Abstract
The hematopoietic system plays a critical role in establishing the proper response against invading pathogens or in removing cancerous cells. Furthermore, deregulations of the hematopoietic differentiation program are at the origin of numerous diseases including leukemia. Importantly, many aspects of blood cell development have been conserved from human to Drosophila. Hence, Drosophila has emerged as a potent genetic model to study blood cell development and leukemia in vivo. In this chapter, we give a brief overview of the Drosophila hematopoietic system, and we provide a protocol for the dissection and the immunostaining of the larval lymph gland, the most studied hematopoietic organ in Drosophila. We then focus on the various paradigms that have been used in fly to investigate how conserved genes implicated in leukemogenesis control blood cell development. Specific examples of Drosophila models for leukemia are presented, with particular attention to the most translational ones. Finally, we discuss some limitations and potential improvements of Drosophila models for studying blood cell cancer.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
11.1 Introduction
Cells of the hematopoietic system are essential for maintaining the homeostasis of the organism , notably by participating in the immune response, removing apoptotic or cancerous cells, and producing various cytokines or clotting factors (Provan and Gribben 2010). Nonetheless, these cells have both protective and pathogenic functions in antimicrobial defense, autoimmune diseases, inflammatory reaction, metabolic disorders, or tumorigenesis. Hence, their development and function have to be tightly regulated. Accordingly mutations affecting blood cell development can lead to various hemopathies including leukemia. This heterogeneous class of malignancies affecting the hematopoietic lineages represents ±3% of all classes of cancers . It is characterized by the presence in the bone marrow and in peripheral tissues of misdifferentiated blood cells with proliferative and/or survival advantage that eventually outnumber normal blood cells, leading to deadly illnesses. The emergence of a leukemic clone is usually associated with the stepwise accumulation of a limited number of genetic mutations in hematopoietic stem or progenitor cells (Ferrando and Lopez-Otin 2017). The identification of the mutated genes and the characterization of their mode(s) of action remain an important issue to decipher the mechanisms of blood cell transformation and develop new therapies.
The development of animal models, in particular mouse, has been instrumental in characterizing how hematopoietic cell fate and function are controlled in vivo under normal and pathological conditions (Kohnken et al. 2017). In addition, it has become clear that researches in Drosophila melanogaster can also provide relevant information to gain insights into these processes. Indeed works from several labs over the last 20 years have revealed that the molecular pathways controlling blood cell production and function are highly conserved from human to Drosophila. Notably, several key transcription factors and signaling pathways implicated in normal and malignant blood cell development in human control hematopoiesis in fly too. Hence, thanks to the outstanding genetic toolbox available in Drosophila and to the development of more and more sophisticated markers and assays to characterize Drosophila blood cell status and functions, this organism can serve as a valuable model to investigate various aspects of blood cell biology relevant to cancer . Here, we will focus on the use of Drosophila to study leukemogenesis. First, we provide a rapid survey of the development of the Drosophila hematopoietic system, together with a protocol to assess blood cell status in the lymph gland, a well-described larval hematopoietic organ. Then, we present the three main approaches that have been developed to gain insights into leukemogenesis using Drosophila: (1) the study of the so-called melanotic tumors, which can arise from leukemic-like processes, (2) the expression in non-hematopoietic cell types of oncogenic variants of genes participating in blood cell transformation in human, and (3) the study of these oncogenic variants or their homologues in the Drosophila hematopoietic system. We present specific examples showing how these various strategies have shed light on blood cell transformation and/or helped tackle the mechanisms of action of specific proteins implicated in leukemia in humans. Finally, we present some possible directions to improve the use of Drosophila in leukemia research.
11.2 Drosophila Hematopoiesis
As the development and regulation of the Drosophila hematopoietic system have been covered extensively in several recent reviews (Gold and Bruckner 2015; Letourneau et al. 2016; El Chamy et al. 2017; Yu et al. 2017), we only provide here a description of its salient features, and we refer interested readers to the aforementioned reviews for further details.
11.2.1 Development of the Drosophila Hematopoietic System
Reminiscent of the situation in vertebrates, Drosophila hematopoiesis takes place in successive waves (Holz et al. 2003 ). First, in the early embryo, a pool of pluripotent blood cell progenitors (called prohemocytes) is specified in the head mesoderm and gives rise to peripheral blood cells, which populate the body cavity (hemocoel) of the larva (Makhijani et al. 2011). A second population of prohemocytes arises later during embryonic development from the lateral/cardiac mesoderm, which generates a specialized larval hematopoietic organ called the lymph gland (Mandal et al. 2004; Jung et al. 2005). Under normal conditions, blood cells produced in the lymph gland are released into the hemolymph only at the end of larval life (Honti et al. 2010; Grigorian et al. 2011). In the adult fly, blood cells generated during these two distinct waves of hematopoiesis are present, with limited blood cell proliferation or differentiation (Holz et al. 2003; Honti et al. 2014; Ghosh et al. 2015). Overall, there are ± 700 hemocytes in late embryos (Tepass et al. 1994), while third instar larvae contain ±8000 peripheral hemocytes (Lanot et al. 2001, Petraki et al. 2015) and 4000–8000 lymph gland hemocytes (Krzemien et al. 2010). The number of blood cells in the adult is difficult to assess but is estimated to ±2000 cells (Lanot et al. 2001), declining with age (Mackenzie et al. 2011; Horn et al. 2014).
It is worth reminding that beside the larval and adult heart tube, which is open at both ends, Drosophila has no proper vascular network (Hartenstein and Mandal 2006), and blood cells travel freely within this open circulatory system. If most of the peripheral larval hemocytes and adult hemocytes are sessile and form patches of cells under the epidermal wall (Braun et al. 1998; Elrod-Erickson et al. 2000; Markus et al. 2009; Makhijani et al. 2011), significant turnaround has been observed between sessile and circulating hemocytes in the larva (Makhijani et al. 2011). In addition, hemocytes are also associated with other tissues such as the eye imaginal discs (Fogarty et al. 2016), the heart (Elrod-Erickson et al. 2000 ; Ghosh et al. 2015 ), the gut (Zaidman-Remy et al. 2012; Ayyaz et al. 2015; Chakrabarti et al. 2016), or the ovaries (Brandt and Schneider 2007; Van De Bor et al. 2015).
11.2.2 Drosophila Blood Cell Lineages
As most metazoans, Drosophila lacks equivalents of the lymphoid lineages, and its mature blood cells, collectively called hemocytes, can be subdivided into three specialized cell types functionally related to vertebrate myeloid cells: the plasmatocytes, the crystal cells, and the lamellocytes (Parsons and Foley 2016). Plasmatocytes are professional phagocytes and represent the vast majority of the differentiated blood cells (>90%); they are functionally similar to mammalian monocytes, macrophages, and neutrophils (Wood and Martin 2017). They recognize and engulf small pathogens as well as apoptotic cells, and they are a major source of extracellular matrix components, thus playing important functions in the innate cellular immune response but also in tissue remodeling and homeostasis. Plasmatocytes are highly motile cells and constitute a popular model to study the conserved mechanisms regulating cell migration in vivo and by extension to gain insights into metastatic processes (Fauvarque and Williams 2011, Wood and Martin 2017). While plasmatocytes are usually considered as a single entity, populations expressing different subsets of markers have been identified (Jung et al. 2005; Honti et al. 2014). Moreover, two plasmatocyte subpopulations with distinct functions in the adult immune response have been identified (Clark et al. 2011). A better assessment of plasmatocyte heterogeneity is thus certainly needed. Crystal cells are involved in melanization, an insect-specific defense response related to clotting (Whitten and Coates 2017). They are named according to the presence of large paracrystalline inclusions in their cytoplasm, which contain some of the enzymes required for melanin production. Upon wounding, melanization limits fluid loss and participates in the fight against infection notably by trapping microbes and producing microbicidal reactive oxygen species. Finally, lamellocytes are large flat cells (30–60 μm) that are absent in healthy larvae but whose production can be massively induced in response to some stresses and immune challenges such as the infection by parasitoid wasp eggs (Lanot et al. 2001; Eslin et al. 2009) but also in several cancer-related conditions (see below). Together with the plasmatocytes, the lamellocytes adhere to the wasp egg and form a multilayered capsule, which eventually melanizes and kills the intruder. In contrast with plasmatocytes and crystal cells, which are observed in the embryo, the larva, and the adult, lamellocytes are only produced during the larval stages (Honti et al. 2014).
The lineage relationship between the three mature blood cell types and the presence of genuine hematopoietic stem cells are still a matter of debate, which is out of the scope of this chapter. In short, the prevailing view is that blood cell progenitors present in the early embryo and in the lymph gland are transient populations, which do not persist in the larva or in the pupa, respectively (Grigorian et al. 2011; Makhijani et al. 2011; Dey et al. 2016). It is not clear whether the “undifferentiated” blood cells described in the adult are long-lasting, multipotent, and capable of self-renewing (Ghosh et al. 2015). Moreover, Drosophila prohemocytes can give rise to plasmatocytes, crystal cells, and lamellocytes (Krzemien et al. 2010), but larval peripheral plasmatocytes can also proliferate (Makhijani et al. 2011; Anderl et al. 2016) and transdifferentiate into crystal cells (Leitao and Sucena 2015) or lamellocytes (Markus et al. 2009; Avet-Rochex et al. 2010; Stofanko et al. 2010; Anderl et al. 2016). Thus, it seems that the production of the different blood cell types can be achieved by various routes.
11.2.3 Control of Drosophila Hematopoiesis
Hematopoietic progenitor maintenance, hemocyte differentiation, and the overall homeostasis of the hematopoietic system are finely tuned by intrinsic factors and by environmental stimuli. These features have been particularly well studied in the larvae. For instance, in the lymph gland, high levels of reactive oxygen species (ROS) (Owusu-Ansah and Banerjee 2009), activation of the wingless signaling pathway (Sinenko et al. 2009), and expression of the EBF transcription factor Collier (Benmimoun et al. 2015; Oyallon et al. 2016) are required in prohemocytes to promote their maintenance. In addition, prohemocyte fate is controlled by local signals from the neighboring heart tube (Morin-Poulard et al. 2016), posterior signaling center (Krzemien et al. 2007; Mandal et al. 2007), and differentiated hemocytes (Mondal et al. 2011; Zhang and Cadigan 2017), as well as by systemic signals released in response to nutrient levels (Benmimoun et al. 2012; Shim et al. 2012) and olfactory stimulations (Shim et al. 2013). Similarly, in peripheral hemocytes , local cues from the peripheral nervous system attract plasmatocytes to subepidermal hematopoietic pockets and promote their survival, their proliferation, and their differentiation into crystal cells (Makhijani et al. 2011, 2017).
