Abstract
Global cereal demand will increase up to 38% by 2025, and to achieve it in a sustainable way, 60% increase in global nitrogen (N) use will be necessary. In cereals ~30 to 50% of the applied N is taken up by the crop, and the rest is lost in the environment causing pollution. Hence, improvement of N use efficiency (NUE) in cereals is really important. The NUE is the total biomass or grain yield produced per unit of applied N fertilizer. Soil and plant management practices play a key role toward enhancing N recovery, but again it greatly depends on environmental conditions. Another option for improvement of NUE is the genetic strategy. Broadly, NUE has two components, N uptake efficiency (NUpE), which is N acquisition by the plant per unit of available N in the soil, and N utilization efficiency (NUtE), which is yield per unit of acquired N by the plant. As NUtE is directly related to the crop yield, it depends on subcomponent N assimilation, remobilization, and finally efficient utilization of assimilated N for starch biosynthesis in the grain. Understanding the mechanisms and gene regulating of these processes, exploiting genotypic variant in each subcomponent (N uptake, assimilation, and remobilization) to find genes and superior alleles is crucial for the improvement of NUE in crop plants. In addition, the studies on starch metabolism during grain filling are an important factor for N utilization. To study this, genotypes with similar background of uptake and assimilation but differing in grain filling should be taken into consideration. Global metabolomic profiling of these genotypes, transcriptome profiling, identification, and mapping of quantitative trait loci (QTLs) in combination with marker-assisted selection (MAS), analyzing mutants defective in their normal response to N limitation, and studying plants that show better growth under N-limiting conditions are different options to study the N-utilization efficiency and gene identification. In the first topic, we have highlighted the N application and its effect on yield in cereals. Introduction of N-responsive genotype during green revolution has enhanced yield, but indiscriminate use of fertilizer mainly N fertilizer has caused severe damage to environment. In the subsequent topic, we have defined NUE as a whole; later the main focus was on biological NUE and their different components. Thereafter we described strategies for genetic improvement to reduce N use without much compromising yield. Primarily we tried to highlight candidate genes and their role in NUE reported in cereals as well as model plant system. We have also described the advance molecular techniques to identify the gene in strategic manner. As a part of molecular breeding, QTL identification and its introgression are described in one of the topics at the last part.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
6.1 Introduction
During last 50 years, the global human population has increased from 5 billion to 9 billion, which primarily compelled to increase the staple foodstuff production. Population in India has increased from 0.36 billion to 1.21 (fourfold). The major two cereals (rice and wheat) production has increased from 21 to 103 million tonnes and from 6 to 88 million tonnes in India (GOI 2012). These two cereals occupy the major cultivated area and are used as staple food throughout the world. Wheat alone provides nearly 55% of the carbohydrate and 20% of the calories consumed globally (Breiman and Graur 1995).
Nitrogen (N) is a key element involved in various life processes of plants like biosynthesis of amino acid, nucleic acid, chlorophyll, cofactors, etc. Plants uptake on average 40–50 mg N to produce 1 kg of dry weight, whose limited supply in soil affects its availability to plant making it a critical limiting element for growth and development (Robertson and Vitousek 2009). External supply of N in the form of chemical fertilizers has become necessarily a need to sustain food production. In the last five decades, cereal production has boosted up to 260%, but this increase in production was at the cost of sevenfold increase in N fertilizer application (Ladha and Chakraborty 2016). As per the input survey data of 2006–2007, the average N fertilizer use of wheat in India is 122 kg ha−1 which makes it the third most N fertilizer consumable crop after sugarcane and cotton. Presently, India stands second in N fertilizer use with 17.2 million tonnes after China. The global N fertilizer demand is expected to increase from a total of 105.3 million tonnes in 2011 to 119 million tonnes in 2018 (FAO 2015). At the same time, global cereal demand is also expected to increase up to 38% by 2025, and to achieve this, 60% increase in global N use will be needed till that time (Dobermann 2005). In case of the wheat crop, the nitrogen use efficiency (NUE) is 40%, whereas in rice, it varies from 20–40% (under submergence condition) to 40–60% (under upland condition) (Raghuram et al. 2007); the remaining N gets lost in the environment and cause pollution. Intensive use of N fertilizers in agricultural production is continuing, leaving the negative consequence to environment. Because of these consequences, the diversity and functioning of nonagricultural system are facing detrimental effect, i.e., eutrophication of freshwater and marine water due to leaching of N (Hirel et al. 2007). In addition to environmental effect, N fertilization also increases input cost because the Haber-Bosch process of N production consumes natural gas as high as 873 m3 for producing 1 metric ton of fertilizer N (Xu et al. 2012). One percentage increase in NUE for cereal production could save around 1731 crore rupees ($234) fertilizer cost (Mosier and Syers 2004). Hence, developing cultivar which can efficiently use N is important to render these problems.
Nitrogen use efficiency has several definitions, but for plant biologists and breeder’s, definition and expression given by Moll et al. (1982) is mostly used. The NUE is calculated by dividing grain weight to total available soil N. Nitrogen use efficiency is the multiplication of two components, N uptake efficiency (NUpE) and N utilization efficiency (NUtE). The NUpE is calculated by dividing N in plant harvest to total available N in soil, whereas NUtE is obtained by dividing grain dry mass to total N in plant harvest. In totality, NUE is defined as the total biomass or grain yield produced per unit of available N, which mostly comes from applied fertilizer in case of cereals.
Genetics and biochemical mechanisms involved in each component of N use have been widely studied in model plants as well as cereal crops to identify many key genes and regulators which are potential targets for improving crop NUE and also understanding genetic basis of NUE.
6.2 Nitrogen Application and Yield Enhancement in Cereals
Nitrogen derives from atmosphere which is subsequently transformed and transported to pedosphere and hydrosphere through the process of biological nitrogen fixation (BNF). Therefore, N is unique among major nutrient elements which mostly form by weathering of rocks and hence also present in less amount in soil. Atmosphere contains a large, well-mixed, biologically non-available N pool, a relatively small part of which is converted into a biologically available or reactive N pool primarily through BNF. Industrial N fixation has, however, become more important in agriculture, since taking care of the developing demand for sustenance has resulted in huge increments in the utilization of N fertilizer.
Nitrogen is one of the nutrients that most often limits crop production, particularly in the major staple cereals. In the tropics, lowland rice produces an approximate yield of 10–15 qha−1 using naturally available N derived from processes like BNF by diazotrophs and mineralization of soil N and wet and dry depositions from the atmosphere. Similarly, wheat and maize yields of 10–15 qha−1 were obtained without any fertilizer application (Janssen et al. 1990; Witt et al. 1999; Pathak et al. 2003). Such agro-systems have been sustained, though with low yields, for numbers of years without external N supplement (Fischer 2000). Whether it is good or bad, environmentally friendly or not, that is a matter of debate, but additional N application is necessary to increase the yield to three- to fourfold to feed the ever-increasing population (Lassaletta et al. 2014).
6.3 Green Revolution and Effect of Nitrogen Fertilizer in Environment
During Green Revolution the major emphasis was on genetic improvement of crop to produce high-yielding varieties (HYVs). Another aspect was intended to shortening of maturity time in cereal to ultimately achieve greater cropping intensity. Rice-wheat system adopted in the Indo-Gangetic Plain is contribution of shorter crop growing period. And lastly improved inputs, including fertilizer, irrigation, and, to a certain extent, pesticides, also contributed significantly in the Green Revolution.
Therefore, this increased N fertilization resulted in many negative impacts on the environment. Added N allows farmers to simplify plant community by displacing need of N fixing plant. Addition of N to soil system also interferes with their inhabiting microbial community and their associated processes like decomposition, nitrification, denitrification, etc. (Robertson and Vitousek 2009). Other than this, losses of fertilizer N to the soil and atmospheric environment cause serious pollution which is the fact why the plant use efficiency of N fertilizers is lower and thus higher cost of cultivation. Environmental pollution includes eutrophication of freshwater (London 2005) and marine ecosystems (Beman et al. 2005) which leads to augmentation of algal growth. Subsequently, after the death and decomposition of algae in water bodies, the content of organic matter in the water increases. In turn, this consumes higher amount of oxygen, causing drop in its level and creating conditions of hypoxia or dead zone. In absence of adequate oxygen availability, the aquatic organisms like fish, crabs, and others die (Rabalais et al. 2002). Some soluble portion of N leaches down as nitrate (\( {\mathrm{NO}}_3^{-} \)) and pollute groundwater reserve (Powlson et al. 2008). In addition to leaching losses, reactive N can also lose to the atmosphere in the form of N-containing gases like ammonia (NH3), nitrogen oxide (N2O), nitric oxide (NO), etc. causing buildup of greenhouse gases (Robertson and Vitousek 2009).
6.4 Definitions of Nitrogen Use Efficiency (NUE)
The simplest definition of NUE is “the ratio of yield either in terms of grain or biomass to the total N (soil N and fertilizer applied)” (Good et al. 2004). However, NUE is a complex concept, and its expression and measurement must be context dependent. Complexity in meaningful definition and expression of NUE is dependent on many factors: (i) nutrient source including soil N, manure/fertilizer, or atmosphere and (ii) factor influencing crop nutrient demand like environmental factor, crop management practices, or genotypic makeup of plant. In addition to these factors, variation in NUE expression also depends on (iii) kind of data available and (iv) scale of interest (for plant breeder, interest may be a single plant, and for policy-making it may be as large as a country). Some of the measurements of NUE are partial factor productivity (PEP), agronomic efficiency (AE), partial nutrient balance (PNB), apparent recovery efficiency (RE), and N physiological use efficiency (NpUE) (Dobermann 2005, 2007; Fixen et al. 2015) which mainly denote efficiency of fertilizer input and are used by agronomist and policy-makers (these are explained briefly below). Plant biologist and breeders are mostly interested in another concept of NUE where it is mainly presented as plant phenotypic trait. Plant NUE (Xu et al. 2012) is a combination of two plant physiological components NUpE and NUtE. This book chapter is mainly focused on plant NUtE, and it is elaborated later in the section.
Partial Factor Productivity (PEP)
It is the expression of production efficiency calculated as amount of crop yield produced per unit of nutrient added. The PEP can be calculated at farm, regional, or national level if proper statistical record of fertilizer input as well as crop yields is available. It is also dependent on cropping system, which mainly explains productivity of a cropping system within a region in comparison to its N input and is a long-term indicator of trend.
Agronomic Efficiency (AE)
It is calculated as unit of grain yield increased per unit of nutrient added. It denotes degree of gain in productivity by nutrient input. Usually short-term impact of applied nutrient on productivity is indicated by AE, but if long-term trials are conducted, then contribution of fertilizer input to crop yield can also be indicated by AE.
