Abstract
Boundary delineation of a hypoechoic layers between the surrounding tissues and the bone structure is a necessary step in order to compute the knee cartilage thickness from ultrasound images. Speckle noise and intensity bias often complicates the segmentation task in the ultrasound images. This paper presents knee cartilage boundary segmentation using locally statistical level set method (LSLSM). Comparing to other methods in segmenting the cartilage, LSLSM produces a more satisfying outcome. Application of LSLSM on a set of 80 images illustrates a significant agreement with Cohen’s \( \kappa \) coefficient equal to \( 0.73 \) for the segmentation quality of the cartilage region rated by two raters. The quantitative evaluation measures of Dice coefficient and Hausdorff distance indicate the overall average values of \( 0.91 \pm 0.01 \) and \( 6.21 \pm 0.59 \) pixels, respectively. These good and consistent segmentation performances indicate that the segmented images can be applied for making the thickness computation in the ultrasound images.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Knee osteoarthritis is a prevalent disease among elderly [5]. Cartilage degeneration is one of the primary features of this disease [6]. Ultrasound imaging is useful for the evaluation of extra-articular structures [6]. It has been applied to quantify the cartilage thickness and diagnose the cartilage degeneration [1] in patients with osteoarthritis, rheumatoid arthritis, [4] and knee pain [6].
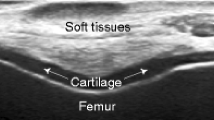
Segmentation is an important task that could significantly affect the accuracy of the thickness measurement [3]. In ultrasound images, the femoral condylar cartilage is shown as a monotonous hypoechoic band between both interfaces of the soft tissue-cartilage and the cartilage-bone as shown in Fig. 1 [6]. Thus, the goal in segmenting the cartilage is to delineate the boundaries between both interfaces. Delineating the cartilage boundary from the adjacent tissues is difficult because the boundary between different tissues is hard to distinguish.
Speckle noise and intensity inhomogeneity occur caused by physical constraint in the ultrasound image acquisition, which often adversely affect the image contrast. If only intensity bias is considered and not by speckle, this problem could be solved similarly to the inhomogeneity correction in magnetic resonance images [10]. The intensity bias correction is often addressed by assuming that intensity inhomogeneity associated with a component of an observed image is modelled as the multiplicative noise model. Furthermore, the multiplicative noise model is associated with the classic reflection imaging equation of ultrasound physics of image formation [10]. It is employed retrospectively in the images and usually incorporated with the segmentation algorithm where level set techniques for simultaneous segmentation and intensity inhomogeneity estimation have been presented [7, 11]. While these intensity-based segmentation methods are in general robust to noise, the usage of local intensity and joint intensity inhomogeneity correction could handle the intensity bias.
In this paper, boundary segmentation and thickness computation methods in two dimensional (2-D) knee cartilage ultrasound images are presented. To locate the cartilage boundary corrupted by speckle noise and intensity bias, the locally statistical level set technique is used using the energy derived from Gaussian distributions of local intensity and multiplicative noise model. Segmentation and computational performances of LSLSM are compared to other level set techniques when segmenting the knee cartilage. In addition, the segmentation results of these level set techniques on the total 80 data sets are evaluated qualitatively and quantitatively using Cohen’s \( \kappa \) coefficient, Dice similarity coefficient, and Hausdorff distance measures, respectively.
2 Materials and Methods
2.1 Data Acquisition
The Toshiba Aplio MX ultrasound system with a 8–12 MHz, 2-D linear array probe (PLT-805AT) was used to capture axial views of the femoral cartilage revolved the knee. The knee joint was \( 120^{ \circ } \) flexed with the subject positioned in the supine posture. The probe was put transversely to the leg and perpendicular to the bone surface above the patella [6, 8]. Total 10 asymptomatic participants (male with age range between 23 and 27 years were registered with the written consent for data collection. The cartilage of both knee joints were acquired four times by repositioning the ultrasound transducer. The image resolution is \( 0.1316 \times 0.1316 \) mm stored in DICOM format. Professional sonographer conducted this musculoskeletal sonography. The ethics approval letter of this study was obtained from UMMC Medical Ethics Committee (MECID No. 20147-396).