The larval hematopoietic system is highly responsive to immune challenges and stresses. The infection of the larva by parasitoid wasp eggs causes lymph gland expansion and premature dispersal, as well as differentiation of lamellocytes from lymph gland progenitors and from peripheral plamatocytes at the expense of crystal cell development (Sorrentino et al. 2002; Crozatier et al. 2004; Markus et al. 2009; Ferguson and Martinez-Agosto 2014; Anderl et al. 2016). Moreover, bacterial infection was recently found to promote blood cell progenitor differentiation in the lymph gland (Khadilkar et al. 2017) and to induce some proliferation in adult hemocytes (Ghosh et al. 2015). Finally, mechanical stress (Petraki et al. 2015), oxygen levels (Mukherjee et al. 2011), nutrition (Benmimoun et al. 2012; Shim et al. 2012), and odors (Shim et al. 2013) can greatly influence larval blood cell homeostasis. By contrast, the development of the embryonic blood cells seems rather stereotypical (Bataille et al. 2005), and it remains to be shown whether it can be influenced by external factors.
11.2.4 Protocol: Immunostaining in the Larval Lymph Gland
The larval lymph gland is currently the most popular system to study hematopoiesis in Drosophila (Letourneau et al. 2016). One of its advantages is the presence of a large pool of blood cell progenitors and of all their differentiated progenies within a confined organ from which they are normally not released in circulation until metamorphosis. Therefore, the lymph gland is well suited to study the control of progenitor blood cell fate and to gain insight into the gene networks regulating blood cell homeostasis . In addition, thanks to the effort of many teams, a large set of well-characterized markers and genetic tools are now available to study lymph gland homeostasis and specifically label or manipulate the different cell types present in this complex organ (Evans et al. 2014). Below, we give a brief presentation of the lymph gland organization and a generic protocol that we use to prepare larval lymph glands for immunostaining.
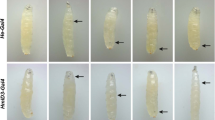
In the larva, the lymph gland is lining the anterior part of the dorsal vessel/cardiac tube, just behind the ring gland and the brain. It is composed of a large pair of anterior lobes followed by 2–4 pairs of posterior lobes. Each lobe is surrounded by a layer of extracellular matrix and separated from its posterior neighbor by a pericardial cell. The lymph gland anterior lobes are specified in the embryo and grow considerably during the larval stages (Jung et al. 2005), with a shift from blood cell progenitor proliferation toward differentiation in the late second larval instar stage (Krzemien et al. 2010). The ontogeny of the posterior lobes is not well characterized, but they are detectable in late first instar larvae. They constitute a large pool of blood cell progenitors that enter differentiation later than those present in the anterior lobes. In mid-third instar larvae (96h after egg laying), the posterior lobes mostly comprise undifferentiated blood cells, while the anterior lobes contain blood cell progenitors in their inner/medullary zone and differentiated hemocytes in their outer/cortical zone (Fig. 11.1). In addition, a small group of ±30 cells located at the posterior tip of each anterior lobe form the so-called posterior signaling center (PSC). The PSC expresses various signaling molecules that regulate blood cell fate, and it exerts a prominent role in the response to infection (Letourneau et al. 2016).
The Drosophila larval lymph gland. Confocal image showing the expression of the plasmatocyte marker P1/NimC1 (red), the prohemocyte marker dome-meso-lacZ (blue), and the posterior signaling center (PSC) marker col-GAL4,UAS-GFP (green). In the anterior lobes, a line demarcates the medullary zone (MZ) from the cortical zone (CZ). PC, pericardial cells. PL, posterior lobes
Even in third instar larvae, the lymph glands are small and fragile organs that are tedious to dissect as compared to other tissues such as the imaginal discs. While the initial steps of the protocol described below are relatively straightforward, some practice is necessary to mount properly the lymph glands before observation, especially to keep the posterior lobes intact or for the observation of first/second instar larva lymph glands. Moreover blood cell number and proliferation/differentiation status evolve significantly during larval life, are sensitive to various external stimuli, and show interindividual variations. It is thus essential to work under well-controlled breeding conditions and to analyze a sufficient (minimum ten) number of stage-matched samples to make sure of the significance of any phenotype.
11.2.4.1 Materials
11.2.4.1.1 Equipment
-
Glass dissection dishes (Electron Microscopy Science).
-
6- or 12-well tissue culture plate (Corning).
-
Forceps (Fine Science Tools, Dumont #5).
-
1 ml syringes (TERUMO).
-
Needles (TERUMO, 0.9*38 mm).
-
Microscope slides and 18x18 mm coverslips.
-
Stereomicroscope (for dissection).
-
Fluorescent microscope (for analysis).
-
Transfer pipette (Sterilin).
-
1.5 ml centrifuge tube (Eppendorf).
11.2.4.1.2 Solutions and Reagents
-
Sterile phosphate-buffered saline (PBS) (e.g., Dulbecco).
-
Bovine serum albumin (BSA).
-
4% formaldehyde (made from 16% stock, Electron Microscopy Sciences) in 1xPBS
-
Wash solution: 1xPBS 0.1% Triton X-100 (PBST).
-
Permeabilization solution: 1xPBS 0.3% Triton X-100.
-
Blocking solution: 1xPBST 1% BSA.
-
Primary antibodies and fluorescent-dye conjugated secondary antibodies (e.g., Alexa Fluor).
-
DNA staining solutions (e.g., DAPI 5 mg/ml in H2O, or TO-PRO-3 1mM in DMSO).
-
Mounting medium (e.g., Vectashield H-1000, Vector Laboratories).
-
Glycerol.
-
Optional: Methanol.
11.2.4.2 Methods
11.2.4.2.1 Larvae Collection
-
1.
Set up the appropriate fly cross(es), and transfer the adults to fresh vials every 12 h. Make sure to avoid overcrowding of the larvae.
-
2.
Delicately transfer third instar wandering larvae (or other appropriately staged larvae) from the vials to a dissecting dish or a 6-well plate containing 1xPBS with a pair of forceps or a paintbrush.
-
3.
Using a transfer pipette, wash the larvae with 1xPBS.
11.2.4.2.2 Larvae Dissection
-
1.
Transfer a single larva in a clean dissection dish containing 1xPBS under a stereomicroscope.
-
2.
Orient the anterior part of the larva to the left (for a right-handed person). Using the left hand, clamp the posterior part of the larva (second or third segment from the end) with a pair of forceps. With the right hand, rip the posterior part of the larva with a second pair of forceps.
-
3.
With both pair of forceps, invert the larvae by pushing the mouth hook through the body. Using the left hand, hold the larva anterior part while placing with your right hand a single tine of the forceps in the mouth hooks. Then roll the larvae inside out by gradually pushing the larva on the forceps tine with your left hand. Extend completely the larva so that the internal organs are fully apparent on the outside and the cuticle stretched on the inside. Remove carefully the stretched inverted larva from the forceps tine.
-
4.
Hold the larval carcass with the left-hand forceps, and using the right-hand forceps, unwind and remove the gut, the proventriculus, and as much as possible of the fat body without damaging the dorsal vessel/lymph gland region (which is lined by two dorsal patches of fat body).
-
5.
Transfer the carcass with the brain , the lymph gland, and the heart/aorta in a 1.5 ml microcentrifuge tube containing 1 ml of 1xPBS on ice.
-
6.
Repeat steps 1–5 to prepare as many larvae as needed. Note: steps 1–6 need to be performed as quickly as possible to limit perturbation of lymph gland homeostasis. Do not exceed 30 min in total before proceeding to step 7/fixation.
-
7.
Replace buffer with 1 ml of freshly prepared fixative solution (4% formaldehyde in 1xPBS), and incubate 30 min on a rocking platform at room temperature.
-
8.
Wash the fixed larvae with 1 ml of 1xPBST (1xPBS 0.1% Triton) for 10 min. Repeat twice.
-
9.
Fixed larvae can be stored at 4 °C overnight, but we usually proceed to the immunostaining straight away. For long-term storage, wash the larvae once in 0.5xPBS- 50% methanol and twice in 100% methanol before storing them in 100% methanol at -20 °C. In this case, serial rehydration steps in PBS of the samples will be needed before use. Note however that some epitopes are sensitive to methanol and may not be detected similarly as on freshly prepared tissues.
11.2.4.2.3 Immunostaining
Unless specified, all washes and incubations are performed at room temperature on a rocking platform.
-
1.
Discard 1xPBST, and permeabilize the tissues by adding 1 ml of 1xPBS, 0.3% Triton for 30 min.
-
2.
Discard permeabilization buffer, and wash twice with 1 ml of 1xPBST.
-
3.
Replace wash buffer with 1 ml blocking solution (1xPBST, 1% BSA) for 30 min.
-
4.
Discard blocking solution, and replace with 1xPBST containing the primary antibody diluted at the appropriate concentration. Flip the tube to mix well, and incubate overnight at 4 °C. Note: a minimal volume of 50 μl of diluted primary antibody can be used for 10–15 larvae.
-
5.
Remove primary antibody, and wash with 1 ml of 1xPBST for 10 min; repeat twice.
-
6.
Discard wash buffer, and add 500 μL of 1xPBST containing the secondary antibody diluted at the appropriate concentration (usually 1:1000). Mix well, and incubate for 4 h in the dark. Overnight incubation at 4 °C is also possible.
-
7.
Discard secondary antibody, and wash with 1 ml of 1xPBST for 10 min in the dark; repeat twice.
-
8.
Replace wash buffer with 1 ml of 1xPBST + DNA stain (TO-PRO-3 or DAPI, 1:1000), and incubate 20 min in the dark.
-
9.
Discard DNA stain, and wash rapidly twice with 1 ml of 1xPBST and twice with 1 ml of 1xPBS, and then transfer to 1xPBS 5% glycerol (the presence of glycerol helps to prevent drying out during the final dissection).
-
10.
Store at 4 °C in the dark, or begin the mounting procedure.