Partial Nutrient Balance (PNB)
It is denoted as nutrient output per unit of nutrient input. It expresses the amount of nutrient being removed from the system in relation to amount of application. So basically, it expresses nutrient recovery rate. If PNB is close to 1, then it is assumed that fertility of soil will be sustained at steady state. However, there is lacuna in this assumption because removal of N from soil by means of losses like leaching and erosions is not considered. It indicates trends in long term and is more advantageous when used along with soil fertility record.
Apparent Recovery Efficiency (RE) or Apparent N Recovery Rate (ANR)
It is the relative amount of nutrient uptake in aboveground parts of fertilized and unfertilized crop plants as a function of quantity of applied nutrient. It tells what amount of fertilizer N is acquired by the plant, hence also indicating potential nutrient loss from that cropping system and management practice.
N Physiological Use Efficiency (NpUE)
It is the ratio of net increased grain weight to net increased N uptake in aboveground plant parts with and without application of fertilizer N. It is more like uptake efficiency denotes ability of plants to transform nutrients acquired by soil and fertilizer to economic yield. This expression also used to denote plant NUE.
NUE as Phenotypic Trait
Many people presented different definitions and evaluation methods for NUE (reviewed in Good et al. 2004; Fageria et al. 2008), but the appropriate way to estimate it depends on the crop, its harvest product, and whether the researcher wants to analyze specific physiological processes involved in NUE. The most common and widely used definition among plant breeders is the definition given by Moll et al. (1982). He defined it as grain production per unit of N available in the soil, and its expression is as follows:
where Gw, grain weight; Ns, N supply (gplant-1); and Nt, total N in plant at maturity. Ratio of Gw and Ns denotes NUE which is the multiplication of its two components NUpE (Nt/Ns) and NUtE (Gw/Nt).
Another definition is usage index (UI). Usage index is calculated by multiplying total plant biomass to the ratio of the total plant biomass and total plant N. For cereals, NUE is to be described as NUEg, which is the grain production per unit of N available and is a more appropriate presentation than UI plant could produce a lot of biomass for every unit N (high UI) without changing over the procured N to seed production; therefore, in spite of having high UI, it should have a low NUEg. Crop improvement mainly focused on the improvement of cereal yield is the main driver to bring economic prosperity in developing countries like India from the last five decades (Conant et al. 2013). Scientific approach to plant breeding accelerated its rate during this period, but this increase in crop production per unit area also owed intensive use of N nutrient input in the form of synthetic N fertilizers. All the selection for high-yielding, lodging-resistant, short-stature cultivars of cereals are carried out in high N environment which results in high N-responsive varieties, which means giving more yields with increased N input. Although cereal boosted the food production by about 260%, the contribution of N fertilizer to N input increased by 45% (Ladha et al. 2016). Annually 100 million tonnes of N fertilizers are applied in cropland and pastures, globally. Approximately half of this N input is taken by three major cereal crops, namely rice, wheat, and maize (Ladha and Chakraborty 2016). By seeing the rate of increase in N fertilizer input in these crops for the last 50 years, it is assumed that to meet 3 million tonnes of global requirement of cereal by 2050 and with 7% increase in crop land area, fertilizer N input will increase by 65% in these three crops with no change in NUE. Synthetic fertilizer production would also increase two times by 2050 (Ladha et al. 2016).
6.5 Components of Nitrogen Use Efficiency (NUE)
6.5.1 Nitrogen Uptake
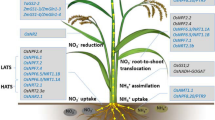
The factors like soil type, pH, temperature, precipitation, and wind affect the N availability and its ionic form in soil both spatially and temporarily. Therefore, the N form to be taken by plant will depend on its adaptability to soil condition. Forest soils are mostly acidic; hence plant adapted to this condition prefers ammonium and amino acids, similarly plants adapted to more aerobic soil and high pH prefer \( {\mathrm{NO}}_3^{-} \) form as in the case of most of crop plants except lowland rice because it grows in anaerobic condition. For N uptake transporters and root system architecture plays a crucial role.
6.5.1.1 Nitrate Transporters
Nitrate form taken by the plants through \( {\mathrm{H}}^{+}/{\mathrm{NO}}_3^{-} \) (hydrogen/nitrate) cotransports mechanism (Ullrich 1987). Many experiments showed \( {\mathrm{NO}}_3^{-} \) uptake causes alkalization of medium due to strong ionic difference (Mistrik and Ullrich 1996); this occurs due to 2:1 stoichiometry between \( {\mathrm{H}}^{+}/{\mathrm{NO}}_3^{-} \) cotransporter. Nitrogen availability in soil fluctuates, and to counteract this plants have developed two types of transporter systems (Epstein 1972). They are called low-affinity transport system (LATS) and high-affinity transport system (HATS). For both the forms, inducible and constitutive types coexist and act coordinately to uptake nutrient from soil through roots and distribute it all over the plants. Low-affinity uptake system acquires nutrients in high external substrate concentration (>0.5 mM for \( {\mathrm{NO}}_3^{-} \)), while the high-affinity system uptakes substrate at low external concentrations (< 0.2 for \( {\mathrm{NO}}_3^{-} \)). Along with these, some dual-affinity transporters are also reported which act as both LATS and HATS (Liu et al. 1999). In model plant Arabidopsis thaliana, transport system for \( {\mathrm{NO}}_3^{-} \) is well studied. Four families, namely, nitrate transporter 2 (NRT2) transporters, NRT 1/peptide transporter family (NPF) transporters, chloride channel family (CLC) transporters, and slow anion-associated channel homolog (SLAC/SLAH) (Orsel et al. 2002a; Negi et al. 2008; Barbier-Brygoo et al. 2011; Krapp et al. 2014; Léran et al. 2014), are responsible for \( {\mathrm{NO}}_3^{-} \) uptake, distribution, and storage. Among these, transporters from NPF and NRT2 family are responsible for \( {\mathrm{NO}}_3^{-} \) uptake from root. In addition to it, some members in NPF and NRT2 are also involved in \( {\mathrm{NO}}_3^{-} \) sensing and signaling. The NRT2 family consists of high-affinity \( {\mathrm{NO}}_3^{-} \) transporters (Krapp et al. 2014). A total of seven members are known from Arabidopsis NRT2 family. The NRT2.1 and NRT2.2 exclusively express in root, while the NRT2.5 and NRT 2.6 express in leaves along with root, whereas NRT2.7 expresses in leaves (Orsel et al. 2002b). Many NRT2 gene expressions are regulated by availability of \( {\mathrm{NO}}_3^{-} \) and some other factors (Zhuo et al. 1999; Orsel et al. 2002b, 2006). AtNRT2.1 interacts with another protein NAR2 and forms two component \( {\mathrm{NO}}_3^{-} \) uptake systems (Orsel et al. 2006). The ZmNRT2.1 and ZmNRT2.2 are the main transports controlling high-affinity \( {\mathrm{NO}}_3^{-} \) uptake in maize plant (Garnett et al. 2013). Lupini et al. (2016) showed ZmNRT2.1 expression and localization influenced by ZmNAR2.1; expression of these two genes was also correlated by \( {\mathrm{NO}}_3^{-} \) influxes. Five members of NRT2 family are characterized in rice (Feng et al. 2011), unlike Arabidopsis each showing different affinities to \( {\mathrm{NO}}_3^{-} \) and N supply dependent regulation also differs. Three NRT2 transporters (NRT2.1, NRT2.2, and NRT2.3a) interact with OsNAR2.1 at messenger RNA (mRNA) as well as protein levels to influence \( {\mathrm{NO}}_3^{-} \) uptake over both high- and low-concentration ranges (Yan et al. 2011).
For low-affinity uptake, one family of \( {\mathrm{NO}}_3^{-} \) transporters, NPF (NRT1), is responsible, but the exception is NRT 1.1/NPF6.3/CHL1 which is a dual-affinity transporter (Wang et al. 1998; Liu et al. 1999); along with it, NRT1.1 also act as \( {\mathrm{NO}}_3^{-} \) sensor (Ho et al. 2009). Fifty-three NRT genes have been reported till date in Arabidopsis, of which 51 gene families may have unique functions of each because its expression is found to be limited to a specific tissue (Tsay et al. 2007). The NRT 1.1 initially discovered as low-affinity transporter by Tsay et al. (1993) was later found to act as high-affinity transporter at low \( {\mathrm{NO}}_3^{-} \) concentration after phosphorylation of Thr101 (Liu and Tsay 2003). The protein complex CIPK23-CBL9 (CIPK, CBL-interacting protein kinase; CBL, calcineurin B-like protein) phosphorylates Thr101 at low \( {\mathrm{NO}}_3^{-} \) condition which results in switching of NRT1.1 to HAT (Liu and Tsay 2003; Ho et al. 2009). Another protein ABI2, a phosphatase, also regulates NRT1.1 and enhances rate of transport by inhibiting CIPK23-CBL1 complex phosphorylation (Léran et al. 2015). The location of NRT1.1 is plasma membrane of epidermis and root tip, because gene expression is observed in these locations along with it in mature part of root; it is also located in the cortex and endodermis (Huang et al. 1999). Another member NRT1.2/NPF4.6/AIT1 is also involved in soil \( {\mathrm{NO}}_3^{-} \) uptake, and it constitutively is expressed in root epidermis, but it comes under LATS (Liu et al. 1999). Interestingly NRT1.2 also transports abscisic acid (ABA) and plays an important role in transpiration and seed dormancy (Kanno et al. 2012). After uptake from soil, \( {\mathrm{NO}}_3^{-} \) is transported and distributed to all tissue. For root-to-shoot transport of \( {\mathrm{NO}}_3^{-} \) across several cell membranes, it is carried out by NRT1.5, NRT1.8, and NPF2.3 (Lin et al. 2008; Li et al. 2010; Taochy et al. 2015). Another NRT1 family member, NPF2.7/NAXT1, involves in \( {\mathrm{NO}}_3^{-} \) efflux to maintain \( {\mathrm{NO}}_3^{-} \) homeostasis (Segonzac et al. 2007).
6.5.1.2 Ammonium Transporter
Nitrogen is also taken up as ammonium ion (\( {\mathrm{NH}}_4^{+} \)) by plants growing under anaerobic condition. The ammonium transporters (AMT) are responsible for uptake and transport of \( {\mathrm{NH}}_4^{+} \). In Arabidopsis, six genes from this family have been reported (Gazzarrini et al. 1999), whereas in rice, which is a species well-adapted to \( {\mathrm{NH}}_4^{+} \) nutrition, ten genes are reported (Sonoda et al. 2003). Analysis of single and multiple mutants from this gene family in Arabidopsis revealed AMT1.1 and AMT1.3 as main transporters conferring 30–35% transport followed by AMT1.2 which confers 18–25%. The AMT 1.5 has lower Km than that of the previous two transporters (AMT 1.1 and 1.3), but in spite of that, it is a low-capacity transporter (Yuan et al. 2007). Spatial organizations of these AMT transporters are also very interesting. Outer root cells and root hairs contain transporters with highest \( {\mathrm{NH}}_4^{+} \) affinity (AMT 1.3, AMT1.5) to take \( {\mathrm{NH}}_4^{+} \) from soil. The electrochemical gradient between the vacuole and cytosol is responsible for NH3 import in exchange of \( {\mathrm{NH}}_4^{+} \) exported to the external side of the vacuole. An intrinsic protein TIP in tonoplast plays an important role in NH3 import to vacuole (Loque et al. 2005).