2.2 Locally Statistical Level Set Method
The two-phase case of the statistical and variational multiphase level set method or referred as the locally statistical level set method (LSLSM) is considered [11]. The energy of LSLSM is obtained from derivation of the Gaussian distributions of local intensity and multiplicative noise model. The energy functions \( e_{i} \) are expressed as
The functions \( e_{i} \) are computed by the equivalent expression as follows
where \( b \), \( c_{i} \), and \( \sigma_{i}^{2} \) for \( i = 1,2 \) are accordingly the restored bias field, the piecewise constants, and the variances. \( * \) is the convolution operation. The function \( {\mathbf{1}}_{K} \) is defined as \( \int K({\mathbf{y}} - {\mathbf{x}})d{\mathbf{y}} \). The kernel function \( K \) chosen in this paper is given by
where \( a \) is a positive constant such that \( \int K({\mathbf{z}})d{\mathbf{z}} = 1 \) and \( \rho \) represents the kernel’s radius.
In the attempt of reducing the overlapping image intensity distribution, only intensities \( I({\mathbf{x}}) \) in the neighborhood of \( {\mathbf{y}} \) are considered in the energy functions \( e_{i} \). The size of the neighborhood depends to the kernel scale. The small neighborhood is able to cope with intensity bias due to the intensities are only involved in the local region [7].
The intensities are estimated by spatially varying means \( bc_{i} \) and variances \( \sigma_{i}^{2} \). To achieve simultaneous segmentation and intensity inhomogeneity estimation, the means are estimated by multiplication between the bias field \( b \) that accounts for intensity bias and the piecewise constants \( c_{i} \) estimating the true image signal in each region. The functions \( e_{i} \) represent an image segmentation and a intensity bias correction. To incorporate these functions to the level set formulation, these functions are combined with membership function \( M_{i} (\phi ) \). Therefore, the energy functional of LSLSM is defined by
where the first term is the regularization term to compute the arc length of the zero level set, which its relative strength is determined by the parameter \( \nu \).
The membership functions defined by \( M_{1} (\phi ) = H(\phi ) \) and \( M_{2} (\phi ) = 1 - H(\phi ) \) represent both regions \( \varOmega_{1} \) and \( \varOmega_{2} \), respectively. The regularized Heaviside function \( H_{\varepsilon } (\phi ) \) and the smoothed Dirac delta function \( \delta_{\varepsilon } (\phi ) \) with \( \varepsilon = 1 \) [2], are defined by
By minimizing the energy function, image partition and bias intensity estimation are accomplished together by approximating the piecewise constants \( c_{i} \), the restored bias field \( b \), the variances \( \sigma_{i}^{2} \), and the membership functions \( M_{i} (\phi ) \). The minimization of the energy functional with respect to each variable \( \phi \), \( c_{i} \), \( b \), and \( \sigma_{i} \) is performed in the iterative process. These variables are obtained from the derivation of the convolution expression of the energy functional. The optimal \( c_{i} \), \( b \), and \( \sigma_{i}^{2} \) are given by
Keeping \( c_{i} \), \( b \), and \( \sigma_{i} \) fixed, the energy functional \( E(\phi ,c_{i} ,b,\sigma_{i} ) \) is minimized with respect to \( \phi \) by solving the gradient flow equation \( \frac{\partial \phi }{\partial t} = - \frac{\partial E}{\partial \phi } \). The Gâteaux derivative \( \frac{\partial E}{\partial \phi } \) can be computed by using calculus of variations. The corresponding gradient flow equation is defined by
For each iteration of Eq. (10), the level set function is diffused by Eq. (11) to keep the level set evolution stable [12].
where \( \phi^{n} \) represents the level set function of the \( n \)-th iteration of Eq. (10), \( \Delta t \) is the diffusion strength, and \( \Delta \) is the Laplacian operator.