11.2.4.2.4 Mounting Lymph Glands for Microscopy
-
1.
Place 10μl of mounting medium on a microscope slide.
-
2.
Using fine forceps, transfer the carcasses next to the drop of mounting medium.
-
3.
Under a stereomicroscope and using syringe needles, carefully separate the lymph glands from the other tissues. The lymph glands are normally still attached to the ring gland and the brain on their anterior side and to the dorsal vessel on their posterior. Separate the ring gland from the brain. You can then use the ring gland or the dorsal vessel to drift the dissected lymph gland into the mounting medium. Align as much as possible the lymph gland lobes along their anterior-posterior axis, anterior to the left and posterior to the right. Proceed similarly with the other larvae. Up to 16 lymph glands can be prepared in 10 μl of mounting medium, but beginners may prefer to split their samples between different slides. Discard the larval carcasses and remaining tissues from the slide.
-
4.
Place the 18 × 18mm coverslip to the left of the lymph glands/drop of mounting medium, and lay it down slowly on the mounting medium to keep the lymph glands well positioned. Note: other sizes of coverslip can be used, in which case the amount of mounting medium needs to be adjusted.
-
5.
Store the slides at 4 °C in the dark until imaging (usually by confocal microscopy).
11.3 Drosophila and Leukemogenesis
The presence of hereditary tumors in Drosophila was reported one century ago (Stark 1919; Wilson 1924), and the participation of blood cells in these tumors was described more than 60 years ago (Oftedal 1953; Rizki 1960). As we shall see below, since then, many studies have used Drosophila first to identify genes whose deregulation causes these tumors and then to characterize the mode of action of conserved genes mutated in human leukemia or to develop specific models for human leukemogenic proteins.
11.3.1 Melanotic Tumor or Drosophila “Leukemia”
Historically, the first strategy to identify and characterize genes regulating blood cell development and potentially involved in leukemia has been to study melanotic tumor formation (Sang and Burnet 1963; Sparrow 1974; Gateff 1978; Gateff 1994; Watson et al. 1994). These melanotic masses, which are easily observable through the cuticle of the larvae (or the adult), are mostly composed of blood cells that have aggregated together or around another tissue and have melanized (Rizki and Rizki 1979, Minakhina and Steward 2006). Their presence is generally associated with increased blood cell numbers, enlarged or precociously ruptured lymph glands, and lamellocyte differentiation. As such, they might represent a model of leukemogenesis in Drosophila, and, indeed, several genetic screens using this phenotype as a readout have been performed to unveil new genes controlling blood cell homeostasis. These screens initially relied on classical mutagenic events such as P-element-mediated insertion or EMS mutagenesis (Hanratty and Ryerse 1981; Watson et al. 1991; Torok et al. 1993; Luo et al. 1997; Wu et al. 2001). More recently the UAS/GAL4 system and the advent of genome-wide UAS-RNAi libraries made it possible to target gain or loss of function specifically in the hematopoietic compartment (Zettervall et al. 2004; Stofanko et al. 2008; Avet-Rochex et al. 2010; McNerney et al. 2013). Importantly, enhancers and suppressors of melanotic tumor genes can then be sought in modifier screens (Luo et al. 1995; Shi et al. 2006; Anderson et al. 2017), leading to further characterization of the pathways regulated by these genes (see below). To date, more than 150 genes have been identified that are associated with melanotic tumor formation. These include loss of function mutations in ribosomal proteins (Watson et al. 1992), which could be related to human ribosomopathies that are associated with predisposition to leukemia (Danilova and Gazda 2015), or activating mutation in the Toll/NF-κB pathway (Qiu et al. 1998) that is also constitutively activated and promotes cell survival in a number of hematological malignancies (Gasparini et al. 2014).
However, one important caveat of the “melanotic tumors” is that they can arise from two distinct origins: on the one hand, they can form as a consequence of immune response to damaged tissues, and on the other hand, they can be caused by cell-autonomous deregulation of the hematopoietic program (Wu et al. 2001; Minakhina and Steward 2006; Avet-Rochex et al. 2010; Zang et al. 2015). Hence, their presence often reflects the spurious activation of the lamellocytes rather than a leukemic-like process, and in-depth follow-up studies are necessary to delineate whether the function of these “melanotic tumor genes” is more relevant to tissue homeostasis, immunity, or blood cell cancer.
11.3.2 From Melanotic Tumor to Human Leukemia: The JAK/STAT Pathway
The most notorious example of “melanotic tumor gene” whose study turned out to be highly significant to human leukemia is certainly hopscotch (hop). Hop encodes the Drosophila homologue of the JAK kinase, a key component of the eponym JAK/STAT pathway, which mediates signaling from various cytokines (such as Unpaired1, 2, and 3 in Drosophila or erythropoietin and granulocyte colony-stimulating factor in human) and their cognate receptors (Amoyel et al. 2014).
The mutation tumorous-lethal, an allele of hop (hop Tum-l), was identified more than 40 years ago as a recessive lethal temperature-sensitive mutation associated with melanotic tumor formation (Corwin and Hanratty 1976; Hanratty and Ryerse 1981). Importantly, the hypertrophic lymph glands of hop Tum-l larvae are neoplastic and can give rise to lethal tumors upon serial transplantations into recipient adult flies (Hanratty and Ryerse 1981, Luo et al. 1995) (Note: while secondary transplantation is standard in mouse to assess blood cell oncogenic transformation, this experiment is unfortunately seldom used in Drosophila). Molecular analyses showed that hop Tum-l encodes a hyperactive Hop kinase due to the G341E substitution in JAK homology domain 4 (JH4) (Harrison et al. 1995; Luo et al. 1995). Overexpression of HopTum-l in the lymph gland is sufficient to induce lymph gland hypertrophy, lamellocyte differentiation, and melanotic tumor formation (Luo et al. 1995). Likewise, the Hop T42 allele, which causes similar phenotypes, also gives rise to a hyperactivated form of Hop due to the E695K substitution in the JH2 domain (Luo et al. 1997). The primary effect of these mutations is to activate in a ligand-independent manner the JAK/STAT pathway by phosphorylating transcription factors of the STAT family, thereby inducing their dimerization and nuclear translocation. Consistent with this idea, decreasing the dosage of the Drosophila STAT factor STAT92E in hop Tum-l larvae is sufficient to reduce melanotic tumor incidence (Hou et al. 1996; Yan et al. 1996; Luo et al. 1997; Shi et al. 2006), whereas the overexpression of an active STAT92E induces melanotic tumor formation (Ekas et al. 2010). These findings thus pointed toward an oncogenic role of the JAK/STAT pathway in leukemia. Strikingly, since 2005, it has been demonstrated that a functionally equivalent activating point mutation in JAK2 (V617F, in the JH2 domain) is one of the most common initiating events in various human myeloproliferative neoplasms (MPN), such as polycythemia vera, essential thrombocytosis, or primary myelofibrosis (Jones et al. 2005; Kralovics et al. 2005; Levine et al. 2005). This mutation accounts for ±70% of MPN, and other point mutations or translocations leading to JAK2 constitutive activation have been identified at lesser frequencies in other hematopoietic malignancies (Kantarcioglu et al. 2015; Vainchenker and Kralovics 2017). Hence, understanding how JAK activation promotes leukemia has become a major issue.
Further studies in Drosophila blood cells have brought to light a number of modulators of the JAK/STAT pathway potentially implicated in leukemia. In particular, different genetic screens for second-site modifiers of hop Tum-l-induced melanotic tumor formation have been performed. For instance, using a set of genetic deficiencies uncovering ± 70% of the Drosophila autosomes, Shi et al. identified more than 30 genes acting as dominant modifiers of hop Tum-l -induced melanotic tumor formation in the adult (Shi et al. 2006). Notably, they showed that JAK overactivation promotes proliferation and tumorigenesis by counteracting heterochromatin gene silencing. This effect seems to involve a noncanonical mechanism whereby the unphosphorylated STAT92E is targeted to the heterochromatin by the linker histone H1 and maintains heterochromatin protein 1 (HP1) localization and heterochromatin stabilization (Shi et al. 2006, Xu et al. 2014). A similar link between JAK/STAT and heterochromatin gene silencing has been observed in human: unphosphorylated STAT5 was found to bind HP1α and stabilize heterochromatin (Hu et al. 2013), while JAK2 activation was shown to displace HP1α from the heterochromatin, potentially by directly phosphorylating histone H3 (Dawson et al. 2009). Moreover, JAK2 promotes the survival of primary mediastinal B cell lymphoma and Hodgkin lymphoma cells by promoting heterochromatin formation in cooperation with the histone demethylase JMJD2C (Rui et al. 2010). It is thus possible that heterochromatin alteration is implicated in MPN development, and the role of unphosphorylated STAT in leukemia certainly deserves further investigations.
Thanks to another deficiency screen for modifiers of hop Tum-l, Anderson et al. recently found that the Hippo signaling pathway is activated by Hop and contributes to melanotic tumor development by inducing blood cell proliferation in peripheral larval hemocytes (Anderson et al. 2017). On the other hand, Terriente-Felix et al. showed that JAK-induced hypertrophy of the lymph gland was mediated by the p38 MAPK pathway (Terriente-Felix et al. 2017). It will thus be interesting to test whether these two pathways are activated in MPN and contribute to blood cell neoplasia.
Another modifiers of JAK/STAT overactivation in Drosophila blood cells is abnormal wing disc (awd) (Zinyk et al. 1993), the homologue of the tumor suppressor Nm23. Awd regulates the endocytosis of several receptors including the JAK/STAT pathway receptor Domeless (Dome) (Nallamothu et al. 2008), and some evidence suggests that Nm23 is implicated in leukemia in human (Lilly et al. 2015). Interestingly, Dome is required for Hop-induced lymph gland hypertrophy (Terriente-Felix et al. 2017), which is consistent with ex vivo experiments showing that JAK2V617F requires a cytokine receptor scaffold for its transforming and signaling activities (Lu et al. 2005). In addition, Hop induces a feed-forward loop by activating the expression of Dome ligand Upd3, which also contributes to lymph gland hypertrophy (Terriente-Felix et al. 2017). Along the same line, JAK2 V617F-induced MPN in a mouse model seems to depend on the expression of thrombopoietin and its receptor MPL (which is also subject to activating mutations in some MPN) (Sangkhae et al. 2014). Hence, cytokine receptors may participate in JAK/STAT-induced blood cell proliferation by several mechanisms.