6.5.1.3 Amino Acid Transporters
Plant also takes up amino acid from soil in very trace amount. Rentsch et al. (2007) reported that at least 5 gene families comprised of a total of 65 genes are considered as putative transporter of amino acid. A member of ATF (amino acid transporter) family, LHT1 (lysine/histidine transporter), is important for uptake of neutral and acidic amino acid by roots. Under high external concentration of amino acids, the uptake of uncharged amino acids is taken care of by AAP1 (amino acid permease 1) (Lee et al. 2007). The AAP5 is responsible for uptake of cationic amino acids (Svennerstam et al. 2008).
6.5.1.4 Root System Architecture
Root system architecture is an important trait which affects the performance of crop in various stresses like drought, nutrient, and mineral toxicity and has implication in providing tolerance from these stresses (Manske and Vlek 2002). Tendency of plants to capture limited resources in soil is greatly influenced by the capacity of its root system to explore and forage. The root system architecture (RSA) mainly comprises of primary roots, lateral roots, and accessory roots. These are the key determinants of NUE and water use efficiency (WUE). At minor scale, root hairs are also included in root system (RS). It alleviates uptake of water and nutrient by increasing surface area. Many factors influence RSA, one of which is the form and concentration of N (Marschner 1995). Localized supply of \( {\mathrm{NH}}_4^{+} \) and \( {\mathrm{NO}}_3^{-} \) is responsible for initiation and elongation of lateral roots, respectively (Zhang and Forde 1998; Lima et al. 2010). Nitrate-dependent root elongation is regulated by miR393 (micro RNA 393) and AFG3 (auxin signaling F-box 3), and AFG3 is induced by \( {\mathrm{NO}}_3^{-} \) itself and miR399 by N metabolites (Vidal et al. 2010). In addition to these, NRT1.1 also regulates lateral root proliferation. It senses external N and also induces N signaling pathway by activating ANR1 (MAD box gene) (Remans et al. 2006; Ho et al. 2009). The \( {\mathrm{NH}}_4^{+} \)-regulated root growth is governed by some AMTs like AMT1.3 and a GMPase (GDP mannose pyrophosphorylase) which is encoded by HSN1 (hypersensitive to \( N{H}_4^{+} \)) (Qin et al. 2008; Lima et al. 2010).
6.5.2 Nitrogen Assimilation
Nitrogen assimilation is a vital process in controlling growth and development of plants. Plants take N from soil in the form of \( {\mathrm{NO}}_3^{-} \), ammonium (NH4), and in small amount as amino acid. Ultimate substrate for amino acid biosynthesis is NH3; therefore \( {\mathrm{NO}}_3^{-} \) form has to first reduce to \( {\mathrm{NH}}_4^{+} \). Reduction of \( {\mathrm{NO}}_3^{-} \) to \( {\mathrm{NH}}_4^{+} \) takes place in two steps. In the first step, \( {\mathrm{NO}}_3^{-} \) reduced to form \( {\mathrm{NO}}_2^{-} \) (nitrite) in cytosol by the action of enzyme nitrate reductase (NR) (Meyer and Stitt 2001). After that, \( {\mathrm{NO}}_2^{-} \) is transported to chloroplast where it gets reduced to \( {\mathrm{NH}}_4^{+} \) by nitrite reductase enzyme (NiR). Inorganic NH3 is then assimilated to amino acid glutamine and glutamate which serve to translocate organic N from source to sink (Peoples and Gifford 1993; Roche et al. 1993; Lam et al. 1996). Enzyme NR is homodimer, and each monomer is attached to three prosthetic groups: (i) flavin adenine dinucleotide [cytochrome b reductase, binding NAD(P)H] (nicotinamide adenine dinucleotide phosphate), (ii) a heme (cytochrome b), and (iii) a molybdenum cofactor (MoCo) (site for \( {\mathrm{NO}}_3^{-} \) binding and reduction). The nitrate reductase activity (NRA) is considered as the rate-limiting step in the \( {\mathrm{NO}}_3^{-} \) assimilating pathway, and different genotypes of a species differ in NRA (Gniazdowska-Skoczek 1997; Bussi et al. 1997; Marwaha 1998). The NRA is induced by light in plants (Li and Oaks 1994; Kronzucker et al. 1995), but effect of light can be replaced by glucose, sucrose, or acetate (Galvan et al. 1996; Pajuelo et al. 1997; Sivasankar et al. 1997; Gniazdowska et al. 1998). Two classes of genes, Nia and Cnx, code for NR apoenzyme and MoCo cofactor, respectively. Most of the reports showed NR localized in cytoplasm, but there is some evidence of its localization in plasma membrane also (maize root and barley) (Ward et al. 1989). The next enzyme of pathway, NiR, is located in chloroplast and encoded by Nii gene whose number varies from one to two in different species (Meyer and Stitt 2001).
Ammonium is further assimilated to form amino acid. Nitrate reduction, photorespiration, or breakdown of amino acid generates \( {\mathrm{NH}}_4^{+} \) which is assimilated in plastid/chloroplast by GS (glutamine synthetase)/GOGAT (glutamine-2-oxoglutarate aminotransferase or glutamate synthase) cycle. The first step of fixation of \( {\mathrm{NH}}_4^{+} \) to amino acid is catalyzed by GS. It fixes \( {\mathrm{NH}}_4^{+} \) group to the δ-carboxylic group of glutamate to form glutamine. This step consumes an ATP. The second enzyme GOGAT transfers δ-amide group of glutamines to 2-oxoglutarate and forms two molecules of glutamate. Therefore, the net outcome of GS/GOGAT cycle is glutamate which further forms another amino acid by transferring its amino group to other carbon (C) skeletons with the help of aminotransferases or transaminases (Forde and Lea 2007). All N-containing molecules like chlorophyll, protein, secondary metabolites, nucleic acid, cytochrome/phytochrome, etc. are subsequently synthesized by a specific amino acid precursor. As we see, N metabolism requires energy and C skeletons, which comes from C-metabolism, so there must be crosstalk between these two pathways. Evidences in support of it are as follows: (a) Reduction of \( {\mathrm{NO}}_3^{-} \) also requires parallel C oxidation to form 2-oxoglutarate through respiratory pathway (Foyer et al. 2011). (b) Correlation between starch and protein contents has been always found in plants (Sulpice et al. 2009). (c) N availability affects partitioning of assimilated C between carbohydrates and organic acids (Foyer et al. 2011).
Enzymes GS and GOGAT exist in the form of different isoenzymes, and individual isoenzymes have been proposed to play a role in three major NH3 assimilation processes: primary N assimilation, re-assimilation of photorespiratory NH3, and re-assimilation of recycled N (Lam et al. 1996). Traditional assignment of GS/GOGAT isoenzyme functions is based on their organ-specific distribution. Chloroplastic GS2 and Fd-GOGAT (ferredoxin-GOGAT) are predominant GS-GOGAT enzymes located in leaves therefore proposed to involve in primary assimilation of NH3 to glutamine and glutamate. Photorespiratory mutants also show they are specifically defective in enzyme GS2, hence the also highlighted role of these two isoforms in photorespiratory NH3 re-assimilation. Catabolic processes include protein breakdown, deamination of amino acid, and biosynthetic pathways which release ammonia (met hionine, isoleucine, phenylpropanoid, and lignin biosynthesis) (Miflin and Lea 1976; Lea 1993). These processes are highest during seed germination and senescence of leaf. Cytosolic GS1 and NADH-GOGAT are involved in these processes (Stewart et al. 1980; Lea et al. 1990). In higher plants, chloroplastic GS2 is coded by a single nuclear gene GLN2 and cytosolic GS1 by multiple GLN1 genes (Peterman and Goodman 1991; Oliveira and Coruzzi 1999). Expression of GLN2 is primarily observed in leaves and is regulated by light via phytochrome (Peterman and Goodman 1991; Oliveira and Coruzzi 1999). By contrast, the GLN1 genes encoding cytosolic GS1 isoenzymes are expressed at higher levels in roots (Oliveira and Coruzzi 1999).
In addition to glutamine synthase and GOGAT, three more enzymes playing important roles in \( {\mathrm{NH}}_4^{+} \) assimilation are cytosolic asparagine synthase (AS), carbamoyl phosphate synthase (CPS), and mitochondrial NADH-glutamate dehydrogenase (GDH). The AS catalyzes ATP-dependent transfer of amide group of glutamines to aspartate and generates glutamate and asparagine. Some evidences also showed utilization ofNH3 as substrate (Lam et al. 2003; Masclaux-Daubresse et al. 2006). Three genes ASN1, ASN2, and ASN3 encode AS. Asparagine is suitable for long-distance transport and storage of fixed N because it has high N/C ratio than glutamine (Rochat and Boutin 1991; Lam et al. 2003), but all four molecules glutamate, glutamine, aspartate, and asparagine have shown to be used for translocation from source to sink in different plant parts (Lea and Miflin 1980; Peoples and Gifford 1993).
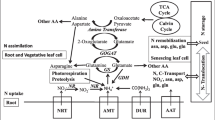
The CPSase synthesizes carbamoyl phosphate, using \( {\mathrm{NH}}_4^{+} \) or amide group of glutamine, bicarbonate, and ATP. Synthesis of carbamoyl phosphate is carried out within plastid which further acts as a precursor of arginine and citrulline. The CPSase is a heteromeric enzyme, and its small and large subunits are encoded by genes CarA and CarB, respectively (Potel et al. 2009). Mitochondrial NADH-glutamate dehydrogenase (GDH) transaminates glutamate reversibly in response to high \( {\mathrm{NH}}_4^{+} \) concentration under stress (Skopelitis et al. 2006). However, glutamate deamination has been shown as major catalytic activity of this enzyme in plant cells (Masclaux-Daubresse et al. 2006) (Fig. 6.1).
