Abstract
Only one closterovirus species (Family: Closteroviridae), Citrus tristeza virus (CTV) is known to occur in India. CTV is one of the most important plant viruses in India and extensive studies have been conducted over the last 60 years. The failure of Malta sweet orange on sour orange root stocks provided the evidence of tristeza disease in India. CTV infects nearly all the citrus species and citrus relatives and hybrids showing variables biological symptoms. Most citrus species and cultivars are susceptible to infection but some are tolerant inducing no obvious symptoms. Citrus orchards in Northeast India are severely affected by citrus decline, and several orchards in this region have been wiped out and many Sweet orange orchards in South and Central India are facing problem of decline. Toxoptera citricida is an efficient vector for the local natural spread of CTV in India. Stem pitting symptoms caused by CTV are not common in India. The Indian CTV isolates are genetically diverse and seven to ten genetic variants have been recognized in India. The complete genome (19,253 nt) of a mandarin decline inducing CTV strain, Kpg3 from the Darjeeling hills was sequenced. This chapter presents the work conducted on CTV in India.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
The family Closteroviridae consists of the three virus genera, Closterovirus (Type species: Beet yellows virus), Ampelovirus (Type species: Grapevine leafroll-associated virus 3) and Crinivirus (Type species: Lettuce infectious yellows virus). The virus particles of the family Closteroviridae are helical, flexuous and filamentous having a size ranging from 650 to 2000 nm in length with 11–12 nm in width. The pitch of the primary helix of the virion is 3.4–3.8 nm, containing ~10 protein subunits per turn of the helix with a central hole of 3–4 nm. The virions have two coat proteins (CPs) as a major CP and a minor CP (CPm). The duplication of the CP gene is the only example in the viruses in the family Closteroviridae among plant viruses with elongated particles. The CPm encapsidates the 5′-terminal 600–700 nt of the viral RNA that coats 75–100 nm of the virus particle resulting in formation a distinct structure, ‘rattlesnake’ in this family. The genome of the family Closteroviridae contain a single molecule of linear, +ve sense, single stranded RNA of 13–19 kb with 7–13 ORFs (Fig. 8.1). The genomic RNAs are 5–6% of the particle weight and the 5′end of the genomic RNA is likely to be capped. The 3′ end do not have poly (A) tract but contains tRNA-like structure. However, the 3′ end has several hairpin structures, and a putative pseudoknot essential for replication. The structural proteins of most of the closterovirids consist of a major CP (22–46 kDa) and CPm (22–46 kDa). A group of ampeloviruses contain a small genome of 13 kb in length that lacks a true CPm. Most of the virus members of this family require CPm for the assembly of the 5′-extremity of the virions.
The family Closteroviriae contains the virus members having the largest genomes among the plant viruses. Because of sequence duplication and acquisition of non-viral coding sequences like protease, HSP70, protein, and genetic recombination, the viruses have largest genome. Recombination is the important cause to make differences in genome organization between the genera and members of the genus. However, the complex ORF-1a to ORF-1b invariably encodes the replication-related proteins, Mtr, Hel, and RdRp. In downstream ORFs at 5–3′ direction, a conserved five-gene module is formed with (i) a 6 K small hydrophobic protein, (ii) the HSP70h (for a homolog of the cellular HSP70 heat shock protein), (iii) the 60 kDa protein, (iv) the CP and (v) CPm.
The HSP70h and the 60 kDa proteins are integral virion components present most of the closteroviruses. The HSP70h in the viruses of the family may function for (i) mediation of cell-to-cell movement through plasmodesmata, (ii) involvement in the assembly of multisubunit complexes for genome replication and/or sgRNAs synthesis, and (iii) assembly of virus particles. The 60 kDa proteins are required for incorporation of both HSP70h and CPm to virion heads. The duplication of the capsid protein gene seems to be the only example among viruses with elongated particles. In general, major CP and their homologs CPm show a significant degree of sequence conservation. But ampeloviruses generally do not possess CPm. The genome expression strategy in the closteroviruses of this family is based on (i) proteolytic processing of the polyprotein encoded by ORF1a, (ii) +1 ribosomal frame shift for the expression of the RdRp domain encoded by ORF-1b and this mechanism is not found in other positive sense RNA plant viruses, (iii) expression of the downstream ORFs via the formation of a nested set of 3′ co-terminal sgRNAs. The dsRNA patterns are very complex and variable among species. Different numbers and sizes of the ORFs are present in individual genomes. In some cases in effective RNAs are existed. Replication of the viruses occurs in the cytoplasm, possibly in association with endoplasmic reticulum-derived membranous vesicles and vesiculated mitochondria. The closterovirruses have probably evolved from the smaller filamentous virus. Under the pressure of further modular evolutionary events, the duplication of the CP gene, acquisition of diverse suppressors of RNA silencing and additional genes are acquired in the closteroviruses.
