Abstract
Neurodegeneration is an alarming problem all over the globe. The significant role of SNCA receptor has been well established in several neurodegenerative disorders including various types of progressive dementia—most commonly in Parkinson’s disorder (PD) and Lewy body dementia. Phytocompounds selected from ayurvedic medicinal plants are thoroughly screened against the SNCA receptor (in silico). The gene receptor responsible for Parkinson’s disorder (PD), SNCA, was taken for this work. Mutated mammalian SNCA implicated as one of the factors responsible for PD was taken from NCBI; templates as retrieved from BLAST were downloaded from PDB. The 3D structure of SNCA was modeled; the 3D structures of phytocompounds (unknown ligands) were taken from various online databases; phytocompounds were virtually screened against the SNCA receptor, and the best ligand was selected.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Background
From very ancient days, different medicinal plants dealing with health care are part and parcel of major populations of India, and other Asian countries.
Parkinson’s disorder (PD) is a movement disorder having well-expressed symptoms like slowing of movement, tremor, rigidity or stiffness, and balancing problems. As per Dr. Dickson, the typical patient with PD has Lewy bodies (aggregates of protein, alpha synuclein or SNCA) in the brain neurons. Research suggests that the above central nervous system (CNS) disorder, PD, is a combination of environmental factors and multi-gene mutation [1,2,3,4,5,6].
In the present study for the preliminary screening, the following medicinal plants have been selected to study the scope and activity of different compounds on CNS using the above-mentioned bioinformatic parameter. These are Hydrocotyle asiatica/Centella asiatica, Bacopa monnieri, Convolvulus pluricaulis, Mucuna pruriens, Ocimum sanctum, Tinospora cordifolia, Curcuma longa, Nardostachys jatamansi, Gloriosa superba, Colchicum autumnale, etc., especially considered in this work.
Mutations in the SNCA (alpha synuclein) receptor and is noted as causal factors for many CNS disorders is used in this work [7,8,9].
SNCA
Alpha synuclein is expressed in the brain and integrates presynaptic signals and membrane traffics, mutations and defects of are implicated in the pathogenesis of PD [10, 11]. Unconventionally spliced transcripts encoding different isoforms have been identified for this gene [12]. Researchers are still much unclear about the function of SNCA, though it is seen it generally maintains a supply of synaptic vesicles in presynaptic terminals by clustering synaptic vesicles and controls the release of dopamine which is an important neurotransmitter which critically controls the voluntary and involuntary movements [13].
In this work, the 3D structure of the SNCA receptor is modeled by homology modeling, using high-throughput screening (HTS), and its ligand is selected from Indian Ayurvedic Herbs.
2 Methodology
The 3D structure of the SNCA receptors was modeled using modeler software [14]. The SNCA receptor’s amino acid sequence is downloaded from NCBI; its homologous templates were selected by BLAST (Table 1). The receptor and their corresponding templates were submitted to modeler software to model their 3D structure. Using Rampage Ramachandran plot server [15], the generated five models generated by modeler were evaluated and the selection of the best model was done (Table 2). The 3D structures of the unknown ligands (phytocompounds as in Table 3) were downloaded from various other online databases, and a phase database is generated [16]. Structure-based pharmacophore model is a unique procedure for generating energy-optimized e-pharmacophores. This was done by selecting the regular features of the 3D structure of SNCA receptor interacting with the known ligands; thus, phores were generated in the 3D structure of the SNCA receptor at the interaction sites with the above known ligands. Hence, these e-pharmacophores were used as queries for virtual screening [17, 18].
Docking was performed by PATCHDOCK server by selecting the best model (model 4) with the ligand selected by pharmacophore modeling, colchicine, to get the docked structure [19, 20].
3 Results and Discussions
3.1 Homology Modeling
The amino acid sequences of SNCA receptor were downloaded from NCBI (Table 1). Its homologous templates were selected by BLAST (Table 1).
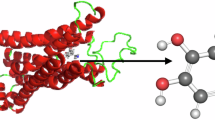
The amino acid sequences of the receptors along with their homologous templates were submitted to modeler software for the generation of the 3D structures of the receptor using the principle of homology modeling [14]. Modeler generated five models for each receptor. The 3D models generated by modeler of SNCA (Table 3) are submitted to Rampage Ramachandran Plot server for model verification [15], and the best (Figs. 1 and 2) model was selected.