Schematic presentation of key enzymes involved in primary N assimilation. Various transporter imports \( {\mathrm{NO}}_3^{-} \), NH3, and trace amount of urea into cytoplasm. Cytoplasm located in NR reduces \( {\mathrm{NO}}_3^{-} \) to \( {\mathrm{NO}}_2^{-} \), and urease converts urea into NH3. \( {\mathrm{NO}}_2^{-} \) and NH3 are transported to plastid where enzyme NiR GS2, fd-GOGAT CPSase, incorporates the NH3 into amino acids. Carbon skeleton for amino acid biosynthesis is obtained by mitochondrial respiratory pathway. Amino acids are transported in the form of glutamine and asparagine through phloem
6.5.3 Nitrogen Remobilization
Recycling of N by-product of various catabolic processes is necessary for efficient utilization of primary assimilated N (Lam et al. 1996). These catabolic processes embody protein catabolism, amino acid deamination, and specific synthesis reactions related to methionine, isoleucine, phenylpropanoid, and lignin biosynthesis (Miflin and Lea 1980; Lea 1993). In plant’s entire growth period, there are two foremost times when re-assimilation of cast-off NH3 to glutamine and glutamate is maximum. First is germination, when seed storage proteins are catabolized and amino acids are transferred to growing seedling (Lea and Miflin 1980), and second is during senescence of leaves where amino acids are transported to developing seed (Miflin and Lea 1976). Increased activity of cytosolic GS1, NADH-GOGAT, AS, and GDH during this process supports involvement of these enzymes in remobilization (Stewart et al. 1980; Lea et al. 1990). Enormous N is obtained during leave senescence due to extensive degradation of photosynthetic proteins of leaves. Plants can tap this N to enhance the nourishment of developing organs, for example, new leaves and seeds. Up to 95% of seed protein is taken from amino acids that are traded to the seed after the degradation of existing proteins in leaves (Taylor et al. 2010). Glutamine and asparagine play key roles in rendering N available for remobilization from the senescing leaves (Masclaux-Daubresse et al. 2008). These amino acids are carried from source tissue to sink via phloem. Rate of phloem loading is determined by activity of amino acid transporters present in sieve element (Tilsner et al. 2005). Phloem loading of amino acids is taken care of by members of AAP which are the nonspecific amino acid transporters (Koch et al. 2003). Uptake of amino acids by root is facilitated by LHT1 transporter which is lysine/histidine transporter with very less Km. This xylem-derived amino acids are supplied to mesophyll cell by the same transporter (Hirner et al. 2006). Induced expression of LHT1 gene during leaf senescence highlights its role in N remobilization. One \( {\mathrm{NH}}_4^{+} \) transport AMT1.1 and a \( {\mathrm{NO}}_3^{-} \) transporter NRT2.5 also showing enhanced expression during leaf senescence suggests that inorganic N might also be mobilized in senescing tissues. A key step for economical N remobilization is phloem loading; however, whether or not it’s limiting for NUE remains to be evidenced. Other processes, like sink strength, may be limiting steps for economical N remobilization from senescing leaves (Fig. 6.2).
Schematic representation of N remobilization process and key enzymes involved in it. During senescence degradation of chloroplast and other proteins occurs, degraded products are transported to vacuole for further degradation via SAV (senescence-associated vacuoles). Similarly recycling of NH3 occurs in different organelles (mitochondria, cytosol) of mesophyll and companion cell. These events generate NH3 which is reduced to glutamine glutamate and asparagine by the action of enzyme GDH, GS1, NADH-GOGAT, and AS. Schematic presentation of key enzymes involved in N assimilation remobilization
6.6 Strategies for Genetic Improvement
6.6.1 Playing with Candidate Genes (CGs)
Nitrogen use efficiency in crop plants is genetically controlled, and therefore natural variation in NUE in different genotype of a crop species is common phenomena and reported in many crops. This variation arises either due to one or more components of NUE including total N uptake, post-anthesis N uptake, N remobilization, and N assimilation. Therefore, genotypes must be screened for its performance for individual traits. Some NUE components are developmental-specific like N uptake, so screening for uptake efficiency must be at different developmental stages. Another important aspect is the screening of NUE variant in different N regimes as different genotypes behave differently in low- and high-N doses. Under present scenario there is a need to find out genotypes which are efficient in N use under low-N dose and also give acceptable yield. Maintaining acceptable yield is important because plant will still be called N use-efficient if it requires less N and also gives less yield (as NUE is ratio between yield and available N). Once genotypes/germplasm for this is identified, it will be exploited to identify the genes and pathways underling NUE, mapping quantitative trait loci (QTLs) and markers associated with NUE, and ultimately introgression of genes and QTLs in the background of elite cultivars. Gene underlying NUE can be identify and validated by various techniques. Here we categorized these techniques into following groups:
-
(i)
Expression-based gene identification: Global expression profile of different genotypes and different N treatment helps to uncover differentially expressed genes which will further be functionally validated for its role in NUE. This technique includes suppression subtractive hybridization (SSH) (Rounsley et al. 1996), serial analysis of gene expression (SAGE) (Velculescu et al. 1995), massively parallel signature sequencing (MPSS) (Brenner et al. 2000), microarray (Katagiri and Glazebrook 2009) and RNA (ribonucleic acid) sequencing (Wang et al. 2009) as discussed below.
-
Suppression subtractive hybridization (SSH): This is a hybridization-based transcriptome analysis technique to identify differentially expressed genes. It allows comparison of two cDNA (complimentary DNA) populations (derived from transcripts) and isolation of a fraction enriched in differentially distributed molecules. An advantage of this technique is that it allows the detection of low-abundance differentially expressed transcript, whereas limitation includes false-positives and possibility of only pairwise comparison.
-
SAGE (serial analysis of gene expression): The main principle is a short sequence tag (10–14 bp) developed from individual mRNA which contains sufficient information to uniquely identify a transcript. Sequence tags are then linked together to form long serial molecules that can be cloned and sequenced. The number of times a particular tag is observed provides the information about expression level of the corresponding transcript.
-
Microarray: Microarray technology is based on the fact that complementary sequences of cDNA can be used to hybridize immobilized cDNA molecules. Here in this technique, probe-target hybridization is usually detected and quantified by detection of fluorophore or chemiluminescence-labeled targets to determine relative abundance of nucleic acid sequences in the target. An advantage of this technique includes large data generation in a single experiment. Its limitation includes cost, escape of rare allele detection, need for sequence information to generate microarray chip, do not detect unique genes, and analysis is tedious.
-
RNA sequencing: RNA sequencing is a comparatively new and high-throughput technique of NGS-based sequencing cDNA. It works by sequencing every RNA molecule and profiling the expression of a particular gene by counting the number of its transcript, thereby correlating phenotype with CGs. The advantages of this technique are high reproducibility, prior sequence information may not be required, discovery of rare alleles, and unique genes are possible, although this technique has complexity of analysis and cost.
-
-
(ii)
Discovery of gene and its functional validation by mutation and transgenic studies: Artificial mutant population and natural variants both can be used to discover a gene which has a role in NUE. In addition, the functional validation of putative NUE-governing gene is also possible by targeted mutation of that gene. After determining a phenotype which is associated with NUE, the very first step is creating mutagenized population and then screening for the plant showing mutated phenotype of desired trait. Mutation can be point or insertion. The insertion mutation can be further divided into insertion leading to loss of function and insertion resulting into gain of function mutation. At last the gene is recovered through map-based cloning approach. The mutagenized population may be created by EMS (ethyl methanesulfonate), T-DNA, or transposon tagging/activation tagging. However, NUE is a complex trait defining a single phenotype for it will be misleading.
-
(iii)
T-DNA/transposon tagging: Insertional mutagenesis is useful since flanking site information; in other words the disrupted gene, can be retrieved easily but limited by gene redundancy, lethal knockouts, and inability to target the inserted element to a specific gene.
-
(iv)
Activation tagging: Through activation tagging, gain of function phenotype can be obtained because it contains four copies of enhancer elements of CaMV 35S (cauliflower mosaic virus 35S) which can mediate transcriptional activation of nearby genes. It is mainly useful for genes having redundant function. However, activation tagging may fail because every gene may not have an overexpression phenotype. After identification of genes, its function should be validated either by disruption of its function or by overexpression. Disruption of gene can be carried out by the following techniques:
-
RNA interference (RNAi): RNA interference (abbreviated RNAi) is a mechanism for RNA-guided regulation of gene expression in which double-stranded ribonucleic acid inhibits the expression of genes with complementary nucleotide sequences. It has several specific advantages over forward genetics. Targeted, requiring only a few transformants per target gene. It facilitates the study of essential genes whose inactivation would lead to lethality. But there are some disadvantages too, like it requires prior sequence information and has leaky expression.
-
-
(v)
Other methods for gene disruption are techniques such as site-directed mutagenesis, gene knockout through homologous recombination, T-DNA insertion, targeted genome modification through TALEN (transcription activator-like effector nucleases), ZFN (zinc finger nucleus), and CRISPR-Cas9 (Cas9-clustered regularly interspaced short palindromic repeats) systems. The overexpression is carried out by expressing the gene with a strong promoter (CaMV 35S).
6.6.1.1 Gene for Nitrogen Acquisition
Many CGs for N uptake and RSA are known which are exploited to improve NUE in the past decades either by overexpressing them or by knockout mutation. The CKX1 gene code for cytokinin oxidase/dehydrogenase is responsible for degradation of cytokinin (negative regulator of root growth). Root-specific overexpression of this gene in Arabidopsis and tobacco leads to increase in root length, branching, and root-to-shoot ratio without affecting shoot growth and development (Werner et al. 2010). N accumulation is not estimated in this study, which highlights the sufficiency of a single dominant gene to alter complex trait like root growth. The ANR1 is a transcription factor involved in \( {\mathrm{NO}}_3^{-} \) signaling pathway to induce \( {\mathrm{NO}}_3^{-} \)-stimulated lateral root growth. The ANR1 overexpression can stimulate lateral root growth, but presence of \( {\mathrm{NO}}_3^{-} \) and posttranscriptional modification of ANR1 is prerequisite (Walch-Liu and Forde 2008). Root-based traits can provide exquisite opportunities for future improvements in NUE for cereals, but direct evidence in terms of gene manipulation is still lacking. Overexpression of \( {\mathrm{NO}}_3^{-} \) and \( {\mathrm{NH}}_4^{+} \) transporter is also documented. The NRT2.7 overexpression in Arabidopsis enhanced \( {\mathrm{NO}}_3^{-} \) accumulation and improved seed germination (Chopin et al. 2007). In rice, OsNRT2.1 overexpression showed improved seedling growth without any effect on N uptake (Katayama et al. 2009). This may be due to absence of required amount of OsNAR2.1, as we know NRT2.1 interacts with NAR2.1 to form functional transporter. The expression of OsNRT2.3b may increase rice yield and total N uptake (Xu et al. 2012). Overexpression of AMT1enhances \( {\mathrm{NH}}_4^{+} \) uptake capacity, but probably due to its toxicity, the plant biomass gets reduced. Therefore, AMT1 can be a potential gene under low-\( {\mathrm{NH}}_4^{+} \) condition (Hoque et al. 2006). Transporter and RSA controlling gene may be the potential candidate to increase NUE, provided utilization of acquired N should be efficient. So, introgression of these genes in the genotypic background efficient in N utilization might be useful to meet the need of NUE genotype.