Citrus tristeza virus (CTV) is the only closterovirus known in India. Another closterovirus species Grapevine leafroll associated virus 2, which known to infect grapevine has been examined in Himachal Pradesh, India, however, it could not be detected (Kumar et al. 2013). In this chapter the research work on the characterization of closteroviruses carried out in India is summarised.
2 CTV in India
CTV, an aphid-transmitted closterovirus, causes devastating ‘Tristeza’ or decline in mostly all the economically grown citrus species in the world. It causes phenomenal economic damage to citrus production globally destroying about 100 million citrus trees over the last 70 years (Moreno et al. 2008). As citrus is cultivated in diverse ecological conditions, it is exposed to several CTV strains/variants resulting in diverse disease syndromes; decline of citrus species grafted on sour orange, yellowing and growth cessation of many citrus species and stunting and stem pitting with poor yield and quality in citrus species regardless of kind of citrus rootstock used. The serious epidemic causing the decline of large new plantings of sweet orange trees on sour orange rootstock was recorded for the first time in 1930 in Argentina that destroyed about 30 million trees in this country. Similar situations were also reported from Brazil (1937), Venezuela (1980), South Africa (1974) Ghana (1938), Spain (1960), California (1939) and Florida (1951) causing death of millions of citrus trees.
CTV, a phloem-limited, flexuous filamentous virus with particle size of 2000 × 11 nm belongs to the genus Closterovirus under the family Closteroviridae. It is predominantly transmitted by brown citrus aphid (BrCA; Toxoptera citricida) in a semipersistent manner (Bar-Joseph et al. 1989). CTV genome is positive sense, ssRNA of 19.3 kb in length and contains 12 ORFs; ORF1a and b and ORFs 2–11 potentially encoding at least 19 putative proteins (Karasev et al. 1995). ORFs 1a and 1b plus the nontranslated termini are all that is required for replication in protoplasts. Ten 3′ ORFs, are expressed by 3′-coterminal subgenomic mRNAs (Karasev et al. 1995) encode proteins, p33, p6, p65, p61, p27 (CPm), p25 (CP), p18, p13, p20 and p23, those are expressed via 3′ co-terminal sub genomic RNAs (sgRNA) (Satyanarayana et al. 2000). These proteins involve in movement and interactions with its insect vectors and hosts. CTV contains two capsid proteins (CP): a major CP of 25 KDa (p25) that encapsidates about 95% and a minor CP (CPm) of 27 KDa (p27) that encapsidates about 5% of total particles (Febres et al. 1996). In addition, the p65 (plant heat shock protein hsp70 homolog) and p61 are required for efficient virion assembly (Satyanarayana et al. 2000). CTV has three RNA silencing suppressors (RSSs), proteins CP (p25), p20 and p23 (Lu et al. 2004). In addition to RSSs activity p23 regulates asymmetrical accumulation of the positive and negative strands RNA during replication (Satyanarayana et al. 2000). CTV requires two proteins p20 and p6 (transmembrane protein) for translocation/systemic infection in citrus host (Tatineni et al. 2011). The p6 homologue in BYV has been shown to be a movement protein and required for systemic invasion of host plants (Peremyslov et al. 2004). Recently it was reported that p33 protein is responsible for superinfection exclusion between genetically related CTV isolates (Folimonova 2012). CTV has three genes (p33, p18, and p13) that are not necessary for infection of most of its hosts, but are needed in different combinations for infection of certain citrus species. These genes apparently were acquired by the virus to extend its host range (Dawsona and Folimonova 2013).