3.2 Structure-Based Pharmacophore
Pharmacophore sites were created in the SNCA receptor (model 4) using the known ligands, viz. phenothiazine, N-acylaminophenothiazine, N-alkylphenothiazine, stimovul, etorphine, propoxyphene, and pentazdine. The above ligands are established ligands for SNCA receptor [21, 22].
Based on preliminary screening, special emphasis is given on the following herbs having known phytocompounds as given in Table 3 were screened.
As per structure-based pharmacophore results, phytocompound having the fitness score 1.089598 was selected as the best fitted ligand (Fig. 3, Table 4) and the further docking studies were done using this phytocompound.
3.3 Molecular Docking
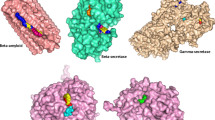
SNCA receptor (model 4) was docked with the phytocompound using software PATCHDOCK [19, 20]. It was seen that SNCA receptor docks with the phytocompound, colchicine, with a docking score of −5566 kcal/mol (Fig. 4, Table 5).
From the above result is compared with the docking interactions of SNCA receptor with known phytocompounds Jatamanin11 from Valeriana jatamansi (Fig. 5) [23, 24] and Bacopaside I from Bacopa monnieri (Fig. 6) [25] which were established as ligands for PD [23, 25] in our previous works.
Docked structure of SNCA receptor with phytocompound bacopaside I [25]
3.4 ADME Screening
ADME is an abbreviation for absorption, distribution, metabolism, and excretion. It is used to calculate the drug-like properties of the molecules [16].
QikProp generated the following output (Tables 6 and 7) [18, 23] for the phytocompound colchicine.
4 Conclusion
As per Rampage Ramachandran Plot analysis, model 4 of SNCA receptor was selected as the best model. Further, pharmacophore and molecular docking studies prove that phytocompound colchicine can be used as ligand for SNCA receptor. Further, in vitro receptor–ligand binding studies can be performed on SNCA receptor with the above ligand to justify the selection of colchicine as the ligand for SNCA receptors and as a remedy for PD. Also, as per ADME studies, colchicine satisfies all drug-like properties which correlate with the results of Bagchi et al. [26].
References
Alexander, G.E., (2004), “Biology of Parkinson’s disease: pathogenesis and pathophysiology of a multisystem neurodegenerative disorder”, Dialogues Clin Neurosci., 6(3):259–280.
Thomas, B. and Beal, M.F., (2007) “Parkinson’s disease, Human Molecular Genetics”, 2007, 16(2): R183–R194.
Saha, A., (2013), “Psychiatric morbidity in Parkinson’s Disease: a case report”, American Journal of Life Sciences, 1(2):27–30.
Djamshidian, A. and Lees, A., (2012), “Impulsive Compulsive Behaviours In Patients With Parkinson’s Disease Treated With Dopamine Agonists”, Focus on Parkinson’s Disease, 23(1):16–21.
Stacy, M., (2009), “Impulse control disorders in Parkinson’s disease”, F1000 Med Rep. 1: 29.
Stacy, M., Galpern, W., Samuel, M., Lang, A., (2008), “Impulse control disorders in Parkinson’s disease abstract supplement”, Movement Disorders, 23(9):1332–1351.
Klein, C. and Westenberger, A., (2012), “Genetics of Parkinson’s Disease”, Cold Spring Harb Perspect Med., 2(1):1–15.
Dauer, W. and Przedborski, S., (2003), “Parkinson’s Disease: Mechanisms and Models”, Neuron, 2003, 39:889–909.
Rachakonda, V., Pan, T.H. and Le, W.D., (2004), “Biomarkers of neurodegenerative disorders: How good are they?”, Cell Research, 14:347–358.
George JM (2002). “The synucleins”. Genome Biol. 3 (1): REVIEWS3002.
Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, et al. (1997). “Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease”. Science. 276 (5321): 2045–7.
Goedert M (July 2001). “Alpha-synuclein and neurodegenerative diseases”. Nat. Rev. Neurosci. 2 (7): 492–501.
Shah, M., Doctoral Thesis, 2013, The cytoskeletal linker protein, Ezrin, inhibits α-synuclein fibrillization and toxicity by a novel mechanism, Freien Universität Berlin.