6.6.1.2 Genes for Nitrogen Utilization
The improvement of N utilization can improve grain yield per unit of N acquired. The first step of \( {\mathrm{NO}}_3^{-} \) reduction by NR has been long known as rate-limiting step. The utility of NR/NiR overexpression is to improve how NUE is limited. Overexpression of NR in tobacco showed retention of NR activity for a longer period and some advantage during water stress (Ferrario-Mery et al. 1998). Similar result showed with NiR overexpression in Arabidopsis, potato, and tobacco. It reduces \( {\mathrm{NO}}_3^{-} \) concentration in plant tissues but does not show effect on seed or tuber yield. This may be due to regulation of these enzymes in posttranscriptional level (Pathak et al. 2009). The GS1 overexpression studies are more numerous than GS2, and in many cases it resulted in higher growth yield in biomass in low N supply (Habash et al. 2001; Oliveira et al. 2002). Overexpression of ASN1in Arabidopsis enhances plant fitness and growth in low N condition and also increases total protein content (Lam et al. 2003). Under anaerobic condition (i.e., flooding), alanine is the major storage amino acid. The AlaAT (alanine aminotransferase) catalyzes the synthesis of alanine and 2-oxoglutarate from pyruvate and glutamate. Expression of barley AlaAT in rice with the help of rice tissue-specific promoter showed improved NUpE, biomass, and yield and hence NUE (Shrawat et al. 2008). In addition to enzymes of N assimilation and amino acid metabolism, the attempts have been made to generate plants modified for expression of transcription factors. This is of particular importance because one transcription can regulate more than one gene in a metabolic pathway, and hence modification in TF expression may achieve modification of more than one gene. The Dof1 (DNA binding with one zinc finger 1) is reported to be one of the regulators for N metabolism by coordinated gene expression involved in TCA (tricarboxylic acid) cycle and hence C-skeleton production, viz., pyruvate kinase, PEP carboxylase (phosphoenolpyruvate carboxylase), citrate synthase, and isocitrate dehydrogenase. In Arabidopsis, expression of ZmDof1 results in enhanced N assimilation and better adaption of transgenic plants under N stress (Yanagisawa 2004). In rice, transgenic plant overexpressing ZmDof1 showed enhanced N assimilation (Kurai et al. 2011). Similarly, in wheat and sorghum, the constitutive expression of ZmDof1 leads to increase in PEPC expression (Peña et al. 2017). These effects suggest that NUE could also be improved by manipulating carbon metabolism pathways. Other potential regulatory proteins which could be subjected to further research are P-II, NPL7, and TOR (target of rifampicin). The P-II is a nuclear-encoded plastid protein which is homologous to bacterial P-II signaling proteins known to be involved in regulation of N metabolism by regulating key enzyme of arginine biosynthesis pathway. N-acetyl glutamate kinase (NAGK. P-II) knockout mutant in Arabidopsis showed accumulation of reduced ornithine, citrulline, and arginine accumulation in response to \( {\mathrm{NH}}_4^{+} \) supply after N starvation (Ferrario-Mery et al. 2006). The NLP7 (NIN-like protein 7) is an important element of the \( {\mathrm{NO}}_3^{-} \) signal transduction pathway. New regulatory protein specific for N assimilation in non-nodulating plants Arabidopsis NLP7-knockout mutants constitutively showed several features of N-starved plants (Castaings et al. 2009). Target of rifampicin (TOR) kinase showed positive regulation of growth in Arabidopsis under environmental stress (Deprost et al. 2007) (Fig. 6.3 and Table 6.1).
6.6.1.3 Genes for Starch Metabolism
Although N utilization components includes N assimilation and N remobilization, if we see the definition of NUE, it explains grain yield per unit of N captured, i.e., how efficiently captured N is utilized to ultimately produce yield. So, in case of cereal, better N utilization means efficient grain filling by starch biosynthesis and accumulation because starch is the main component of grain in cereals. Good N utilization in cereals does not mean better partitioning of N to the grain. Better N partitioning may enhance protein quality of grain, but grain protein and cereal yield have inverse relation, so more protein/N accumulation in grain will actually lead to less grain filling due to less starch biosynthesis which results in low NUE. Therefore, the role of N in grain filling and enhancement of yield is more important in cereal to achieve NUE. The effect of different levels of N fertilization on dry weight partitioning, grain filling, and starch metabolism activities has been investigated in wheat. Results indicated increase of N input within a certain extent could increase dry matter weights of stem and sheath at the heading and harvest stages. Activities of key enzymes of starch synthesis, namely, soluble starch synthase (SS), ADP (adenosine diphosphate) glucose pyrophosphorylase (AGPase), and soluble starch branching enzymes (SBE), are also influenced by N dose during grain filling. The study highlighted higher export and transform percentages of stem and sheath and rate of grain filling, and activities of key enzymes of wheat grain are the physiological basis for higher yield under an appropriate N level (Wang et al. 2013). Xiong et al. (2014) provided visual evidence of N effect on starch granules (SGs) in wheat endosperm. The results suggest that increased N fertilizer application mainly increased the numbers of small SGs and decreased the numbers of large SGs but that the results varied in different regions of the wheat endosperm. These observations give some hint of N effect on starch, but very less study is done so far. N assimilation is estimated only in terms of protein content, and amino acid content, parameters like yield and grain N, is and indicator of plant NUE so far. But in the cereals, to estimate utilization efficiency of N, it is more appropriate to focus on the factors which are responsible for starch synthesis and grain filling to give more yield per unit of plant N. To identify these factors and genes underlying efficient grain filling and starch biosynthesis, it is important to screen out the genotype which does not differ in terms of N uptake, assimilation, and remobilization capacity, but they differ in grain filling, mainly due to starch accumulation. By comparing genes and alleles related to starch transport and metabolism, regulatory genes for starch metabolic in these genotypes will help to uncover few underlying CGs which will be responsible for efficient grain filling and therefore yield per-unit N in cereals.
The process of starch biosynthesis and genes involved is explained here. Phosphoglucose isomerase and plastidial phosphoglucomutase forms glucose-1-phospate (G-1-P) from fructose-6-phosphate derived from the Calvin cycle. The G-1-P forms ADP glucose by the action of AGPase enzyme. The ADP glucose is the substrate of starch synthases (SSs) and granule-bound starch synthase (GBSS). The GBSS synthesizes amylose, while soluble SSs (different types of starch synthetase are summarized in Table 6.2), branching enzymes (BEs), and isoamylase-type debranching enzyme (ISA), pollunase collectively synthesize amylopectin (Pfister and Zeeman 2016).
The SS4 is proposed to play a role in generating glucan primers required to initiate starch granule synthesis (Roldán et al. 2007). The SS4 loss-of-function mutant showed a smaller number of granules per plastid but larger in size. Protein targeting to starch (PTST1) is a protein which functions to localize GBSS to starch granules for normal amylose synthesis. PTST2 and PTST3 control starch granule initiation (Seung et al. 2017). Along with these other enzymes like D-enzyme, phosphorylase, glucan water dikinase is also important. Studies focusing on these genes and its regulator including miRNA should be taken up in the genotypes differing only in grain filling under the same background (uptake and assimilation).
6.6.2 Discovery of QTLs and Genes by Mapping Studies
Mapping approaches include (a) linkage analysis in biparental mapping population and (b) association mapping in naturally existing population. They may not need any sequence information, but based on markers, they identify the position of the gene. Linkage analysis involves creating biparental populations, genotyping and phenotyping of segregating progeny in the populations, and testing if sequence variations in the CGs/marker co-segregate or co-localize with the loci controlling the trait in the populations. The main advantage of biparental mapping through linkage analysis is that it offers high level of confidence. Among disadvantages, it needs to construct mapping population, so it requires more time and resource and also cannot offer high resolution. Association mapping, also known as “linkage disequilibrium mapping,” is a method of mapping QTLs taking the advantage of linkage disequilibrium to link phenotype to genotype. It actually discovers linked markers associated with gene controlling the trait by exploiting diverse lines from natural population or germplasm collection. It also has own advantages as well as disadvantages. The prominent advantages over biparental mapping is (1) higher resolution, (2) lesser time, and that (3) it can discover additional genes or rare alleles for trait of interest. But it has somewhat lesser confidence because it may show association with spurious marker.
The QTL generally falls in two groups of genes; one is a major gene having large effect. These QTLs contribute to larger variation in highly heritable trait and called major QTLs, whereas minor QTLs have lesser effect. In this group, each QTL explains a small portion of total trait variation. Many agronomically important quantitative traits have small number of moderate effect QTLs and very large number of small effect QTLs to derive its genetic variation (Robertson 1967; Kearsey and Farquhar 1998). For a given study, the number of QTLs detected also depends on various factors like mapping population size and type, trait of interest, and effect of environment on that trait, in which environment phenotyping has been done as well as genome coverage. Major gene effect can be studied by means of segregation analysis, as properly as evolutionary history; however, when several genes with minor effect decide a trait (like in case of NUE), it is a good deal of great challenge, as they commonly can’t be investigated individually. Despite of these challenges, NUE trait has been mapped in many crop plants. Obara et al. (2001) investigated QTL association with NUE in rice. The main focus of their investigation is to see co-segregation of NUE with GS1 and NADH-GOGAT and identify seven and six loci for co-segregation with GS1 and NADH-GOGAT, respectively. Also, in GS2 spanning chromosomal region in wheat and rice has been mapped for number of QTLs for NUE, yield, and other agronomic traits (Han et al. 2015). Therefore, introgression of this chromosomal region to NUE-inefficient background in rice and wheat may be useful to develop cultivar with improved NUE and agronomic traits. The QTL association with NUE in maize has been carried out in segregating maize population in numerous studies (Gallais and Hirel 2004). A meta-analysis of QTLs for yield and yield components was carried out in low and optimum N regimes to identify meta-QTLs and characterize its map position. It reveled 22 QTLs under low N condition (Liu et al. 2012). But many researchers do not consider it the analysis for NUE as the selection was for yield at low N. Due to multiple QTLs for the trait and very low contribution of each QTL for variation, these studies point out many challenges. Genome-wide association mapping in 196 accession of wheat for yield and yield components reveled 23 N-responsive genomic regions which may be useful for breeding for N responsiveness (Bordes et al. 2013). Traits such as enzyme activity for N metabolism and assay of N metabolites are incorporated with QTL studies. It gave some metabolic QTLs (mQTLs) (Habash et al. 2007) advantage from mQTL as it provides evidence to link agronomic traits with potential gene(s) underlying the QTLs (Hill et al. 2013). But a disadvantage is that such traits are vulnerable to environmental cues. Nitrogen use efficiency and grain protein content in wheat and barley are controlled by a common QTL known as Gpc-B1. These loci control N remobilization, hence resulting in significant difference in NUE. The NILs (near-isogenic lines) with Gpc-B1 showed increase GPC without decreasing grain yield (Uauy et al. 2006; Heidlebaugh et al. 2008).