The genome expression strategy in closteroviruses is based on (i) proteolytic processing of the polyprotein by ORF1a, (ii) +1 ribosomal frame shift for the expression of the RdRp domain encoded by ORF1b, (iii) expression of the downstream ORFs via the formation of a nested set of 3′ co-terminal sgRNAs. Replication of the viruses occurs in the cytoplasm, possibly in association with endoplasmic reticulum-derived membranous vesicles and vesiculated mitochondria. From an evolutionary point of view, the closteroviruses might have evolved from a smaller filamentous virus.
Genetic diversity in CTV in different citrus growing regions of the world has been reported earlier. Analysis with several complete genomes of CTV isolates shows extensive sequence variation in CTV genome and determines at least seven CTV genotypes; T36, T3, VT, T30, B165, HA16-5 and RB (resistance breaking) occurring in citrus growing countries in the world (Melzer et al. 2010; Biswas et al. 2012a; Harper 2013). Of them, genotypes VT is biologically severe, T30 as mild and T36 as intermediate strains of CTV (Anonymous 2012).
CTV infect all the citrus species, and citrus relatives and hybrids and a non-rutaceous host Passiflora sp. (Moreno et al. 2008). The CTV epidemics are experienced in countries where sour orange is used as rootstock and BrCA vector, T. citricida is common and active. This virus is transmitted by aphids in a semi-persistent manner. CTV can be transmitted to other hosts by inoculation of sap, although the sap transmission is very difficult. It can be transmitted through dodder (Cuscuta sp.). None of the closteroviruses is transmitted through seeds. Virions are usually found in the phloem tissues. The virus is localized only in phloem tissue and sieve tubes, occasionally in the mesophyll and epidermis of infected trees. The necrosis and degeneration below the bud union result in the decline of trees (Schneider 1959). The virus is transported through sieve elements and occasionally enters an adjacent companion or phloem parenchyma cell where virus replication occurs; in some plants this is followed by cell-to-cell movement into only a small cluster of adjacent cells, while in others there is no cell-to-cell movement.
3 History of CTV in India
First indication in occurrence of Tristeza or citrus decline disease in Indian subcontinent was given by Brown in 1920, when he observed the failure of Malta sweet orange (C. sinensis) on sour orange (C. aurantium) root stock in Peshawar (now in Pakistan). After that, decline of citrus orchard was reported from Bombay State (Nagpal 1959; Capoor 1961, 1963; Vasudeva and Capoor 1968) and subsequently from North India (Nariani et al. 1965). Tristeza was shown to be widely spread in almost all the citrus growing regions of India (Raychaudhuri et al. 1977). Transmission of citrus tristeza disease through Toxoptera citricidus was demonstrated by Vasudeva and Capoor (1968). It infects all the commercially grown citrus species, cultivars and hybrids of mandarin (Citrus reticulata), sweet orange (C. sinensis), acid lime (C. aurantifolia), sweet lime/lemon (C. limettoides/limon) grown in India that has killed more than one million citrus trees (Ahlawat 1997; Biswas 2008). CTV was detected in infected citrus trees for the first time India using ELISA using CTV specific antisera by Chakroborty et al. (1992). Using monoclonal antibodies like MCA 13 and MABs3DF1, the CTV isolate of India has been reported to be different from CTV isolate of USA (Chakroborty et al. 1992). The CTV particle measuring about 2000 × 11 nm in size was observed under electron microscopy for the first time in India by Ahlawat et al. (1992). Cloning and sequencing of CP gene of four South Indian isolates were reported by Manjunath et al. (1993). It was reported that the citrus tristeza disease destroyed about a million of citrus trees in India (Ahlawat 1997).
4 Incidence of CTV
Citrus is cultivated in all the four geographical zones of India; Northeast, Northwest, Central and South India and CTV occurs in nearly all the commercial citrus species in India (Ahlawat 1997; Biswas 2008, 2010). CTV is reported to be a century old problem in India but it was un-recognized earlier (Ahlawat 1997). Northeast India is considered to be one of the most important centres of origin of different citrus species (Ghosh 2007) and CTV is an important factor to cause citrus decline in this region (Ahlawat 1997; Bhagabati et al. 1989; Borah et al. 2012). In the Northeast India, CTV is a major problem as occurence of the efficient aphid vector; BrCA is very common. Mandarin orchards in the Darjeeling hills are severely being affected, many of them are been wiped out, due to severe infection of CTV that causes huge economic losses in the citrus industry in this region (Ahlawat 1997; Biswas 2008). Assam, and Meghalaya of Northeast India produce important citrus fruits like Khasi mandarin (C. reliculata), Kagzi lime and Assam lemon (C. lemon) and CTV was reported to be one of the major factors to cause citrus decline in these Northeast states of India (Bhagabati et al. 1989; Chakroborty et al. 1992).