Beside these in vivo studies, it is worth mentioning that Drosophila blood cell lines (such as Kc167 or S2 cells) are particularly well suited for genome-wide RNAi screens . Using this approach, two studies identified more than 100 genes regulating JAK/STAT-dependent transactivation of a reporter gene (Baeg et al. 2005; Muller et al. 2005), including BRDWD3 and Ptp61F, which also genetically interacted with hop Tum-l in vivo (Muller et al. 2005). Besides, transcriptomic profiling in Kc167 cells and in larval tissue led to the identification of JAK/STAT target genes, some of which, like G protein a 73B, chinmo, or eukaryotic initiation factor 1A, contribute to hop Tum-l-induced hematopoietic tumor formation (Myrick and Dearolf 2000; Bina et al. 2010; Flaherty et al. 2010; Bausek and Zeidler 2014). Whether homologues of these genes are implicated in JAK/STAT signaling and leukemia in human certainly warrants further investigation. All together these data illustrate how a variety of approaches in Drosophila can highlight multiple levels of regulation and of action of the JAK/STAT pathway relevant to blood cell transformation.
11.3.3 Study of Leukemogenic Proteins in Drosophila Non-Hematopoietic Tissues
The development of transgenic or knock-in animal models expressing a human leukemogenic protein has been instrumental to decipher how these proteins interfere with the normal functions of the cells. Of course, mouse remains the prevalent model for such studies (Kohnken et al. 2017), but Drosophila offers a cost- and time-effective surrogate to assess in vivo the function(s) and mode(s) of action of human proteins involved in leukemia. While targeting their expression in hematopoietic cells may seem most suitable (see below), “ectopic” expression in unrelated tissue can also present some advantages (e.g., tissue accessibility, previous knowledge of the system, available tools, etc.), and this approach has been used in a few cases in Drosophila.
Actually, the first human leukemogenic protein studied in Drosophila was BCR-ABL, the product of the notorious Philadelphia chromosome , which is responsible for almost all cases of chronic myeloid leukemia (CML) and some cases of acute lymphoid leukemia (ALL) (Mughal et al. 2016). BCR-ABL is generated by a balanced translocation between c-Abelson (Abl) on chromosome 9 and the breakpoint cluster region (bcr) on chromosome 22. Depending on the location of the breakpoint within bcr, two main fusion proteins are generated: p210 in most CML and p185 in most ALL. In both proteins, the dimerization domain coded by bcr induces the constitutive activation of the tyrosine kinase ABL. To gain insight into the respective mode of action of these two isoforms, Fogerty et al. generated transgenic flies expressing p210 or p185 human/fly chimeras (Fogerty et al. 1999): BCR and the N-terminal ABL were derived from human, whereas the divergent C-terminal tail of ABL was from Drosophila. Both p210 and p185 rescued the lethality of dAbl mutant flies and activated ABL signaling pathway. Yet, their overexpression generated distinct phenotypes and ectopically activated some pathways not employed by ABL (Fogerty et al. 1999; Stevens et al. 2008). Further work using this model may thus help to identify components of the BCR-ABL signaling cascades and the differences underlying the distinct clinical features of p210- and p185-associated leukemia.
A similar strategy was employed to study two different leukemogenic fusions involving mixed lineage leukemia (MLL, also known as KMT2A for lysine-specific methyltransferase 2A), which is translocated in 5–10% of patients with acute myeloid or lymphoid leukemia (AML/ALL) (Slany 2016, Yokoyama 2017). Mll is the homologue of Drosophila trithorax. It is the target of more than 100 different chromosomal rearrangements that result in the expression of a fusion protein between MLL, deprived of its PHD and SET domains, and the C-terminus of its partner. The two most common translocation products are MLL-AF9 in AML and MLL-AF4 in ALL. AF4 and AF9 interact with each other and also directly recruit other transcriptional coactivator complexes. Using various drivers (including some blood cell drivers), it was shown that the expression of MLL-AF4 or MLL-AF9 but not MLL causes larval to pupal lethality (Muyrers-Chen et al. 2004). In addition, these two fusions had different effects on proliferation and chromosome condensation in larval brains and displayed largely nonoverlapping binding patterns on polytene chromosomes. These findings thus suggested that the C-terminal partners of the MLL fusion proteins may modify differentially MLL activity notably by regulating its targeting to distinct set of genes. Consistent with this idea, recent ChIP-seq experiments in human leukemia cell lines showed that MLL-AF4 and MLL-AF9 have distinct binding site repertoires (Prange et al. 2017).
Finally, a recent study has developed a transgenic fly model for the transactivator Tax-1 (Shirinian et al. 2015). Tax-1 is encoded by the human T cell leukemia virus type 1 (HTLV-1), a retrovirus that causes an aggressive adult T cell leukemia/lymphoma in ± 5% of infected individuals (Bangham and Ratner 2015). Tax-1 is essential for HTLV-1 oncogenic properties, and several lines of evidence indicate that the binding of IKK kinases by Tax-1 and the ensuing activation of the NF-κB pathway are critical for T cell transformation. The expression of Tax-1 in the Drosophila eye or in the plasmatocytes, respectively, caused a rough eye phenotype and an increase in larval blood cell number (Shirinian et al. 2015). In contrast, the expression of Tax-2, which is encoded by the genetically related but non-oncogenic retrovirus HTLV-2, did not alter eye development or hemocyte number. Moreover, further experiments demonstrated that the deleterious function of Tax-1 in the Drosophila eye was mediated by activation of the NF-κB pathway. These findings thus established that Drosophila could be used as a genetic model to investigate the mode of action of Tax-1 in cell transformation.
The above three examples illustrate how works in non-hematopoietic tissues of Drosophila can help to describe the activity of human leukemogenic proteins. In each case, the expression of the oncogene gave rise to a robust phenotype (rough eye for BCR-ABL and Tax-1, larval/pupal lethality for MLL-AF9 and MLL-AF4) that could be used as readouts in a modifier screen . Hence, further experiments exploiting the genetic tools available in Drosophila could undoubtedly bring interesting insights into the mode(s) of action of these factors in vivo.
11.3.4 Study of Leukemogenic Proteins in Drosophila Hematopoietic Cells
A seemingly more relevant approach is to study the function of human leukemogenic protein or that of their Drosophila counterparts, directly in the fly hematopoietic system.
The best example here is provided by studies of the transcription factor RUNX1 and of its oncogenic derivative RUNX1-ETO. RUNX1 is a key regulator of several steps of blood cell development in vertebrates (de Bruijn and Dzierzak 2017). Recurrent point mutations or translocations affecting RUNX1 are among the most frequent genetic abnormalities in human leukemia (Sood et al. 2017). For instance, the prototypical t(8;21) translocation, which accounts for ±10% of all cases of AML, gives rise to a fusion protein between RUNX1 DNA-binding domain and the transcriptional corepressor ETO (also known as RUNX1T1). The resulting chimera, RUNX1-ETO, chiefly acts by interfering directly with the regulation of RUNX1 target genes. Understanding how deregulation of RUNX1 activity leads to hematological malignancies is thus a field of intense investigation, and several RUNX1-ETO animal models have been developed in mouse, zebrafish, and fly. Of note, in Drosophila, the RUNX1 homologue Lozenge (Lz) is expressed in the crystal cell lineage and is absolutely required for the development of this blood cell type (Waltzer et al. 2010). In addition, Lz is well known for its function in the eye where it regulates photoreceptor and cone cell fate (Canon and Banerjee 2000). Besides an early study where RUNX1-ETO was expressed in the Drosophila eye and found to interfere with Lz function by acting as a constitutive transcriptional repressor of two Lz target genes (Wildonger and Mann 2005), two concurrent studies investigated RUNX1-ETO impact on the development of Drosophila hematopoietic system (Osman et al. 2009, Sinenko et al. 2010).
In one case, RUNX1-ETO expression was induced in the majority of the circulating larval blood cells using the hml-GAL4 driver (Sinenko et al. 2010). Reminiscent of the phenotypes observed in mouse models, RUNX1-ETO expression caused a sharp increase in the number of circulating hemocytes along with an expansion of the immature blood cell population. This “leukemic” phenotype required RUNX1 DNA-binding activity and its interaction with its cofactor CBFß as well as different domains of ETO known to interact with transcriptional corepressors. Further analyses revealed that elevated ROS levels were crucial for RUNX1-ETO-induced expansion of the immature blood cells in Drosophila, suggesting that a similar mechanism might participate in t(8;21)+ AML. Actually increased ROS levels are observed in many cases of leukemia (Udensi and Tchounwou 2014). In addition, RUNX1-ETO expression was associated with the development of melanotic tumors (Sinenko et al. 2010). By screening a panel of 231 deficiencies and 1500 automosal insertional mutations, 10 suppressors and 12 enhancers of RUNX1-ETO-induced melanotic tumor formation/blood cell number increase were identified. It will be thus of particular interest to test whether the homologues of these genes also interfere with or promote RUNX1-ETO oncogenic activity in human.
In a second case, RUNX1-ETO was expressed in the Lz+ blood cell lineage using the lz-GAL4 driver to mimic more closely the t(8;21) situation (Osman et al. 2009). As in mammals, it was found that RUNX1-ETO interferes with the function of the endogenous RUNX protein Lz. This led to the accumulation of a high number of Lz+ cells that failed to differentiate in crystal cells. However, RUNX1-ETO did not solely behave as a repressor on Lz target genes in the hematopoietic system. Indeed, experiments in human t(8;21)+ AML cell have since then revealed that RUNX1-ETO binding can lead both to activation and repression of its target genes (Ptasinska et al. 2012). In addition, lz-driven expression of RUNX1-ETO caused pupal lethality. Using an in vivo RNAi-based screen strategy, more than 2000 genes were individually knocked down in RUNX1-ETO-expressing cells to identify genes that are cell-autonomously required for its activity. Among the nine suppressors of RUNX1-ETO-induced lethality, the protease calpain B and the AAA+ ATPase RUVBL1/Pontin were studied in more detail. Interestingly both are required for Lz+ blood cell number increase and differentiation blockade caused by RUNX1-ETO, while their knockdown or mutation does not affect Lz+ blood cell development in a wild-type situation (Osman et al. 2009; Breig et al. 2014). It appears that calpain B is required for RUNX1-ETO stability in Lz+ cells (Osman et al. 2009). Strikingly, in human, calpain inhibition also causes RUNX1-ETO degradation and specifically impairs the viability and clonogenic growth of t(8;21)+ AML cells, which are known to depend on RUNX1-ETO expression. Therefore, the regulation of RUNX1-ETO by calpains seems conserved, and calpain inhibitors might be used as therapeutic agents in leukemia. Similarly, works in human cell lines showed that Pontin expression is activated by RUNX1-ETO and cooperates with this oncogene to sustain leukemic blood cell proliferation and survival (Breig et al. 2014). As RUNX1-ETO-expressing cells appear sensitized to Pontin knockdown, recently developed chemical inhibitors of its ATPase activity might be useful therapeutic agents in t(8;21)+ AML.