As we discussed the main objective of a mapping studies is to identify CG responsible for the trait, an identification of marker which can further be used to track trait of interest and cloning of favorable allele for the trait. Identification of CGs for complex trait like NUE faces challenges in spite of precision of these mapping studies because QTLs identified have many other genes that also co-segregate. Hence, further narrowing of the mapping interval to a place of the chromosome that carries a practical wide variety of genes may constrain. Therefore, identification of genes that would possibly have an effect on NUE is nonetheless based totally on our information of the gene’s characteristic like gene for N uptake, assimilation, remobilization, etc. But there are many evidences that other types of genes completely unrelated to N metabolism are also associated with NUE. For example, various studies describe involvement of phenology gene with NUE traits, which are semi-dwarf gene Rht and Ppd, and Vrn (Habash et al. 2007; Laperche et al. 2007).
6.7 Conclusion
Nitrogen use efficiency is a complex trait regulated by many environmental factors. The manipulation of NUE is a difficult process in spite of genetic potential. Recent years have seen a tremendous increase in the number of genes found to be involved in the mechanisms of inorganic N uptake and utilization in plants, but maximum study is limited to model plant, and there is need to extend it to crop plants. The first component of NUE is uptake; many genes of transports are identified in Arabidopsis, and some of it is also utilized to develop transgenic, but none of it resulted in NUE phenotype so far. Improvement of N uptake may be helpful in counteracting N loss from the environment, but it will not necessarily generate NUE phenotype. Nitrogen utilization is utilizing of the acquired N to produce grain yield. Traditionally it consists of N assimilation and remobilization, but for cereal, utilization of N to produce starch is also important. Several genes in N assimilation and remobilization have been reported, and some are used to overexpress in crop or model plant to achieve NUE phenotype. Genes of starch metabolism, which potentially affect grain filling in N limitation, need to identify in cereals which may further be exploited in creating NUE cereal.
Abbreviations
- AAP:
-
Amino acid permease
- ABA:
-
Abscisic acid
- ADP:
-
Adenosine diphosphate
- AE:
-
Agronomic efficiency
- AFG:
-
Auxin signaling F-box
- AGPase:
-
ADP glucose pyrophosphorylase
- AlaAT:
-
Alanine aminotransferase
- AMT:
-
Ammonium transporter
- ARE:
-
Apparent recovery efficiency
- AS:
-
Asparagine synthase
- ATF:
-
Amino acid transporter
- BE:
-
Branching enzyme
- BNF:
-
Biological nitrogen fixation
- C:
-
Carbon
- CaMV:
-
Cauliflower mosaic virus
- cDNA:
-
Complimentary DNA
- CGs:
-
Candidate genes
- CLC:
-
Chloride channel family
- CPSase:
-
Carbamoyl phosphate synthase
- CRISPR-Cas9:
-
Clustered regularly interspaced short palindromic repeats
- DNA:
-
Deoxyribonucleic acid
- Dof:
-
DNA binding with one zinc finger
- EMS:
-
Ethyl methanesulfonate
- G-1-P:
-
Glucose-1-phospate
- GBSS:
-
Granule-bound starch synthase
- GDH:
-
Glutamate dehydrogenase
- GMPase:
-
GDP mannose pyrophosphorylase
- GOGAT:
-
Glutamine-2-oxoglutarate aminotransferase or glutamate synthase
- GS:
-
Glutamine synthetase
- H+ :
-
Hydrogen
- HAT:
-
High-affinity transport system
- HSN1:
-
Hypersensitive to \( {\mathrm{NH}}_4^{+} \)
- HYVs:
-
High-yielding varieties
- ISA:
-
Isoamylase
- LATS:
-
Low-affinity transport
- LHT:
-
Lysine/histidine transporter
- MAS:
-
Marker-assisted selection
- miR:
-
micro RNA
- MPSS:
-
Massively parallel signature sequencing
- mQTLs:
-
Metabolic QTLs
- N:
-
Nitrogen
- N2O:
-
Nitrogen oxide
- NAD(P)H:
-
Nicotinamide adenine dinucleotide phosphate
- NAGK:
-
N-acetyl glutamate kinase
- NH3 :
-
Ammonia
- NIL:
-
Near-isogenic lines
- NiR:
-
Nitrite reductase
- NO:
-
Nitric oxide
- \( {\mathrm{NO}}_3^{-} \) :
-
Nitrate
- NPF:
-
Peptide transporter family
- NpUE:
-
N physiological use efficiency
- NR:
-
\( {\mathrm{NO}}_3^{-} \) reductase
- NRA:
-
\( {\mathrm{NO}}_3^{-} \) reductase activity
- NRT:
-
\( {\mathrm{NO}}_3^{-} \) transporter
- NUE:
-
Nitrogen use efficiency
- NUpE:
-
N uptake efficiency
- NUtE:
-
N utilization efficiency
- PEP:
-
Partial factor productivity
- PEPC:
-
Phosphoenolpyruvate carboxylase
- PNB:
-
Partial nutrient balance
- PTST:
-
Protein targeting to starch
- QTL:
-
Quantitative trait loci
- RNA:
-
Ribonucleic acid
- RNAi:
-
RNA interference
- RS:
-
Root system
- RSA:
-
Root system architecture
- SAGE:
-
Serial analysis of gene expression
- SAV:
-
Senescence associated vacuoles
- SBE:
-
Soluble starch branching enzymes
- SG:
-
Starch granules
- SLAC/SLAH:
-
Slow anion-associated channel homolog
- SS:
-
Starch synthase
- SSH:
-
Suppression subtractive hybridization
- T-DNA:
-
Transfer DNA
- TALEN:
-
Transcription activator-like effector nucleases
- TCA:
-
Tricarboxylic acid
- TIP:
-
Tonoplast intrinsic protein
- TOR:
-
Target of rifampicin
- UI:
-
Usage index
- WUE:
-
Water use efficiency
- ZFN:
-
Zinc finger nucleus
References
Barbier-Brygoo H, DeAngeli A, Filleur S, Frachisse JM, Gambale F, Thomine S et al (2011) Anion channels/transporters in plants: from molecular bases to regulatory networks. Annu Rev Plant Biol 62:25–51
Beman JM, Arrigo K, Matson PM (2005) Agricultural runoff fuels large phytoplankton blooms in vulnerable areas of the ocean. Nature 434:211–214
Bordes J, Ravel C, Jaubertie JP, Duperrier B, Gardet O et al (2013) Genomic regions associated with the N limitation response revealed in a global wheat core collection. Theor Appl Genet 126:805–822
Breiman A, Graur D (1995) Wheat evolution. Isr J Plant Sci 43:85–98
Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D et al (2000) Gene expression analysis by massively parallel signature sequencing (MPSS) on micro bead arrays. Nat Biotechnol 18:630–634
Bussi C, Gojon A, Passama L (1997) In situ nitrate reductase activity in leaves of adult peach trees. J Hortic Sci 72:347–353
Castaings L, Camargo A, Pocholle D et al (2009) The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J 57:426–435
Chopin F, Orsel M, Dorbe MF, Chardon F, Truong HN et al (2007) The Arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell 19:1590–1602
Conant RT, Berdanier AB, Grace PR (2013) Patterns and trends in N use and N recovery efficiency in world agriculture. Glob Biogeochem Cycles 27:558–566
Deprost D, Yao L, Sormani R et al (2007) The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep 8:864–870
Dobermann A (2005) Nutrient use efficiency – state of the art. In: IFA International Workshop on Enhanced-Efficiency Fertilizers. International Fertilizer Industry Association (IFA), Frankfurt/Paris, pp 1–16
Dobermann A (2007) Nutrient use efficiency – measurement and management. In: Fertilizer best management practices. General principles, strategy for their adoption and voluntary initiatives vs regulations. IFA International Workshop on Fertilizer Best Management Practices. International Fertilizer Industry Association (IFA), Brussels; Paris, pp 1–28
Epstein E (1972) Mineral nutrition of plants: principles and perspectives. Wiley, New York
Fageria NK, Baligar VC, Li YC (2008) The role of nutrient efficient plants in improving crop yields in the twenty first century. J Plant Nutr 31:1121–1157
FAO (2015) Current world fertilizer trends and outlook to 2015. Food and Agriculture Organization of the United Nations, Rome
Feng HM, Yan M, Fan XR, Li BZ, Shen QR et al (2011) Spatial expression and regulation of rice high-affinity nitrate transporters by N and carbon status. J Exp Bot 62:2319–2332
Ferrario-Mery S, Valadier M, Foyer C (1998) Overexpression of nitrate reductase in tobacco delays drought-induced decreases in nitrate reductase activity and mRNA. Plant Physiol 117:293–302
Ferrario-Mery S, Besin E, Pichon O, Meyer C, Hodges M (2006) The regulatory PII protein controls arginine biosynthesis in Arabidopsis. FEBS Lett 580:2015–2020
Fischer KS (2000) Frontier project on N fixation in rice: looking ahead. In: Ladha JK, Reddy PM (eds) The quest for N fixation in rice. International Rice Research Institute, Los Banos, pp 25–31
Fixen P, Brentrup F, Bruulsema TW, Garcia F, Norton R, Zingore S (2015) Nutrient/fertilizer use efficiency: measurement, current situation and trends. In: Drechsel P, Heffer P, Magen H, Mikkelsen R, Wichelns D (eds) Managing water and fertilizer for sustainable agricultural intensification. Copyright 2015 IFA, IWMI, IPNI and IPI, Paris, pp 8–38
Forde BG, Lea PJ (2007) Glutamate in plants: metabolism, regulation and signalling. J Exp Biol 58:2339–2358
Foyer CH, Noctor G, Hodges M (2011) Respiration and N assimilation: targeting mitochondria associated metabolism as a means to enhance N use efficiency. J Exp Bot 62:1467–1482
Gallais A, Hirel B (2004) An approach to the genetics of N use efficiency in maize. J Exp Bot 55:295–306
Galvan A, Quesada A, Fernandez E (1996) The use of mutants to study nitrate assimilation in green microalgae. Sci Mar 60:191–194
Garnett T, Conn V, Plett D, Conn S, Zanghellini J, Mackenzie N et al (2013) The response of the maize nitrate transport system to N demand and supply across the lifecycle. New Phytol 198:82–94
Gazzarrini S, Lejay L, Gojon A, Ninnemann O, Frommer WB, von Wiren N (1999) Three functional transporters for constitutive, diurnally regulated, and starvation induced uptake of ammonium into Arabidopsis roots. Plant Cell 11:937–947
Gniazdowska A, Mikulska M, Rychter AM (1998) Growth, nitrate uptake and respiration rate in bean roots under phosphate deficiency. Biol Plant 41:217–226