3 Results and Discussion
3.1 Comparison with Other Level Set Methods
Several relevant level set techniques in segmenting a real knee cartilage ultrasound image are compared in this subsection. The other two level set techniques without and with multiplicative noise estimation are summarized as follows. First, the local Gaussian distribution fitting (LGDF) model [9] considers a Gaussian distribution with locally varying mean and variance similar to LSLSM. Because LGDF does not approximate bias field, it can be used for segmentation purpose only. Meanwhile, LSLSM can be applied for simultaneous segmentation and bias correction. Second, the locally weighted K-means variational level set (WKVLS) method is considered [7]. WKVLS does not consider the variance component which helps LSLSM to differentiate the boundary from surrounding tissues more satisfactorily. Both WKVLS and LSLSM are essentially designed for simultaneous segmentation and intensity inhomogeneity correction.
In this experiment, all the methods were implemented in MATLAB R2014a in an Intel (R) Xeon (R), 2.00 GHz, 32 GB RAM using the settings as follows. The kernel’s scale \( \rho = 5 \) was set to be small to produce more accurate segmentation result. The parameter \( \nu \) was chosen as small as \( 0.001 \times 255^{2} \) for images with intensity range in \( \left[ {0,255} \right] \) when capturing objects of any size. The time steps for level set evolution \( \Delta t_{1} \) and for regularization \( \Delta t_{2} \) were set as \( \Delta t_{1} = 0.01 \) for LGDF, \( \Delta t_{1} = 0.1 \) and \( \Delta t_{2} = 0.1 \) for WKVLS, and \( \Delta t_{1} = 0.01 \) and \( \Delta t_{2} = 0.01 \) for LSLSM. The image size is of 420 \( \times \) 150 pixels.
Figure 2 depicts segmentation performances of the three different level set methods when employed to the cartilage boundary segmentation. The initial contour is in circle shape with 10 pixels radius and positioned around the middle of the images. In general, these three level set methods were able to delineate the desired object in the image corrupted by speckle noise and intensity inhomogeneity. This is because the local intensity defined in the local neighborhood that reduces the overlapped intensity distribution. With the joint bias field estimation, WKVLS and LSLSM could suppress the intensity bias therefore delineate the boundaries between surrounding tissues satisfactorily as depicted in Fig. 2c, d. Without the joint bias field estimation, LGDF produces some misclassified and unnecessary contours inside and around the object as seen in Fig. 2b. Both methods yield satisfactory segmentation outcomes, while LSLSM that takes into account the variance component achieved a more desirable segmentation outcome than WKVLS.
The validation metrics of DSC and HD were computed from the manual outline and the isolated cartilage area as illustrated in Fig. 3. The connected-component labeling was used to extract the cartilage region and remove the other adjacent tissues in the final contours. This is to ensure that the DSC and HD metrics are computed based on the cartilage area only and unaffected by other tissue regions. The first, second, and third rows of the matrices \( \left[ {\begin{array}{*{20}c} {0.9027} \\ {0.9148} \\ {0.9423} \\ \end{array} } \right] \) and \( \left[ {\begin{array}{*{20}c} {6.8557} \\ 7 \\ {6.3246} \\ \end{array} } \right] \) summarized DSC and HD measures for the segmentation outcomes of LGDF, WKVLS, and LSLSM in Fig. 2b–d, respectively. LSLSM obtained DSC value higher than WKVLS and LGDF. Meanwhile, LSLSM obtained HD value smaller than WKVLS and LGDF. Moreover, LGDF, WKVLS, and LSLSM spent the total computational time of 54.82, 13.77, and 12.97 s for 500 iterations, respectively.