Incidentally, other studies in Drosophila blood cells have revealed a mechanism of regulation of RUNX activity relevant to human AML. Myeloid leukemia factor (MLF) was found to be required for Lz-induced transactivation in a genome-wide RNAi screen in Kc167 cells (Gobert et al. 2010; Bras et al. 2012). In human, MLF1 was identified as the target of a rare translocation in patients with myelodysplastic syndrome and AML (Yoneda-Kato et al. 1996) and more recently as a tumor suppressor in infant T cell acute lymphoblastic leukemia (Mansur et al. 2015). Yet the function and mode of action of this conserved family of protein remain largely unknown (Gobert et al. 2012). Analyzing mlf function in Drosophila revealed that it regulates Lz+ cell number by stabilizing Lz (Bras et al. 2012). Interestingly, the expression of human MLF1 can rescue mlf mutant defects, indicating that MLF function is conserved through evolution (Martin-Lanneree et al. 2006; Bras et al. 2012). Moreover, MLF1 appears to be required for RUNX1-ETO stable expression and the growth of t(8;21)+ AML cells (Bras et al. 2012). At the molecular level, MLF acts at least in part as a component of a conserved Hsp70/DnaJ chaperon complex to promote RUNX protein stability (Dyer et al. 2017, Miller et al. 2017). Whether this chaperon complex is also involved in RUNX-dependent AML remains to be assessed. Still it is interesting to note that haploinsufficient mutations in RUNX1 are associated with AML, indicating that a tight regulation of its level is critical to prevent leukemogenesis (Sood et al. 2017). In fly, reducing Lz level impairs Lz-mediated repression of Notch and causes a myelodysplastic-like phenotype due to sustained overactivation of the Notch pathway (Miller et al. 2017). Given the importance of activated Notch signaling in hematological malignancies (Gu et al. 2016), a similar functional relationship between RUNX1, MLF1, and Notch might be at stake in human blood cell transformation.
A slightly different strategy was used to dissect leukemia-associated isocitrate dehydrogenase (IDH) mutants in Drosophila. In that case, rather than using the human oncoproteins, the authors used a transgenic approach to drive the expression of their Drosophila counterpart carrying homologous mutations. In human, somatic mutations in conserved arginine residues within the active sites of IDH1 and IDH2 are very frequent in AML and in gliomas (Gagne et al. 2017). While IDH normally converts isocitrate into alpha-ketoglutarate (αKD), the mutated forms produce D2-hydroxyglutarate (D2-HG), an oncometabolite that accumulates at high levels in cancer tissues and inhibits the activity of αKD-dependent enzymes, some of which are implicated in chromatin compaction, DNA methylation, collagen modification, or response to hypoxia. To develop a genetically tractable model for IDH-associated leukemia, Reitman et al. generated transgenic lines expressing mutated forms of the single Idh Drosophila gene (Reitman et al. 2015). The overexpression of these IDH mutants in circulating larval hemocytes using the hml-GAL4 driver caused an increase in blood cell number, lamellocyte differentiation, and melanotic tumor formation but also a rise in undifferentiated blood cells, similar to what is observed in mouse models of IDH-mutated leukemia. Interestingly, the severity of the phenotypes was correlated with the level of D2-HG production. Moreover, using a UAS-based co-expression strategy, enhancers and suppressors of IDH mutant-associated phenotypes (melanotic tumor formation or wing expansion defects) were sought. This screen suggested that the phenotypes of IDH mutants are caused by increased levels of ROS, as observed in the case of RUNX1-ETO (Sinenko et al. 2010). In contrast to wild-type IDH that reduces NADP+ to NADPH to produce αKG, mutant IDH consumes NADPH to produce D2-HG (Gagne et al. 2017). Therefore a decreased level of NADPH (an important source of reducing power) in IDH1/2-mutated cells could interfere with ROS detoxification. It would thus be interesting to further study the contribution of ROS to IDH-associated leukemia in humans.
Finally a recent report investigated the consequences of expressing the human NUP98-HOXA9 (NA9) fusion protein in the Drosophila lymph gland (Baril et al. 2017). Several HOX transcription factors and their cofactors MEIS and PBX are deregulated in AML (Alharbi et al. 2013). Similarly, recurrent chromosomal translocations between NUP98, which encodes a nucleoporin, and several partner genes, including some HOX, occur in AML (Gough et al. 2011). Among them, the rare and high-risk t(7;11)(p15;p15) translocation generates a fusion between the N-terminal of NUP98 and the DNA-binding and heterodimerization domains of HoxA9. In Drosophila, hml-driven expression of NA9 in differentiating larval hemocytes caused a massive increase in circulating blood cells as well as lymph gland hyperplasia, reminiscent of the myeloproliferative disease induced by NA9 in mouse models (Baril et al. 2017). Also, as in mammals, NA9 activity required interaction with DNA and with PBX mediated by HoxA9 moiety. Interestingly, NA9 expression in differentiating hemocytes not only increased the number of differentiated cells at the expanse of the blood cell progenitors but also altered PSC size and morphology, indicating that NA9 affects lymph gland niche/microenvironment nonautonomously. These phenotypes were due to defective PVR (PDGF/VEGF related) signaling, a receptor tyrosine kinase (RTK) related to several mammalian RTK involved in leukemia (Gough et al. 2011). In mammals, remodeling of the bone marrow niche by AML cells is thought to promote leukemia at the expanse of normal hematopoiesis (Shafat et al. 2017). It is tempting to speculate that NA9 could hijack the hematopoietic stem cell niche by interfering with RTK signaling. Importantly too, these findings also highlight that Drosophila can be used to investigate cross-regulatory interactions between leukemic cells and their microenvironment.
11.4 Conclusion
The various examples above show that works in Drosophila can contribute to our understanding of leukemia pathogenesis. Drosophila seems particularly well suited to decipher the mechanisms of action of proteins involved in human leukemia that have clear homologues in fly. The use of genetic interaction screen in Drosophila is an extremely powerful means to identify regulators or mediators of these leukemogenic proteins. In addition, the relative simplicity of the fly hematopoietic system offers a good opportunity to study its different components and the interaction between them at the cellular, developmental, and molecular levels. Yet, the lack of clearly identified hematopoietic stem cells, the limited functional homologies between mammalian and Drosophila blood cell types, or the more restricted set of genes known to control Drosophila hematopoiesis also hamper the development of leukemia models in this system or at least restrain their heuristic value. Still, there are means to take further advantage of Drosophila hematopoietic system to develop more refined models of leukemia. Leukemia arises as a consequence of a limited number of mutations. While most models in fly have focused on the expression of a single oncogenic factor, it is possible to generate Drosophila avatars where the expression or function of multiple genes is modified in specific blood cell lineages and/or in a temporal series. This could bring important information as to the mechanisms of oncogenic cooperation that drives blood cell transformation. Besides somatic mutations, germline mutations, for instance, in RUNX1 or GATA2, are associated with predisposition to develop leukemia. The CRISPR/Cas9 technology could be used to generate relevant knock-in as well as allelic series to study these familial cases of leukemia. In addition, a wider use of cell sorting or cell transplantation assays could help define the characteristics of Drosophila leukemic blood cells. A better knowledge of the Drosophila adult hematopoietic system could also favor the analysis of leukemia models past the larval stage. Indeed, it would be particularly interesting to follow the evolution of “leukemic clones” over a longer time period and during aging. In addition, the interactions between “leukemic blood cells” and their surrounding cellular environment can easily be studied in fly, and a great deal of knowledge could be drawn from such analyses. Furthermore, Drosophila provides a simple model to investigate exogenous factors (nutrition, microbiota, infection, etc.) susceptible to influence leukemia development. It could also easily be used to perform chemical screens for compounds interfering with leukemogenic proteins function, which would open avenues for new therapeutic development. Finally, and perhaps most importantly, a genuine effort is still needed to transfer more effectively the knowledge obtained from Drosophila models toward human leukemia. Indeed, except for a few cases, the discoveries made in fly have not been tested thereafter in human leukemic cells or in mammalian models. Nonetheless, several features of leukemia have been recapitulated in Drosophila, and new insights into leukemic proteins mode(s) of action have been gained thanks to this model. In light of these promising results, we should endeavor to take a step further by translating these findings to human and eventually to clinic.
References
Alharbi RA, Pettengell R, Pandha HS, Morgan R. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia. 2013;27(5):1000–8.
Amoyel M, Anderson AM, Bach EA. JAK/STAT pathway dysregulation in tumors: a Drosophila perspective. Semin Cell Dev Biol. 2014;28:96–103.
Anderl I, Vesala L, Ihalainen TO, Vanha-Aho LM, Ando I, Ramet M, Hultmark D. Transdifferentiation and proliferation in two distinct hemocyte lineages in Drosophila melanogaster Larvae after Wasp Infection. PLoS Pathog. 2016;12(7):e1005746.
Anderson AM, Bailetti AA, Rodkin E, De A, Bach EA. A genetic screen reveals an unexpected role for yorkie signaling in JAK/STAT-Dependent hematopoietic malignancies in Drosophila melanogaster. G3 (Bethesda). 2017;7(8):2427–38.
Avet-Rochex A, Boyer K, Polesello C, Gobert V, Osman D, Roch F, Auge B, Zanet J, Haenlin M, Waltzer L. An in vivo RNA interference screen identifies gene networks controlling Drosophila melanogaster blood cell homeostasis. BMC Dev Biol. 2010;10:65.
Ayyaz A, Li H, Jasper H. Haemocytes control stem cell activity in the Drosophila intestine. Nat Cell Biol. 2015;17(6):736–48.