Gniazdowska-Skoczek H (1997) Properties of nitrate reductase from seedling leaves of selected barley genotypes. Acta Physiol Plant 19:137–145
GOI (2012) Press information bureau, government of India. Ministry of Agriculture & Farmers Welfare. http://pib.nic.in/newsite/PrintRelease.aspx?relid=80083
Good AG, Shrawat AK, Muench DG (2004) Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci 9:597–605
Habash DZ, Massiah AJ, Rong HI, Wallsgrove RM, Leigh RA (2001) The role of cytosolic glutamine synthetase in wheat. Ann Appl Biol 138:83–89
Habash DZ, Bernard S, Schondelmaier J, Weyen J, Quarrie SA (2007) The genetics of N use in hexaploid wheat: N utilisation, development and yield. Theor Appl Genet 114:403–419
Han M, Okamoto M et al (2015) The genetics of N use efficiency in crop. Plants Annu Rev Genet 49:269–289
Heidlebaugh NM, Trethewey BR, Jukanti AK, Parrott DL, Martin JM, Fischer AM (2008) Effects of a barley (Hordeum vulgare) chromosome 6 grain protein content locus on whole-plant N reallocation under two different fertilisation regimes. Funct Plant Biol 35:619–632
Hill C, Taylor J, Edwards J, Mather D (2013) Whole-genome mapping of agronomic and metabolic traits to identify novel quantitative trait loci in bread wheat grown in a water-limited environment. Plant Physiol 162:1266–1281
Hirel B, Le Gouis J, Ney B, Gallais A (2007) The challenge of improving N use efficiency in crop plants: towards a more central role for genetic variability and quantitative genetics within integrated approaches. J Exp Bot 58:2369–2387
Hirner A, Ladwig F, Stransky H, Okumoto S, Keinath M, Harms A, Frommer WB, Koch W (2006) Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. Plant Cell 18:1931–1946
Ho CH, Lin SH, Hu HC, Tsay YF (2009) CHL1 functions as a nitrate sensor in plants. Cell 138:1184–1194
Hoque MS, Masle J, Udvardi MK, Ryan PR, Upadhyaya NM (2006) Over-expression of the rice OsAMT1-1 gene increases ammonium uptake and content, but impairs growth and development of plants under high ammonium nutrition. Funct Plant Biol 33:153–163
Huang NC, Liu KH, Lo HJ, Tsay YF (1999) Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11:1381–1392
Janssen BH, Guiking FCT, Van Der Eijk D, Smaling EMA, Wolf J, Reuler H (1990) A system for quantitative evaluation of the fertility of tropical soils (QUEFTS). Geoderma 46:299–318
Kanno Y, Hanada A, Chiba Y, Ichikawa T, Nakazawa M, Matsui M et al (2012) Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc Natl Acad Sci 109:9653–9658
Katagiri F, Glazebrook J (2009) Overview of mRNA expression profiling using DNA microarrays. Curr Protoc Mol Biol 22:22
Katayama H, Mori M, Kawamura Y, Tanaka T, Mori M, Hasegawa H (2009) Production and characterization of transgenic rice plants carrying a high-affinity nitrate transporter gene (OsNRT2.1). Breed Sci 59:237–243
Kearsey MJ, Farquhar AGL (1998) QTL analysis in plants; where are we now? Heredity 80:137–142
Koch W, Kwart M, Laubner M, Heineke D, Stransky H, Frommer WB, Tegeder M (2003) Reduced amino acid content in transgenic potato tubers due to antisense inhibition of the leaf H+/amino acid symporter StAAP1. Plant J 33:211–220
Krapp A, David LC, Chardin C, Girin T, Marmagne A, Leprince AS et al (2014) Nitrate transport and signaling in Arabidopsis. J Exp Bot 65:789–798
Kronzucker H, Glass ADM, Siddiqi MY (1995) Nitrate induction in spruce: an approach using compartmental analysis. Planta 196:683–690
Kurai T, Wakayama M, Abiko T, Yanagisawa S, Aoki N, Ohsugi R (2011) Introduction of the ZmDof1 gene into rice enhances carbon and N assimilation under low-N conditions. Plant Biotechnol J 9:826–837
Ladha JK, Chakraborty D (2016) Solutions to improve N use efficiency for the world. In: Proceedings of the international N initiative conference, pp 4–8. December 2016, Melbourne, Australia
Ladha JK et al (2016) Agronomic improvements can make future cereal systems in South Asia far more productive and result in lower environmental footprint. Glob Chang Biol 22:1054–1074
Lam HM, Coschigano KT, Oliveira IC, Melo-Oliveira R, Coruzzi GM (1996) The molecular-genetics of N assimilation into amino acids in higher plants. Annu Rev Plant Physiol Plant Mol Biol 47:569–593
Lam HM, Wong P, Chan HK et al (2003) Overexpression of the ASN1 gene enhances N status in seeds of Arabidopsis. Plant Physiol 132:926–935
Laperche A, Brancourt-Hulmel M, Heumez E, Gardet O, Hanocq E et al (2007) Using genotype × N interaction variables to evaluate the QTL involved in wheat tolerance to N constraints. Theor Appl Genet 115:399–415
Lassaletta L, Billen G, Grizzetti B, Anglade J Garnier J (2014) 50 year trends in N use efficiency of world cropping systems: the relationship between yield and N input to cropland. Environ Res Lett 9:105011
Lea PJ (1993) N metabolism. In: Lea PJ, Leegood RC (eds) Plant biochemistry and molecular biology. Wiley, New York, pp 155–180
Lea PJ, Miflin B (1980) Transport and metabolism of asparagine and other N compounds within the plant, vol 40, pp 569–607
Lea PJ, Robinson SA, Stewart GR (1990) The enzymology and metabolism of glutamine, glutamate, and asparagine. In: Miflin BJ, Lea PJ (eds) The biochemistry of plants, vol 16. Academic, New York, pp 121–159
Lee YH, Foster J, Chen J, Voll LM, Weber AP, Tegeder M (2007) AAP1 transports uncharged amino acids into roots of Arabidopsis. Plant J 50:305–319
Léran S, Varala K, Boyer JC, Chiurazzi M, Crawford N, Daniel-Vedele F et al (2014) A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci 19:5–9
Léran S, Edel KH, Pervent M, Hashimoto K, Corratge-Faillie C, Offenborn JN et al (2015) Nitrate sensing and uptake in Arabidopsis are ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Sci Signal 8:43. https://doi.org/10.1126/scisignal.aaa4829
Li XZ, Oaks A (1994) Induction and turnover of nitrate reductase in Zea mays: influence of light. Plant Physiol 106:1145–1149
Li JY, Fu YL, Pike SM, Bao J, Tian W, Zhang Y et al (2010) The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22:1633–1646
Lima JE, Kojima S, Takahashi H, von Wiren N (2010) Ammonium triggers lateral root branching in Arabidopsis in an AMMONIUM TRANSPORTER1;3-dependent manner. Plant Cell 22:3621–3633
Lin SH, Kuo HF, Canivenc G, Lin CS, Lepetit M, Hsu PK et al (2008) Mutation of the Arabidopsis NRT1.5nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20:2514–2528
Liu HS, Tsay YF (2003) Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J 22:1005–1013
Liu KH, Huang CY, Tsay YF (1999) CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11:865–874
Liu R, Zhang H, Zhao P, Zhang Z, Liang W et al (2012) Mining of candidate maize genes for N use efficiency by integrating gene expression and QTL data. Plant Mol Biol Rep 30:297–308
London JG (2005) N study fertilizes fears of pollution. Nature 433:791
Loque D, Ludewig U, Yuan L, von Wiren N (2005) Tonoplast intrinsic proteins AtTIP2;1 and AtTIP2;3 facilitate NH3 transport into the vacuole. Plant Physiol 137:671–680
Lupini A, Mercati F, Araniti F, Miller AJ, Sunseri F, Abenavoli MR (2016) NAR2.1/NRT2.1 functional interaction with NO3− and H+ fluxes in high-affinity nitrate transport in maize root regions. Plant Physiol Biochem 102:107–114
Manske GG, Vlek PL (2002) Root architecture–wheat as a model plant. In: Waisel Y, Eshel A, Beeckman T, Kafkafi U (eds) Roots: the hidden half. CRC, Boca Raton, pp 249–259
Marschner H (1995) Mineral nutrition of higher plants, 2nd edn. Academic, London
Marwaha RS (1998) Nitrate assimilation in potato cultivars during plant growth. Indian J Plant Physiol 3:147–151
Masclaux-Daubresse C, Reisdorf-Cren M, Pageau K et al (2006) Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source N cycle in tobacco. Plant Physiol 140:444–456
Masclaux-Daubresse C, Reisdorf-Cren M, Orsel M (2008) Leaf N remobilisation for plant development and grain filling. Plant Biol 10:23–36
Meyer C, Stitt M (2001) Plant N. In: Lea PJ, Goudry JFM (eds) Nitrate reduction and signaling. Springer, Berlin/Heidelberg, pp 37–59
Miflin BJ, Lea PJ (1976) The pathway of N assimilation in plants. Phytochemistry 15:873–885
Miflin BJ, Lea PJ (1980) Ammonia assimilation. In: Miflin BJ (ed) The biochemistry of plants: amino acids and derivatives, vol 5. Academic, New York, pp 169–202
Mistrik I, Ullrich CI (1996) Mechanism of anion uptake in plant roots: quantitative evaluation of H+/NO3− and H+/H2PO4− stoichiometries. Plant Physiol Biochem 34:629–636
Moll RH, Kamprath EJ, Jackson WA (1982) Analysis and interpretation of factors which contribute to efficiency of N utilisation. Agronomy 74:562–564
Mosier A, Syers JK (2004) Agriculture and the N cycle: assessing the impacts of fertilizer use on food production and the environment, vol 65. Island Press, Washington, DC
Negi J, Matsuda O, Nagasawa T, Oba Y, Takahashi H, Kawai-Yamada M et al (2008) CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452:483–486
Obara M, Kajiura M, Fukuta Y, Yano M, Hayashi M et al (2001) Mapping of QTLs associated with cytosolic glutamine synthetase and NADH-glutamate synthase in rice (Oryza sativa L.). J Exp Bot 52:1209–1217
Oliveira IC, Coruzzi GM (1999) Carbon and amino acids reciprocally modulate the expression of glutamine synthetase in Arabidopsis. Plant Physiol 121:301–310
Oliveira IC, Brears T, Knight TJ, Clark A, Coruzzi GM (2002) Overexpression of cytosolic glutamate synthetase. Relation to N, light, and photorespiration. Plant Physiol 129:1170–1180
Orsel M, Filleur S, Fraisier V, Daniel-Vedele F (2002a) Nitrate transport in plants: which gene and which control? J Exp Bot 53(370):825–833
Orsel M, Krapp A, Daniel-Vedele F (2002b) Analysis of the NRT2 nitrate transporter family in Arabidopsis. Structure and gene expression. Plant Physiol 129:886–896