In the South and Central zones of India, CTV is a chronic problem that occurs in mixed infections with huanglongbing (citrus greening disease) (Ahlawat 1997; Ghosh et al. 2003, 2009a). Tirupati region of South India produces many economically important citrus fruit like, sweet orange, sweet lime (C. limettoides) and Kagzilime (C. aurantifolia) and association of CTV for causing decline of citrus trees has been reported in this region (Ahlawat 1997; Tarafdar et al. 2013). Recently, based on field survey and detection by ELISA and PCR, overall disease incidence caused by CTV in India has been reported; 26.3% in Central India (Maharastra), 47.1–56.0% in NE India (Assam, Meghalaya, Sikkim and the Darjeeling hills), 36–50% in South India (Andhra Pradesh and Karnataka) and 16–60% in North-Northwest India (Uttarakhand, Delhi, Punjab, Rajasthan) (Biswas et al. 2014a, b).
5 Transmission of CTV
Dispersal of CTV is taken place through virus infected planting materials like buds or grafted- or seedling-plants those are responsible for introduction of this virus in new growing areas. Insect vector transmission, subsequently, is important for local spread in many parts of India, as presence of the efficient aphid vector BrCA, is common in most of the citrus growing areas in India, particularly in Northeast India (Ahlawat 1997; Biswas 2008). Transmission studies of citrus decline disease in India began in early sixties of nineteenth century. It was demonstrated that BrCA (T. citricidus) is one of the important vectors to transmit the citrus decline disease in citrus trees (Vasudeva et al. 1959; Raychaudhuri et al. 1977). The transmission of this disease through other aphid vectors, Aphis gossypii and Myzus persicae has also been reported by Verma et al. (1965), and through A. craccivora and Dactynotus jaccae by Verma et al. (1965). The virus was reported to be non-persistently transmitted from citrus to citrus. Like other citrus growing counties of the world, T. citiricidus is the most efficient vector of CTV in India (Verma et al. 1965; Capoor and Rao 1967; Biswas 2008). It has also been reported that single BrCA can efficiently transmit multiple genotypes of CTV resulting in changed population dynamics and multiple infection in infected trees (Biswas et al. 2004).
6 Symptoms, Host Range, Biological Indexing and Host Resistance
CTV infects nearly all the citrus species and citrus relatives and hybrids in India. Mexican lime/Kagzi lime (C. aurantifolia) is commonly used as an indicator host, and trifoliate orange is used to filter tristeza from mixed infection of other citrus viruses (Tanaka et al. 1971). Some citrus relatives are highly resistant or immune to CTV infection (Garnsey et al. 1987). Trifoliate orange (Poncirus trifoliata) and its relative are immune to CTV isolates. Many CTV tolerant citrus root stock, P. trifoliata and its hybrid, citrumelo (Sweet orange X P. trifoliata) and Rangpur lime (C. limolina) have been identified (Moreno et al. 2008).
In India, most citrus species and cultivars are susceptible to infection but some are tolerant and do not show obvious symptoms of the disease. Rough lemon (C. jambhiri) and trifoliate hybrid (Rangpur lime X P. trifoliata) were immune when tested with the CTV isolates in the Darjeeling hills (Biswas 2008). Although, Rangpur lime has reported to be tolerant to CTV, it shows vein clearing and vein corking symptoms after inoculation of a particular CTV isolate of the Darjeeling hills. All the cultivated lime/lemons/Kagzilime, Assam lemon (C. lemon), Tahiti lime (C. latifolia) and Sweet lime (C. limettioides) are reported to be infectde by CTV (Biswas et al. 2012b). Acid limes (C.aurantifolia) are most susceptible and show vein clearing, stem pitting and stunting when infected by most isolates of CTV. Earlier it has been reported that mandarin is tolerant or resistant to CTV (Ahlawat 1997). But except Kinnow mandarin, all the cultivated mandarins in India: Darjeeling mandarin, Sikkim mandarin, Khasi mandarin, Nagpur mandarin, Mudkhed mandarin and Coorg mandarin are infected by CTV in field and as well as in greenhouse experimentally (Biswas et al. 2016). In Delhi condition acid lime cv Kagzi Kalan, pumello (C. paradisi), Kinnow mandarin were free from CTV. All the Sweet orange orchard in Delhi was highly susceptible to CTV and three CTV variants were reported from these Sweet orange orchard (Sharma et al. 2012).