Baeg GH, Zhou R, Perrimon N. Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes Dev. 2005;19(16):1861–70.
Bangham CR, Ratner L. How does HTLV-1 cause adult T-cell leukaemia/lymphoma (ATL)? Curr Opin Virol. 2015;14:93–100.
Baril C, Gavory G, Bidla G, Knaevelsrud H, Sauvageau G, Therrien M. Human NUP98-HOXA9 promotes hyperplastic growth of hematopoietic tissues in Drosophila. Dev Biol. 2017;421(1):16–26.
Bataille L, Auge B, Ferjoux G, Haenlin M, Waltzer L. Resolving embryonic blood cell fate choice in Drosophila: interplay of GCM and RUNX factors. Development. 2005;132(20):4635–44.
Bausek N, Zeidler MP. Galpha73B is a downstream effector of JAK/STAT signalling and a regulator of Rho1 in Drosophila haematopoiesis. J Cell Sci. 2014;127(Pt 1):101–10.
Benmimoun B, Polesello C, Haenlin M, Waltzer L. The EBF transcription factor Collier directly promotes Drosophila blood cell progenitor maintenance independently of the niche. Proc Natl Acad Sci U S A. 2015;112(29):9052–7.
Benmimoun B, Polesello C, Waltzer L, Haenlin M. Dual role for Insulin/TOR signaling in the control of hematopoietic progenitor maintenance in Drosophila. Development. 2012;139(10):1713–7.
Bina S, Wright VM, Fisher KH, Milo M, Zeidler MP. Transcriptional targets of Drosophila JAK/STAT pathway signalling as effectors of haematopoietic tumour formation. EMBO Rep. 2010;11(3):201–7.
Brandt SM, Schneider DS. Bacterial infection of fly ovaries reduces egg production and induces local hemocyte activation. Dev Comp Immunol. 2007;31(11):1121–30.
Bras S, Martin-Lanneree S, Gobert V, Auge B, Breig O, Sanial M, Yamaguchi M, Haenlin M, Plessis A, Waltzer L. Myeloid leukemia factor is a conserved regulator of RUNX transcription factor activity involved in hematopoiesis. Proc Natl Acad Sci U S A. 2012;109(13):4986–91.
Braun A, Hoffmann JA, Meister M. Analysis of the Drosophila host defense in domino mutant larvae, which are devoid of hemocytes. Proc Natl Acad Sci U S A. 1998;95(24):14337–42.
Breig O, Bras S, Martinez Soria N, Osman D, Heidenreich O, Haenlin M, Waltzer L. Pontin is a critical regulator for AML1-ETO-induced leukemia. Leukemia. 2014;28(6):1271–9.
Canon J, Banerjee U. Runt and Lozenge function in Drosophila development. Semin Cell Dev Biol. 2000;11(5):327–36.
Chakrabarti S, Dudzic JP, Li X, Collas EJ, Boquete JP, Lemaitre B. Remote control of intestinal stem cell activity by haemocytes in Drosophila. PLoS Genet. 2016;12(5):e1006089.
Clark RI, Woodcock KJ, Geissmann F, Trouillet C, Dionne MS. Multiple TGF-beta superfamily signals modulate the adult Drosophila immune response. Curr Biol. 2011;21(19):1672–7.
Corwin HO, Hanratty WP. Characterization of a unique lethal tumorous mutation in Drosophila. Mol Gen Genet. 1976;144(3):345–7.
Crozatier M, Ubeda JM, Vincent A, Meister M. Cellular immune response to parasitization in Drosophila requires the EBF orthologue collier. PLoS Biol. 2004;2(8):E196.
Danilova N, Gazda HT. Ribosomopathies: how a common root can cause a tree of pathologies. Dis Model Mech. 2015;8(9):1013–26.
Dawson MA, Bannister AJ, Gottgens B, Foster SD, Bartke T, Green AR, Kouzarides T. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461(7265):819–22.
de Bruijn M, Dzierzak E. Runx transcription factors in the development and function of the definitive hematopoietic system. Blood. 2017;129(15):2061–9.
Dey NS, Ramesh P, Chugh M, Mandal S, Mandal L. Dpp dependent Hematopoietic stem cells give rise to Hh dependent blood progenitors in larval lymph gland of Drosophila. Elife. 2016;5:26.
Dyer JO, Dutta A, Gogol M, Weake VM, Dialynas G, Wu X, Seidel C, Zhang Y, Florens L, Washburn MP, Abmayr SM, Workman JL. Myeloid leukemia factor acts in a chaperone complex to regulate transcription factor stability and gene expression. J Mol Biol. 2017;429(13):2093–107.
Ekas LA, Cardozo TJ, Flaherty MS, McMillan EA, Gonsalves FC, Bach EA. Characterization of a dominant-active STAT that promotes tumorigenesis in Drosophila. Dev Biol. 2010;344(2):621–36.
El Chamy L, Matt N, Reichhart JM. Advances in Myeloid-Like cell origins and functions in the model organism Drosophila melanogaster. Microbiol Spectrp. 2017;5(1)
Elrod-Erickson M, Mishra S, Schneider D. Interactions between the cellular and humoral immune responses in Drosophila. Curr Biol. 2000;10(13):781–4.
Eslin P, Prevost G, Havard S, Doury G. Immune resistance of Drosophila hosts against Asobara parasitoids: cellular aspects. Adv Parasitol. 2009;70:189–215.
Evans CJ, Liu T, Banerjee U. Drosophila hematopoiesis: markers and methods for molecular genetic analysis. Methods. 2014;68(1):242–51.
Fauvarque MO, Williams MJ. Drosophila cellular immunity: a story of migration and adhesion. J Cell Sci. 2011;124(Pt 9):1373–82.
Ferguson GB, Martinez-Agosto JA. Yorkie and Scalloped signaling regulates Notch-dependent lineage specification during Drosophila hematopoiesis. Curr Biol. 2014;24(22):2665–72.
Ferrando AA, Lopez-Otin C. Clonal evolution in leukemia. Nat Med. 2017;23(10):1135–45.
Flaherty MS, Salis P, Evans CJ, Ekas LA, Marouf A, Zavadil J, Banerjee U, Bach EA. chinmo is a functional effector of the JAK/STAT pathway that regulates eye development, tumor formation, and stem cell self-renewal in Drosophila. Dev Cell. 2010;18(4):556–68.
Fogarty CE, Diwanji N, Lindblad JL, Tare M, Amcheslavsky A, Makhijani K, Bruckner K, Fan Y, Bergmann A. Extracellular reactive oxygen species drive Apoptosis-Induced proliferation via Drosophila macrophages. Curr Biol. 2016;26(5):575–84.
Fogerty FJ, Juang JL, Petersen J, Clark MJ, Hoffmann FM, Mosher DF. Dominant effects of the bcr-abl oncogene on Drosophila morphogenesis. Oncogene. 1999;18(1):219–32.
Gasparini C, Celeghini C, Monasta L, Zauli G. NF-kappaB pathways in hematological malignancies. Cell Mol Life Sci. 2014;71(11):2083–102.
Gateff E. Malignant neoplasms of genetic origin in Drosophila melanogaster. Science. 1978;200(4349):1448–59.
Gateff E. Tumor-suppressor genes, hematopoietic malignancies and other hematopoietic disorders of Drosophila melanogaster. Ann N Y Acad Sci. 1994;712:260–79.
Ghosh S, Singh A, Mandal S, Mandal L. Active hematopoietic hubs in Drosophila adults generate hemocytes and contribute to immune response. Dev Cell. 2015;33(4):478–88.
Gobert V, Haenlin M, Waltzer L. Myeloid leukemia factor: a return ticket from human leukemia to fly hematopoiesis. Transcription. 2012;3(5):250–4.
Gobert V, Osman D, Bras S, Auge B, Boube M, Bourbon HM, Horn T, Boutros M, Haenlin M, Waltzer L. A genome-wide RNA interference screen identifies a differential role of the mediator CDK8 module subunits for GATA/RUNX-activated transcription in Drosophila. Mol Cell Biol. 2010;30(11):2837–48.
Gold KS, Bruckner K. Macrophages and cellular immunity in Drosophila melanogaster. Semin Immunol. 2015;27(6):357–68.
Gough SM, Slape CI, Aplan PD. NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood. 2011;118(24):6247–57.
Grigorian M, Mandal L, Hartenstein V. Hematopoiesis at the onset of metamorphosis: terminal differentiation and dissociation of the Drosophila lymph gland. Dev Genes Evol. 2011;221(3):121–31.
Gu Y, Masiero M, Banham AH. Notch signaling: its roles and therapeutic potential in hematological malignancies. Oncotarget. 2016;7(20):29804–23.
Hanratty WP, Ryerse JS. A genetic melanotic neoplasm of Drosophila melanogaster. Dev Biol. 1981;83(2):238–49.
Harrison DA, Binari R, Nahreini TS, Gilman M, Perrimon N. Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 1995;14(12):2857–65.
Hartenstein V, Mandal L. The blood/vascular system in a phylogenetic perspective. Bioessays. 2006;28(12):1203–10.
Holz A, Bossinger B, Strasser T, Janning W, Klapper R. The two origins of hemocytes in Drosophila. Development. 2003;130(20):4955–62.
Honti V, Csordas G, Kurucz E, Markus R, Ando I. The cell-mediated immunity of Drosophila melanogaster: hemocyte lineages, immune compartments, microanatomy and regulation. Dev Comp Immunol. 2014;42(1):47–56.
Honti V, Csordas G, Markus R, Kurucz E, Jankovics F, Ando I. Cell lineage tracing reveals the plasticity of the hemocyte lineages and of the hematopoietic compartments in Drosophila melanogaster. Mol Immunol. 2010;47(11-12):1997–2004.
Horn L, Leips J, Starz-Gaiano M. Phagocytic ability declines with age in adult Drosophila hemocytes. Aging Cell. 2014;13(4):719–28.
Hou XS, Melnick MB, Perrimon N. Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell. 1996;84(3):411–9.
Hu X, Dutta P, Tsurumi A, Li J, Wang J, Land H, Li WX. Unphosphorylated STAT5A stabilizes heterochromatin and suppresses tumor growth. Proc Natl Acad Sci U S A. 2013;110(25):10213–8.