Orsel M, Chopin F, Leleu O, Smith SJ, Krapp A, Daniel-Vedele F, Miller AJ (2006) Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiology and protein protein interaction. Plant Physiol 142(3):1304–1317
Pajuelo PE, Pajuelo B, Forde G, Marquez AJ (1997) Regulation of the expression of ferredoxin-glutamate synthase in barley. Planta 203:517–525
Pathak H, Aggarwal PK, Roetter R, Kalra N, Bandyopadhaya SK, Prasad S, Keulen HV (2003) Modeling the quantitative evaluation of soil nutrient supply, nutrient use efficiency, and fertilizer requirements of wheat in India. Nutr Cycl Agroecosyst 65:105–113
Pathak RR, Ahmad A, Lochab S, Raghuram N (2009) Molecular physiology of plant N use efficiency and biotechnological options for its enhancement. Curr Sci 94:1394–1403
Peña PA, Quach T, Sato S et al (2017) Expression of the maize Dof1 transcription factor in wheat and sorghum. Front Plant Sci 8:434. https://doi.org/10.3389/fpls.2017.00434
Peoples MB, Gifford RM (1993) Plant physiology, biochemistry and molecular biology. In: Dennis DT, Turpin DH (eds) Long distance transport of carbon and N from sources to sinks in higher plants. Wiley, New York, pp 434–447
Peterman TK, Goodman HM (1991) The glutamine synthetase gene family of Arabidopsis thaliana: light regulation and differential expression in leaves, roots and seeds. Mol Gen Genet 230:145–154
Pfister B, Zeeman SC (2016) Formation of starch in plant cells. Cell Mol Life Sci 73:2781–2807
Potel F, Valadier MH, Ferrario-Mery S et al (2009) Assimilation of excess ammonium into amino acids and N translocation in Arabidopsis thaliana – roles of glutamate synthases and carbamoylphosphate synthetase in leaves. FEBS J 276:4061–4076
Powlson DS, Addiscott TM, Benjamin N, Cassman KG, DeKok TM (2008) When does nitrate become a risk for humans? J Environ Qual 37:291–295
Qin C, Qian W, Wang W, Wu Y, Yu C et al (2008) GDP-mannose pyrophosphorylase is a genetic determinant of ammonium sensitivity in Arabidopsis thaliana. Proc Natl Acad Sci U S A 105:18308–18313
Rabalais NN, Turner RE, Wiseman WJ (2002) Gulf of Mexico hypoxia, aka “the dead zone.”. Annu Rev Ecol Syst 33:235–263
Raghuram N, Sachdev MS, Abrol YP (2007) Towards an integrative understanding of reactive N. In: Agricultural N use and its environmental implications, pp 1–6
Remans T, Nacry P, Pervent M, Filleur S, Diatloff E et al (2006) The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc Natl Acad Sci 103:19206–19211
Rentsch D, Schmidt S, Tegeder M (2007) Transporters for uptake and allocation of organic N compounds in plants. FEBS Lett 581:2281–2289
Robertson A (1967) The nature of quantitative genetic variation. In: Brink R (ed) Heritage from Mende, Univ. Wisc. E Styles. University of Wisconsin Press, Madison, pp 265–280
Robertson GP, Vitousek PM (2009) N in agriculture: balancing the cost of an essential resource. Annu Rev Environ Resour 34:97–125
Rochat C, Boutin JP (1991) Metabolism of phloem-borne amino acids in maternal tissues of fruit of nodulated or nitrate-fed pea plants (Pisum sativum L.). J Exp Bot 42:207–214
Roche D, Temple SJ, Sengupta GC (1993) Two classes of differentially regulated glutamine synthetase genes are expressed in the soybean nodule: a nodule specific and a constitutively expressed class. Plant Mol Biol 22:971–983
Roldán I, Wattebled F, Mercedes Lucas M, Delvallé D, Planchot V, Jiménez S, Pérez R et al (2007) The phenotype of soluble starch synthase IV defective mutants of Arabidopsis thaliana suggests a novel function of elongation enzymes in the control of starch granule formation. Plant J 49:492–504
Rounsley SD, Glodek A, Sutton G, Adams MD, Somerville CR, Venter JC et al (1996) The construction of Arabidopsis expressed sequence tag assemblies. A new resource to facilitate gene identification. Plant Physiol 112:1177–1183
Segonzac C, Boyer JC, Ipotesi E, Szponarski W, Tillard P, Touraine B et al (2007) Nitrate efflux at the root plasma membrane: identification of an Arabidopsis excretion transporter. Plant Cell 19:3760–3777
Seung D, Boudet J, Monroe J, Schreier TB, David LC et al (2017) Homologs of protein targeting to starch control starch granule initiation in Arabidopsis leaves. Plant Cell 29:1657–1677
Shrawat AK, Carroll RT, DePauw M, Taylor GJ, Good AG (2008) Genetic engineering of improved N use efficiency in rice by the tissue-specific expression of alanine aminotransferase. Plant Biotechnol J 6:722–732
Sivasankar S, Rothstein S, Oaks A (1997) Regulation of the accumulation and reduction of nitrate by N and carbon metabolites in maize seedlings. Plant Physiol 114:583–589
Skopelitis D, Paranychianakis N, Paschalidis K, Pliakonis E, Delis I, Yakoumakis D, Kouvarakis A, Papadakis A, Stephanou E, Roubelakis-Anfelakis K (2006) Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine. Plant Cell 18(10):2767–2781
Sonoda Y, Ikeda A, Saiki S, Yamaya T, Yamaguchi J (2003) Feedback regulation of the ammonium transporter gene family AMT1 by glutamine in rice. Plant Cell Physiol 44:1396–1402
Stewart GR, Mann AF, Fentem PA (1980) Enzymes of glutamate formation: glutamate dehydrogenase, glutamine. In: Miflin BJ (ed) The biochemistry of plants: comprehensive treatise, vol. 5. Amino acids and derivatives. Academic, New York, pp 271–327
Sulpice R, Pyl ET, Ishihara H, Trenkamp S, Steinfath M et al (2009) Starch as a major integrator in the regulation of plant growth. Proc Natl Acad Sci 106:10348–10353
Svennerstam H, Ganeteg U, Nasholm T (2008) Root uptake of cationic amino acids by Arabidopsis depends on functional expression of amino acid permease 5. New Phytol 180:620–630
Taochy C, Gaillard I, Ipotesi E, Oomen R, Leonhardt N, Zimmermann S et al (2015) The Arabidopsis root stele transporter NPF2.3 contributes to nitrate translocation to shoots under salt stress. Plant J 83:466–447
Taylor L, Nunes-Nesi A, Parsley K, Leiss A, Leach G, Coates S, Wingler A, Fernie AR, Hibberd JM (2010) Cytosolic pyruvate, orthophosphatedikinase functions in N remobilization during leaf senescence and limits individual seed growth and N content. Plant J 62(4):642–652
Tilsner J, Kassner N, Struck C, Lohaus G (2005) Amino acid content and transport in oilseed rape (Brassica napus L.) under different N conditions. Planta 221:328–338
Tsay YF, Schroeder JI, Feldmann KA, Crawford NM (1993) The herbicide sensitivity gene CHL1of Arabidopsis encodes anitrate-inducible nitrate transporter. Cell 72:705–713
Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK (2007) Nitrate transporters and peptide transporters. FEBS Lett 581:2290–2300
Uauy C, Brevis JC, Dubcovsky J (2006) The high grain protein content gene Gpc-B1 accelerates senescence and has pleiotropic effects on protein content in wheat. J Exp Bot 57:2785–2794
Ullrich WR (1987) Nitrate and ammonium uptake in green algae and higher plants: mechanism and relationship with nitrate metabolism. In: Ullrich WR, Aparicio PJ, Syrett PJ, Castillo F (eds) Inorganic N metabolism. Springer, Berlin/Heidelberg/New York, pp 32–38
Velculescu VE, Zhang L, Vogelstein B, Kinzler KW (1995) Serial analysis of gene expression. Science 270:484–487
Vidal EA, Araus V, Lu C, Parry G, Green PJ et al (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci 107:4477–4482
Walch-Liu P, Forde BG (2008) Nitrate signaling mediated by the NRT1.1 nitrate transporter antagonizes l-glutamate-induced changes in root architecture. Plant J 54:820–828
Wang R, Liu D, Crawford NM (1998) The Arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proc Natl Acad Sci 95:15134–15139
Wang Z, Gartein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10(1):57–63
Wang H et al (2013) Regulating effect of N fertilization on grain filling and activities of enzymes involved in starch synthesis of Yumai49-198. Plant Nutr Fertil Sci 19:288–296
Ward M, Grimes H, Huffaker R (1989) Latent nitrate reductase activity is associated with the plasma membrane of corn roots. Planta 177:470–475
Werner T, Nehnevajova E, Kollmer I, Novak O, Strnad M, Kramer U, Schmulling T (2010) Root specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 22:3905–3920
Witt CA, Dobermann S, Abdulrachman HC, Gines W, Guanghuo R, Nagarajan S, Satawathananont TT, Son PS, Tan L, Van Tiem G, Simbahan Olk DC (1999) Internal nutrient efficiencies in irrigated lowland rice of tropical and subtropical Asia. Field Crop Res 63:113–138
Xiong et al. (2014) Effect of N fertilizer on distribution of starch granules in different regions of wheat endosperm. Crop J 46:54
Xu G, Fan X, Miller AJ (2012) Plant N assimilation and use efficiency. Annu Rev Plant Biol 63:153–182
Yan M, Fan XR, Feng HM, Miller AJ, Shen QR, Xu GH (2011) Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant Cell Environ 34:1360–1372
Yanagisawa S (2004) Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol 45:386–391
Yuan L, Loque D, Ye F, Frommer WB, von Wiren N (2007) N-dependent posttranscriptional regulation of the ammonium transporter AtAMT11. Plant Physiol 143:732–744
Zhang H, Forde BG (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279:407–409
Zhuo D, Okamoto M, Vidmar JJ, Glass AD (1999) Regulation of a putative high-affinity nitrate transporter (AtNrt2.1) in roots of Arabidopsis thaliana. Plant J 17(5):563–568
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Bharati, A., Mandal, P.K. (2020). Strategies for Identification of Genes Toward Enhancing Nitrogen Utilization Efficiency in Cereals. In: Meena, R. (eds) Nutrient Dynamics for Sustainable Crop Production. Springer, Singapore. https://doi.org/10.1007/978-981-13-8660-2_6
Download citation
DOI: https://doi.org/10.1007/978-981-13-8660-2_6
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-8659-6
Online ISBN: 978-981-13-8660-2
eBook Packages: Earth and Environmental ScienceEarth and Environmental Science (R0)