Four CTV isolates of the Darjeeling hills, Kpg1, 2, 3 and 4 were tested for symptom production on different citrus species (Biswas 2010). All the CTV isolates infected Kagzi lime, Darjeeling mandarin and Mosambi sweet orange with a variety of biological reactions. The major symptoms were vein clearing, vein flecking, vein corking and stunting, depending on the hosts. On Mosambi sweet orange, all of the isolates induced stunting, but Kpg1 additionally induced vein corking. Rangpur lime was not infected by three of the four CTV isolates but Kpg2 induced vein clearing and veincorking in Rangpur lime, indicating that Rangpur lime may not be immune to all the CTV isolates. Rough lemon was infected by Kpg1 and Kpg2, inducing vein clearing and vein corking, but was not infected by Kpg3 and Kpg4; showing rough lemon is not immune to all the CTV isolates. Although stem pitting caused by Indian isolates CTV-B and CTV-P has been reported previously on Mosambi sweet orange and Kagzi lime (Roy et al. 2005), but none of the Darjeeling CTV isolates studied did not produce stem pitting symptoms on this host (Biswas 2010).
The virus titer in CTV infected citrus hosts in different agroclimatic zones of India has been studied (Tarafdar et al. 2013). The Kagzilime and sweet orange plants contain higher CTV titer compared to mandarin plant, thus Kagzilime and Sweet orange plants are as ideal indicator hosts for CTV in India. The symptom severity in Kagzilime could not be correlated with the high virus titer value in CTV infected tree (Tarafdar et al. 2013). CTV is not evenly distributed in all the infected plant parts of different citrus species. The tender bark, petiole and mid rib of new leaves, and apical bud contain maximum virus titer. The barks obtained from 6 months to 1 year old twigs have higher CTV titer, upto eightfolds in infected Darjeeling mandarin, Sweet orange and Kagzilime trees. Old bark obtained from more than 2 years old twigs contains very less or no virus titre (Tarafdar et al. 2012). The petioles and mid ribs of all kinds of leaves contain higher amount of CTV titer (7 to 20-fold). The apical buds of all the citrus hosts tested contain huge virus titer ranging from 14 to 17-folds. CTV can persists up to 180 days in crude sap in Phosphate buffer (0.05 M; pH 7.0) kept at 4 °C and only up to 2–4 days at 25–32 °C. Accumulation of CTV titer varied in growing seasons; lower titer value, 3.6 to 4.4-folds in the month of February to May and higher titer value, sevenfolds in the month of September to October were estimated (Tarafdar et al. 2012).
7 Genetic Diversity, Distribution, Intra-farm Diversity of CTV
Extensive genetic diversity in CTV in citrus growing regions of the world including India has been reported time to time (Rubio et al. 2001; Biswas et al. 2012b; Tarafdar et al. 2013). Complete genome analysis identified seven distinct CTV genotypes internationally and they are VT, T36, T30, T3, B165, HA16-5 and RB (resistance breaking) (Roy and Brlansky 2010; Melzer et al. 2010; Biswas et al. 2012a; Harper 2013). Genetic recombination is a major phenomenon in the evolution of CTV variants (Martin et al. 2009; Biswas et al. 2012a, b; Tarafdar et al. 2013). Genetic diversity and factors responsible for the origin of CTV variants in India have been examined and reported by many different workers time to time (Roy et al. 2005; Ghosh et al. 2009a; Biswas 2010; Sharma et al. 2012; Biswas et al. 2012a; Singh et al. 2013; Tarafdar et al. 2013; Chander et al. 2015). Based on biological indexing, multiple molecular marker (MMM) analysis, heteroduplex mobility assay (HMA) and sequence analysis using CP gene and 5′ORF1a fragment of CTV genome, three CTV variants sharing 89–97% nt identity identified in the Darjeeling hills of the Northeastern Himalayan regions (Biswas 2010).