3.2 Knee Cartilage Ultrasound Image Segmentation
An application of the three level set techniques in segmenting a set of 80 cartilage images is presented in this subsection. The data sets consist of the real knee cartilage ultrasound images scanned four times each from both knee joints of the ten participants. Figure 4 illustrates a subset of ten segmentation outcomes achieved by LSLSM from both left and right knee cartilages of a subset of five participants. Qualitative and quantitative evaluations are performed to the total 80 segmentation outcomes obtained by LGDF, WKVLS, and LSLSM. While Cohen’s \( \kappa \) statistics is employed to validate the segmentation outcomes qualitatively, DSC and HD measures are used to assess the segmentation results quantitatively. The manual segmentation results as gold standard were compared against the isolated cartilage area extracted by the level set methods to be examined qualitatively and quantitatively. The cartilage are delineated manually by the expert from each cartilage ultrasound scan. The connected-component labeling was employed to isolate the cartilage area depicted in Fig. 3 from the adjacent tissues in the final segmentation contours.
Left and right columns comprise of the segmentation outcomes achieved by LSLSM from left and right knee cartilages of a subset of five subjects, respectively. The red circles with ten pixels radius put around the middle of the image represent the initial contours. The final contours are depicted by the green lines
The qualitative assessment of the segmentation results was performed by differentiating the boundaries between the both interfaces of the soft tissue-cartilage and the cartilage-bone with the following observations. From the observed agreements of \( 67 \) images (\( 83.75\% \) of the observations), \( 39 \) images (\( 48.75\% \)) are as grade 1 (excellent), \( 21 \) images (\( 26.25\% \)) are as grade 2 (good), \( 5 \) images (\( 6.25\% \)) are as grade 3 (poor), \( 2 \) images (\( 2.5\% \)) are as grade 4 (bad). The number of agreement due to chance is \( 32.05 \) images. Cohen’s \( \kappa = 0.73 \) shows a significant agreement for the overall cartilage segmentation quality rated by two raters.
Figure 5 depicts segmentation results of LGDF, WKVLS, and LSLSM evaluated by DSC and HD measures on a set of 80 cartilage images. DSC values of LGDF, WKVLS, and LSLSM computed from 80 images illustrated in Fig. 5a are ranging from \( 0.84 \) to \( 0.94 \), \( 0.29 \) to \( 0.95 \), and \( 0.82 \) to \( 0.95 \), respectively. A good agreement in size and location of the two comparing contours, which correspond to more accurate segmentation outcomes is indicated by the higher value of DSC. Figure 5b shows HD values of LGDF, WKVLS, and LSLSM fall in the range between \( 4.47 \) and \( 8.83 \), \( 5.39 \) and \( 19.10 \), and \( 4.69 \) and \( 8.25 \) pixels, respectively. The minimal shape difference between the contour pair corresponds to the smaller HD values.
Table 1 summarizes the average values, standard deviations, and p-values for DSC and HD measures of the three methods computed from the total data sets of 80 images. It indicates that the means of DSC values of LSLSM is larger than of LGDF and WKVLS. Moreover, LSLSM obtained smaller means of HD values than LGDF and WKVLS. It can be implied that LSLSM produces an overall satisfying segmentation performance on all set of data illustrated by a good area similarity and the least shape difference of the compared contours. In addition, while the means of LGDF and LSLSM are statistically significant from WKVLS, the means of LGDF is not significantly different from LSLSM.
The overlapping intensity distributions between surrounding tissues caused the segmentation errors. The boundary between the adjacent soft tissue and the bone surface is hard to distinguish. The variance component in the Gaussian distributions considered in LGDF and LSLSM contributes in locating the cartilage boundary more accurately. Although WKVLS take into account the bias field estimation, it has a tendency to misclassify the two interfaces because it does not consider the variance component. DSC measures lower than \( 0.8 \) and HD measures higher than \( 7 \) pixels in the graph indicates the less satisfactory segmentation result caused by the high variety of intensity bias between the scanned images.
4 Conclusion
The knee cartilage boundary segmentation in the 2-D ultrasound axial view is a challenging task. LSLSM has obtained a more satisfactory outcome than other level set techniques in capturing the cartilage. A significant agreement of the segmentation quality rated by two raters was indicated by Cohen’s \( \kappa \) coefficient. A consistent segmentation performance was indicated by DSC and HD measures computed from all available datasets. These segmentation results suggest that the cartilage thickness computation can be made using the segmented cartilage images.