Jones AV, Kreil S, Zoi K, Waghorn K, Curtis C, Zhang L, Score J, Seear R, Chase AJ, Grand FH, White H, Zoi C, Loukopoulos D, Terpos E, Vervessou EC, Schultheis B, Emig M, Ernst T, Lengfelder E, Hehlmann R, Hochhaus A, Oscier D, Silver RT, Reiter A, Cross NC. Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood. 2005;106(6):2162–8.
Jung SH, Evans CJ, Uemura C, Banerjee U. The Drosophila lymph gland as a developmental model of hematopoiesis. Development. 2005;132(11):2521–33.
Kantarcioglu B, Kaygusuz-Atagunduz I, Uzay A, Toptas T, Tuglular TF, Bayik M. Myelodysplastic syndrome with t(9;22)(p24;q11.2), a BCR-JAK2 fusion: case report and review of the literature. Int J Hematol. 2015;102(3):383–7.
Khadilkar RJ, Vogl W, Goodwin K, Tanentzapf G. Modulation of occluding junctions alters the hematopoietic niche to trigger immune activation. Elife. 2017;6:e28081.
Kohnken R, Porcu P, Mishra A. Overview of the use of murine models in leukemia and lymphoma research. Front Oncol. 2017;7:22.
Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M, Skoda RC. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352(17):1779–90.
Krzemien J, Dubois L, Makki R, Meister M, Vincent A, Crozatier M. Control of blood cell homeostasis in Drosophila larvae by the posterior signalling centre. Nature. 2007;446(7133):325–8.
Krzemien J, Oyallon J, Crozatier M, Vincent A. Hematopoietic progenitors and hemocyte lineages in the Drosophila lymph gland. Dev Biol. 2010;346(2):310–9.
Lanot R, Zachary D, Holder F, Meister M. Postembryonic hematopoiesis in Drosophila. Dev Biol. 2001;230(2):243–57.
Leitao AB, Sucena E. Drosophila sessile hemocyte clusters are true hematopoietic tissues that regulate larval blood cell differentiation. Elife. 2015;4:1–38.
Letourneau M, Lapraz F, Sharma A, Vanzo N, Waltzer L, Crozatier M. Drosophila hematopoiesis under normal conditions and in response to immune stress. FEBS Lett. 2016;590(22):4034–51.
Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, Adelsperger J, Koo S, Lee JC, Gabriel S, Mercher T, D'Andrea A, Frohling S, Dohner K, Marynen P, Vandenberghe P, Mesa RA, Tefferi A, Griffin JD, Eck MJ, Sellers WR, Meyerson M, Golub TR, Lee SJ, Gilliland DG. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7(4):387–97.
Lilly AJ, Khanim FL, Bunce CM. The case for extracellular Nm23-H1 as a driver of acute myeloid leukaemia (AML) progression. Naunyn Schmiedebergs Arch Pharmacol. 2015;388(2):225–33.
Lu X, Levine R, Tong W, Wernig G, Pikman Y, Zarnegar S, Gilliland DG, Lodish H. Expression of a homodimeric type I cytokine receptor is required for JAK2V617F-mediated transformation. Proc Natl Acad Sci U S A. 2005;102(52):18962–7.
Luo H, Hanratty WP, Dearolf CR. An amino acid substitution in the Drosophila hopTum-l Jak kinase causes leukemia-like hematopoietic defects. EMBO J. 1995;14(7):1412–20.
Luo H, Rose P, Barber D, Hanratty WP, Lee S, Roberts TM, D'Andrea AD, Dearolf CR. Mutation in the Jak kinase JH2 domain hyperactivates Drosophila and mammalian Jak-Stat pathways. Mol Cell Biol. 1997;17(3):1562–71.
Gagne LM, Boulay K, Topisirovic I, Huot ME, Mallette FA. Oncogenic activities of IDH1/2 mutations: from epigenetics to cellular signaling. Trends Cell Biol. 2017;27(10):738–52.
Mackenzie DK, Bussiere LF, Tinsley MC. Senescence of the cellular immune response in Drosophila melanogaster. Exp Gerontol. 2011;46(11):853–9.
Makhijani K, Alexander B, Rao D, Petraki S, Herboso L, Kukar K, Batool I, Wachner S, Gold KS, Wong C, O'Connor MB, Bruckner K. Regulation of Drosophila hematopoietic sites by Activin-beta from active sensory neurons. Nat Commun. 2017;8:15990.
Makhijani K, Alexander B, Tanaka T, Rulifson E, Bruckner K. The peripheral nervous system supports blood cell homing and survival in the Drosophila larva. Development. 2011;138(24):5379–91.
Mandal L, Banerjee U, Hartenstein V. Evidence for a fruit fly hemangioblast and similarities between lymph-gland hematopoiesis in fruit fly and mammal aorta-gonadal-mesonephros mesoderm. Nat Genet. 2004;36(9):1019–23.
Mandal L, Martinez-Agosto JA, Evans CJ, Hartenstein V, Banerjee U. A Hedgehog- and Antennapedia-dependent niche maintains Drosophila haematopoietic precursors. Nature. 2007;446(7133):320–4.
Mansur MB, van Delft FW, Colman SM, Furness CL, Gibson J, Emerenciano M, Kempski H, Clappier E, Cave H, Soulier J, Pombo-de-Oliveira MS, Greaves M, Ford AM. Distinctive genotypes in infants with T-cell acute lymphoblastic leukaemia. Br J Haematol. 2015;171(4):574–84.
Markus R, Laurinyecz B, Kurucz E, Honti V, Bajusz I, Sipos B, Somogyi K, Kronhamn J, Hultmark D, Ando I. Sessile hemocytes as a hematopoietic compartment in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2009;106(12):4805–9.
Martin-Lanneree S, Lasbleiz C, Sanial M, Fouix S, Besse F, Tricoire H, Plessis A. Characterization of the Drosophila myeloid leukemia factor. Genes Cells. 2006;11(12):1317–35.
McNerney ME, Brown CD, Wang X, Bartom ET, Karmakar S, Bandlamudi C, Yu S, Ko J, Sandall BP, Stricker T, Anastasi J, Grossman RL, Cunningham JM, Le Beau MM, White KP. CUX1 is a haploinsufficient tumor suppressor gene on chromosome 7 frequently inactivated in acute myeloid leukemia. Blood. 2013;121(6):975–83.
Miller M, Chen A, Gobert V, Auge B, Beau M, Burlet-Schiltz O, Haenlin M, Waltzer L. Control of RUNX-induced repression of Notch signaling by MLF and its partner DnaJ-1 during Drosophila hematopoiesis. PLoS Genet. 2017;13(7):e1006932.
Minakhina S, Steward R. Melanotic mutants in Drosophila: pathways and phenotypes. Genetics. 2006;174(1):253–63.
Mondal BC, Mukherjee T, Mandal L, Evans CJ, Sinenko SA, Martinez-Agosto JA, Banerjee U. Interaction between differentiating cell- and niche-derived signals in hematopoietic progenitor maintenance. Cell. 2011;147(7):1589–600.
Morin-Poulard I, Sharma A, Louradour I, Vanzo N, Vincent A, Crozatier M. Vascular control of the Drosophila haematopoietic microenvironment by Slit/Robo signalling. Nat Commun. 2016;7:11634.
Mughal TI, Radich JP, Deininger MW, Apperley JF, Hughes TP, Harrison CJ, Gambacorti-Passerini C, Saglio G, Cortes J, Daley GQ. Chronic myeloid leukemia: reminiscences and dreams. Haematologica. 2016;101(5):541–58.
Mukherjee T, Kim WS, Mandal L, Banerjee U. Interaction between Notch and Hif-alpha in development and survival of Drosophila blood cells. Science. 2011;332(6034):1210–3.
Muller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 2005;436(7052):871–5.
Muyrers-Chen I, Rozovskaia T, Lee N, Kersey JH, Nakamura T, Canaani E, Paro R. Expression of leukemic MLL fusion proteins in Drosophila affects cell cycle control and chromosome morphology. Oncogene. 2004;23(53):8639–48.
Myrick KV, Dearolf CR. Hyperactivation of the Drosophila Hop jak kinase causes the preferential overexpression of eIF1A transcripts in larval blood cells. Gene. 2000;244(1-2):119–25.
Nallamothu G, Woolworth JA, Dammai V, Hsu T. Awd, the homolog of metastasis suppressor gene Nm23, regulates Drosophila epithelial cell invasion. Mol Cell Biol. 2008;28(6):1964–73.
Oftedal P. The histogenesis of a new tumor in Drosophila melanogaster, and a comparison with tumors of five other stocks. Z Indukt Abstamm Vererbungsl. 1953;85(3):408–22.
Osman D, Gobert V, Ponthan F, Heidenreich O, Haenlin M, Waltzer L. A Drosophila model identifies calpains as modulators of the human leukemogenic fusion protein AML1-ETO. Proc Natl Acad Sci U S A. 2009;106(29):12043–8.
Owusu-Ansah E, Banerjee U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature. 2009;461(7263):537–41.
Oyallon J, Vanzo N, Krzemien J, Morin-Poulard I, Vincent A, Crozatier M. Two independent functions of Collier/Early B cell factor in the control of drosophila blood cell homeostasis. PLoS One. 2016;11(2):e0148978.
Parsons B, Foley E. Cellular immune defenses of Drosophila melanogaster. Dev Comp Immunol. 2016;58:95–101.
Petraki S, Alexander B, Bruckner K. Assaying blood cell populations of the Drosophila melanogaster larva. J Vis Exp. 2015;105:e52733.
Prange KHM, Mandoli A, Kuznetsova T, Wang SY, Sotoca AM, Marneth AE, van der Reijden BA, Stunnenberg HG, Martens JHA. MLL-AF9 and MLL-AF4 oncofusion proteins bind a distinct enhancer repertoire and target the RUNX1 program in 11q23 acute myeloid leukemia. Oncogene. 2017;36(23):3346–56.
Provan D, Gribben J. Molecular hematology. 3rd ed: Wiley Online Library/Blackwell Publishing Ltd; Hoboken, NJ, USA, 2010.
Ptasinska A, Assi SA, Mannari D, James SR, Williamson D, Dunne J, Hoogenkamp M, Wu M, Care M, McNeill H, Cauchy P, Cullen M, Tooze RM, Tenen DG, Young BD, Cockerill PN, Westhead DR, Heidenreich O, Bonifer C. Depletion of RUNX1/ETO in t(8;21) AML cells leads to genome-wide changes in chromatin structure and transcription factor binding. Leukemia. 2012;26(8):1829–41.
