielsen, L., & Ingmer, H. (2015). Review and phylogenetic analysis of qac genes that reduce susceptibility to quaternary ammonium compounds in Staphylococcus species. European Journal of Microbiology & Immunology, 1;5(1), 44–61.
Wendlandt, S., Kadlec, K., Feßler, A. T., & Schwarz, S. (2015). Identification of ABC transporter genes conferring combined pleuromutilin–lincosamide–streptogramin A resistance in bovine methicillin-resistant Staphylococcus aureus and coagulase-negative staphylococci. Veterinary Microbiology, 177(3–4), 353–358.
Wilkens, S. (2015). Structure and mechanism of ABC transporters. F1000Prime Rep, 7, 14.
Wink, M. (2012). Secondary metabolites from plants inhibiting ABC transporters and reversing resistance of cancer cells and microbes to cytotoxic and antimicrobial agents. Frontiers in Microbiology, 23(3), 130.
World Health Organization. (2014). Antimicrobial resistance: Global report on surveillance. Geneva: World Health Organization.
Yamaguchi, A., Nakashima, R., & Sakurai, K. (2015). Structural basis of RND-type multidrug exporters. Frontiers in Microbiology, 6, 327.
Yao, H., Shen, Z., Wang, Y., Deng, F., Liu, D., Naren, G., Dai, L., Su, C. C., Wang, B., Wang, S., & Wu, C. (2016). Emergence of a potent multidrug efflux pump variant that enhances Campylobacter resistance to multiple antibiotics. MBio, 7(5), e01543–e01516.
Yin, C. C., Aldema-Ramos, M. L., Borges-Walmsley, M. I., Taylor, R. W., Walmsley, A. R., Levy, S. B., & Bullough, P. A. (2000). The quarternary molecular architecture of TetA, a secondary tetracycline transporter from Escherichia coli. Journal Molecular Microbiology, 38(3), 482–492.
Yoshida, K. I., Nakayama, K., Ohtsuka, M., Kuru, N., Yokomizo, Y., Sakamoto, A., Takemura, M., Hoshino, K., Kanda, H., Nitanai, H., & Namba, K. (2007). MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 7: Highly soluble and in vivo active quaternary ammonium analogue D13-9001, a potential preclinical candidate. Bioorg Med Chem, 15(22), 7087–7097.
Yu, J. L., Grinius, L., Hooper, D. C. (2002, March 1). NorA functions as a multidrug efflux p rotein in both cytoplasmic membrane vesicles and reconstituted proteoliposomes. Journal of Bacteriology, 184(5), 1370–1377.
Zhang, C. Z., Chen, P. X., Yang, L., Li, W., Chang, M. X., & Jiang, H. X. (2018). Coordinated expression of acrAB-tolC and eight other functional efflux pumps through activating ramA and marA in Salmonella enterica serovar Typhimurium. Microbial Drug Resistance, 24(2), 120–125.
Acknowledgement
Samreen is thankful to UGC, New Delhi, for providing Non-Net Fellowship.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Samreen, Ahmad, I., Qais, F.A., Maheshwari, M., Rumbaugh, K.P. (2019). Efflux Pump Inhibitors and Their Role in the Reversal of Drug Resistance. In: Ahmad, I., Ahmad, S., Rumbaugh, K. (eds) Antibacterial Drug Discovery to Combat MDR. Springer, Singapore. https://doi.org/10.1007/978-981-13-9871-1_12
Download citation
DOI: https://doi.org/10.1007/978-981-13-9871-1_12
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-9870-4
Online ISBN: 978-981-13-9871-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)