References
Aisen, A.M., McCune, W.J., MacGuire, A., Carson, P.L., Silver, T.M., Jafri, S.Z., Martel, W.: Sonographic evaluation of the cartilage of the knee. Radiology 153(3), 781–784 (1984)
Chan, T.F., Vese, L.A.: Active contours without edges. IEEE Trans. Image Process. 10(2), 266–277 (2001)
Fripp, J., Crozier, S., Warfield, S.K., Ourselin, S.: Automatic segmentation and quantitative analysis of the articular cartilages from magnetic resonance images of the knee. IEEE Trans. Med. Imag. 29(1), 55–63 (2010)
Iagnocco, A., Coari, G., Zoppini, A.: Sonographic evaluation of femoral condylar cartilage in osteoarthritis and rheumatoid arthritis. Scand. J. Rheumatol. 21(4), 201–203 (1992)
Jackson, D., Simon, T., Aberman, H.: Symptomatic articular cartilage degeneration: the impact in the new millennium. Clin. Orthop. Relat. Res. 391, 14–15 (2001)
Kazam, J.K., Nazarian, L.N., Miller, T.T., Sofka, C.M., Parker, L., Adler, R.S.: Sonographic evaluation of femoral trochlear cartilage in patients with knee pain. J. Ultrasound Med. 30(6), 797–802 (2011)
Li, C., Huang, R., Ding, Z., Gatenby, C., Metaxas, D.N., Gore, J.C.: A level set method for image segmentation in the presence of intensity inhomogeneities with application to MRI. IEEE Trans. Image Process. 20(7), 2007–2016 (2011)
Naredo, E., Acebes, C., Moller, I., Canillas, F., de Agustin, J.J., de Miguel, E., Filippucci, E., Iagnocco, A., Moragues, C., Tuneu, R., Uson, J., Garrido, J., Delgado-Baeza, E., Saenz-Navarro, I.: Ultrasound validity in the measurement of knee cartilage thickness. Ann. Rheum. Dis. 68, 1322–1327 (2009)
Wang, L., He, L., Mishra, A., Li, C.: Active contours driven by local Gaussian distribution fitting energy. Signal Process. 89(12), 2435–2447 (2009)
Xiao, G., Brady, M., Noble, J.A., Zhang, Y.: Segmentation of ultrasound B-mode images with intensity inhomogeneity correction. IEEE Trans. Med. Imag. 21(1), 48–57 (2002)
Zhang, K., Zhang, L., Lam, K.M., Zhang, D.: A level set approach to image segmentation with intensity inhomogeneity. IEEE Trans. Cybern. 46(2), 546–557 (2016)
Zhang, K., Zhang, L., Song, H., Zhang, D.: Reinitialization-free level set evolution via reaction diffusion. IEEE Trans. Image Process. 22(1), 258–271 (2013)
Acknowledgements
This work was supported in part by the University of Malaya Research Grant (RP020A-13AET), in part by the International Graduate Research Assistantship Scheme, and in part by the Postgraduate Research Grant (PG003-2014B).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Science+Business Media Singapore
About this paper
Cite this paper
Faisal, A., Ng, SC., Goh, SL., Lai, K.W. (2018). Knee Cartilage Ultrasound Image Segmentation Using Locally Statistical Level Set Method. In: Ibrahim, F., Usman, J., Ahmad, M., Hamzah, N., Teh, S. (eds) 2nd International Conference for Innovation in Biomedical Engineering and Life Sciences. ICIBEL 2017. IFMBE Proceedings, vol 67. Springer, Singapore. https://doi.org/10.1007/978-981-10-7554-4_48
Download citation
DOI: https://doi.org/10.1007/978-981-10-7554-4_48
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-7553-7
Online ISBN: 978-981-10-7554-4
eBook Packages: EngineeringEngineering (R0)