Qiu P, Pan PC, Govind S. A role for the Drosophila Toll/Cactus pathway in larval hematopoiesis. Development. 1998;125(10):1909–20.
Reitman ZJ, Sinenko SA, Spana EP, Yan H. Genetic dissection of leukemia-associated IDH1 and IDH2 mutants and D-2-hydroxyglutarate in Drosophila. Blood. 2015;125(2):336–45.
Rizki MT. Melanotic tumor formation in Drosophila. J Morphol. 1960;106:147–57.
Rizki RM, Rizki TM. Cell interactions in the differentiation of a melanotic tumor in Drosophila. Differentiation. 1979;12(3):167–78.
Rui L, Emre NC, Kruhlak MJ, Chung HJ, Steidl C, Slack G, Wright GW, Lenz G, Ngo VN, Shaffer AL, Xu W, Zhao H, Yang Y, Lamy L, Davis RE, Xiao W, Powell J, Maloney D, Thomas CJ, Moller P, Rosenwald A, Ott G, Muller-Hermelink HK, Savage K, Connors JM, Rimsza LM, Campo E, Jaffe ES, Delabie J, Smeland EB, Weisenburger DD, Chan WC, Gascoyne RD, Levens D, Staudt LM. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell. 2010;18(6):590–605.
Sang JH, Burnet B. Physiological genetics of melanotic tumors in Drosophila melanogaster. I. the effects of nutrient balance on tumor penetrance in the Tu strain. Genetics. 1963;48(2):235–53.
Sangkhae V, Etheridge SL, Kaushansky K, Hitchcock IS. The thrombopoietin receptor, MPL, is critical for development of a JAK2V617F-induced myeloproliferative neoplasm. Blood. 2014;124(26):3956–63.
Shafat MS, Gnaneswaran B, Bowles KM, Rushworth SA. The bone marrow microenvironment – Home of the leukemic blasts. Blood Rev. 2017;31(5):277–86.
Shi S, Calhoun HC, Xia F, Li J, Le L, Li WX. JAK signaling globally counteracts heterochromatic gene silencing. Nat Genet. 2006;38(9):1071–6.
Shim J, Mukherjee T, Banerjee U. Direct sensing of systemic and nutritional signals by haematopoietic progenitors in Drosophila. Nat Cell Biol. 2012;14(4):394–400.
Shim J, Mukherjee T, Mondal BC, Liu T, Young GC, Wijewarnasuriya DP, Banerjee U. Olfactory control of blood progenitor maintenance. Cell. 2013;155(5):1141–53.
Shirinian M, Kambris Z, Hamadeh L, Grabbe C, Journo C, Mahieux R, Bazarbachi A. A transgenic Drosophila melanogaster model to study Human T-Lymphotropic virus oncoprotein Tax-1-Driven transformation In Vivo. J Virol. 2015;89(15):8092–5.
Sinenko SA, Hung T, Moroz T, Tran QM, Sidhu S, Cheney MD, Speck NA, Banerjee U. Genetic manipulation of AML1-ETO-induced expansion of hematopoietic precursors in a Drosophila model. Blood. 2010;116(22):4612–20.
Sinenko SA, Mandal L, Martinez-Agosto JA, Banerjee U. Dual role of wingless signaling in stem-like hematopoietic precursor maintenance in Drosophila. Dev Cell. 2009;16(5):756–63.
Slany RK. The molecular mechanics of mixed lineage leukemia. Oncogene. 2016;35(40):5215–23.
Sood R, Kamikubo Y, Liu P. Role of RUNX1 in hematological malignancies. Blood. 2017;129(15):2070–82.
Sorrentino RP, Carton Y, Govind S. Cellular immune response to parasite infection in the Drosophila lymph gland is developmentally regulated. Dev Biol. 2002;243(1):65–80.
Sparrow JC. The genetics of some second chromosome melanotic tumour mutants of Drosophila melanogaster. Genet Res. 1974;23(1):13–21.
Stark MB. A benign tumor that is hereditary in Drosophila. Proc Natl Acad Sci U S A. 1919;5(12):573–80.
Stevens TL, Rogers EM, Koontz LM, Fox DT, Homem CC, Nowotarski SH, Artabazon NB, Peifer M. Using Bcr-Abl to examine mechanisms by which abl kinase regulates morphogenesis in Drosophila. Mol Biol Cell. 2008;19(1):378–93.
Stofanko M, Kwon SY, Badenhorst P. A misexpression screen to identify regulators of Drosophila larval hemocyte development. Genetics. 2008;180(1):253–67.
Stofanko M, Kwon SY, Badenhorst P. Lineage tracing of lamellocytes demonstrates Drosophila macrophage plasticity. PLoS One. 2010;5(11):e14051.
Tepass U, Fessler LI, Aziz A, Hartenstein V. Embryonic origin of hemocytes and their relationship to cell death in Drosophila. Development. 1994;120(7):1829–37.
Terriente-Felix A, Perez L, Bray SJ, Nebreda AR, Milan M. Drosophila model of myeloproliferative neoplasm reveals a feed-forward loop in the JAK pathway mediated by p38 MAPK signalling. Dis Model Mech. 2017;10(4):399–407.
Torok T, Tick G, Alvarado M, Kiss I. P-lacW insertional mutagenesis on the second chromosome of Drosophila melanogaster: isolation of lethals with different overgrowth phenotypes. Genetics. 1993;135(1):71–80.
Udensi UK, Tchounwou PB. Dual effect of oxidative stress on leukemia cancer induction and treatment. J Exp Clin Cancer Res. 2014;33:106.
Vainchenker W, Kralovics R. Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms. Blood. 2017;129(6):667–79.
Van De Bor V, Zimniak G, Papone L, Cerezo D, Malbouyres M, Juan T, Ruggiero F, Noselli S. Companion blood cells control ovarian stem cell Niche microenvironment and homeostasis. Cell Rep. 2015;13(3):546–60.
Waltzer L, Gobert V, Osman D, Haenlin M. Transcription factor interplay during Drosophila haematopoiesis. Int J Dev Biol. 2010;54(6-7):1107–15.
Watson KL, Johnson TK, Denell RE. Lethal(1) aberrant immune response mutations leading to melanotic tumor formation in Drosophila melanogaster. Dev Genet. 1991;12(3):173–87.
Watson KL, Justice RW, Bryant PJ. Drosophila in cancer research: the first fifty tumor suppressor genes. J Cell Sci Suppl. 1994;18:19–33.
Watson KL, Konrad KD, Woods DF, Bryant PJ. Drosophila homolog of the human S6 ribosomal protein is required for tumor suppression in the hematopoietic system. Proc Natl Acad Sci U S A. 1992;89(23):11302–6.
Whitten MMA, Coates CJ. Re-evaluation of insect melanogenesis research: views from the dark side. Pigment Cell Melanoma Res. 2017;30(4):386–401.
Wildonger J, Mann RS. The t(8;21) translocation converts AML1 into a constitutive transcriptional repressor. Development. 2005;132(10):2263–72.
Wilson IT. Two new hereditary tumors in Drosophila. Genetics. 1924;9(4):343–62.
Wood W, Martin P. Macrophage functions in tissue patterning and disease: new insights from the fly. Dev Cell. 2017;40(3):221–33.
Wu LP, Choe KM, Lu Y, Anderson KV. Drosophila immunity: genes on the third chromosome required for the response to bacterial infection. Genetics. 2001;159(1):189–99.
Xu N, Emelyanov AV, Fyodorov DV, Skoultchi AI. Drosophila linker histone H1 coordinates STAT-dependent organization of heterochromatin and suppresses tumorigenesis caused by hyperactive JAK-STAT signaling. Epigenetics Chromatin. 2014;7:16.
Yan R, Small S, Desplan C, Dearolf CR, Darnell JE Jr. Identification of a Stat gene that functions in Drosophila development. Cell. 1996;84(3):421–30.
Yokoyama A. Transcriptional activation by MLL fusion proteins in leukemogenesis. Exp Hematol. 2017;46:21–30.
Yoneda-Kato N, Look AT, Kirstein MN, Valentine MB, Raimondi SC, Cohen KJ, Carroll AJ, Morris SW. The t(3;5)(q25.1;q34) of myelodysplastic syndrome and acute myeloid leukemia produces a novel fusion gene, NPM-MLF1. Oncogene. 1996;12(2):265–75.
Yu S, Luo F, Jin LH. The Drosophila lymph gland is an ideal model for studying hematopoiesis. Dev Comp Immunol. 2017;83:60–9.
Zaidman-Remy A, Regan JC, Brandao AS, Jacinto A. The Drosophila larva as a tool to study gut-associated macrophages: PI3K regulates a discrete hemocyte population at the proventriculus. Dev Comp Immunol. 2012;36(4):638–47.
Zang Y, Wan M, Liu M, Ke H, Ma S, Liu LP, Ni JQ, Pastor-Pareja JC. Plasma membrane overgrowth causes fibrotic collagen accumulation and immune activation in Drosophila adipocytes. Elife. 2015;4:e07187.
Zettervall CJ, Anderl I, Williams MJ, Palmer R, Kurucz E, Ando I, Hultmark D. A directed screen for genes involved in Drosophila blood cell activation. Proc Natl Acad Sci U S A. 2004;101(39):14192–7.
Zhang CU, Cadigan KM. The matrix protein Tiggrin regulates plasmatocyte maturation in Drosophila larva. Development. 2017;144(13):2415–27.
Zinyk DL, McGonnigal BG, Dearolf CR. Drosophila awdK-pn, a homologue of the metastasis suppressor gene nm23, suppresses the Tum-1 haematopoietic oncogene. Nat Genet. 1993;4(2):195–201.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Boulet, M., Miller, M., Vandel, L., Waltzer, L. (2018). From Drosophila Blood Cells to Human Leukemia. In: Yamaguchi, M. (eds) Drosophila Models for Human Diseases. Advances in Experimental Medicine and Biology, vol 1076. Springer, Singapore. https://doi.org/10.1007/978-981-13-0529-0_11
Download citation
DOI: https://doi.org/10.1007/978-981-13-0529-0_11
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-0528-3
Online ISBN: 978-981-13-0529-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)