Sali, A. and Blundell, T.L., (1993), “Comparative protein modelling by satisfaction of spatial restraints” J. Mol. Biol., 234:779–815.
Laskoswki, R.A., MacArthur, M.W., Moss, D.S. & Thorton, J.M., (1993), “Procheck: a program to check the stereochemical quality of protein structures”, J. Appl. Cryst. 26:283–291.
Schrödinger Suite (2010) Protein Preparation Guide, SiteMap 2.4; Glide version 5.6, LigPrep 2.4, QikProp 3.3, Schrödinger, LLC, New York, NY.
Taha MO, Dahabiyeh LA, Bustanji Y, Zalloum H, Saleh S, (2008)Combining Ligand-Based Pharmacophore Modeling, Quantitative Structure-Activity Relationship Analysis and in Silico Screening for the Discovery of New Potent Hormone Sensitive Lipase Inhibitors. J Med Chem 51:6478–6494.
Singh KhD, Kirubakaran P, Nagarajan S, Sakkiah S, Muthusamy K, Velmurgan D, Jeyakanthan J (2012) Homology modeling, molecular dynamics, e-pharmacophore mapping and docking study of Chikungunya virus nsP2 protease. J Mol Model 18: 39–51.
Duhovny D, Nussinov R, Wolfson HJ, (2002), “Efficient Unbound Docking of Rigid Molecules”, In Gusfield et al., Ed. Proceedings of the 2’nd Workshop on Algorithms in Bioinformatics (WABI) Rome, Italy, Lecture Notes in Computer Science 2452, pp. 185–200, Springer Verlag.
Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ, (2005), “PatchDock and SymmDock: servers for rigid and symmetric docking”, Nucl. Acids. Res. 33: W363–367.
Yu L, Cui J, Padakanti PK, Engel L, Bagchi DP, Kotzbauer PT, and Tua Z, (2012), “Synthesis and in vitro evaluation of α-synuclein ligands”, Bioorg Med Chem., 20(15): 4625–4634.
Jayaraj RL, Ranjani V, Manigandan K And Elangovan N, (2013), “Insilico Docking Studies To Identify Potent Inhibitors Of Alpha-Synuclein Aggregation In Parkinson Disease”, Asian Journal of Pharmaceutical and Clinical Research, 6(4): 127–131.
Preenon Bagchi, Waheeta Hopper 2011 “Virtual Screening of compounds from Valeriana jatamansi with α-Synuclein” IPCBEE, 5:11–14.
Preenon Bagchi, Somashekhar. R and Ajit Kar, 2015, “Scope of some Indian medicinal plants in the management of a few neuro-degenerative disorders in-silico: a review”, International Journal of Public Mental Health & Neurosciences, 2(1):41–57.
Preenon Bagchi, Anuradha M, Ajit Kar, 2017, “Ayur-Informatics: Establishing an Ayurvedic Medication for Parkinson’s Disorder” Int’l Journal of Advances in Chemical Engg., & Biological Sciences, 4(1): 21–25.
Bagchi P, Venkatramana DK, Mahesh. M, Somashekhar. R and Kar A, 2014, “Identification of Novel Drug Leads for Receptors Implicated in Migraine from Traditional Ayurvedic Herbs Using in silico and in vitro Methods”, Journal of Neurological Disorders, 2(6):2–6 ISSN: 2329-6895.
Acknowledgements
This work was supported by SERB-NPDF for Preenon Bagchi, author reference number PDF/2015/000047.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Bagchi, P., Anuradha, M., Kar, A. (2018). Pharmacophore Modeling and Docking Studies of SNCA Receptor with Some Active Phytocompounds from Selected Ayurvedic Medicinal Plants Known for their CNS Activity. In: Bhateja, V., Nguyen, B., Nguyen, N., Satapathy, S., Le, DN. (eds) Information Systems Design and Intelligent Applications. Advances in Intelligent Systems and Computing, vol 672. Springer, Singapore. https://doi.org/10.1007/978-981-10-7512-4_1
Download citation
DOI: https://doi.org/10.1007/978-981-10-7512-4_1
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-7511-7
Online ISBN: 978-981-10-7512-4
eBook Packages: EngineeringEngineering (R0)