Near about 114 CTV isolates covering all the citrus growing-geographical zones of India (Table 8.1) have been characterized based on sequencing of complete CP gene and 5′ORF1a (L-Pro domain) fragment gene (Ghosh et al. 2009a; Biswas 2010; Sharma et al. 2012; Biswas et al. 2012b; Singh et al. 2013; Tarafdar et al. 2013; Palchoudhury et al. 2017). Indian CTV isolates are genetically extremely diverse sharing 80–99% identity for 5′ORF1a and 89–99% identity for CP genes analyzed till today (Biswas et al. 2012b; Tarafdar et al. 2013; Palchoudhury et al. 2017). Homologous and non-homologous recombination may be frequent phenomena in the evolution of all of the known CTV genotypes. Identification of several potential recombination events among Indian CTV isolates has been determined (Biswas et al. 2012a; Sharma et al. 2012; Singh et al. 2013). Recombination phenomena among CTV isolates is responsible for evolution of extensive diversity of CTV in India.
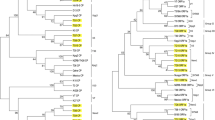
Based on sequence analysis of 5’ORF1a gene fragment, occurrence of eight CTV variants in citrus growing areas of India has been reported earlier (Biswas et al. 2012b). Later on, 5′ORF1a of more number of CTV isolates of Northeast (Assam, Manipur, Meghalaya, the Darjeeling hills and Sikkim hills) and South India (Tirupati) were included and analysed and occurrence of overall ten CTV variants; Kpg3/VT, K5 (distinct), AR1(distinct), BAN-1 (distinct), D13(distinct), K38/T3, Kpg2/T30, AG-28/HA16-5, K10/B165 and BAN-2/T36/RB-G90; were determined in India (Fig. 8.2a) (Tarafdar et al. 2013; Palchoudhury et al. 2015, 2017). Based on analysis of CP genes, Biswas et al. (2012b) reported occurrence of six CTV variants in India. Recently, including and analyzing CP genes more numbers of CTV isolates collecting from Northeast and South India , overall seven CTV variants; B165/VT, P14/T36, Kpg2/RB-G90, TP6 (distinct), K10/T3, K5 (distinct) and Kpg3/HA16-5 are determined (Fig. 8.2b) (Tarafdar et al. 2013; Palchoudhury et al. 2015, 2017). Therefore, it is concluded that seven to ten CTV variants are prevalent in citrus growing regions of India. Genetically most of the Indian isolates are similar to the Israel severe CTV isolate VT (Biswas et al. 2012a; Tarafdar et al. 2013; Palchoudhury et al. 2015, 2017). The distribution of CTV genotypes in citrus growing different geographical regions of India have also been determined and mentioned in Table 8.1.
Phylogenetic grouping of Indian CTV isolates based on 5′ORF1a region (a) and CP gene (b) using maximum likelihood method (Tarafdar et al. 2013). Significance of the nodes was estimated with 1000 bootstrap repetitions
Occurrence of several CTV variants in citrus growing areas of Northeastern Himalayan hill region of India has been reported. During the year of 2007 and 2008, analysis of CP genes and 5’ORF1a gene fragment of 26 CTV isolates from this region, four to five CTV variants were reported in this region (Biswas et al. 2012b). Recently, nine more number of CTV isolates of Mirik region of the Darjeeling hills and Rumtek area of the Sikkim hills were analysed and overall five CTV variants; Kpg3/VT, K10/B165, Kpg2/T30, K5 (distinct) and K38/T3 type in the Darjeeling hills and its surrounding areas of Northeastern Himalayan hill regions of India are reported (Palchoudhury et al. 2017). CTV isolates of Manipur state of Northeast India have been characterized and occurrence of Kpg3/VT and T3 genotypes is common in Manipur state (Palchoudhury et al. 2015).
The p23 gene (ORF11, 630 nt;) of Indian CTV isolates (two from Arunachal Pradesh, two from Nagaland, three from Manipur, six from Assam, three from Delhi, two from Vidarbha, one from the Darjeeling hills and two from South India) was characterized. Extensive sequence diversity ranging from 88–100% nucleotide to 87–100% amino acid identity among the p23 gene were found and made five genetic clusters (Chander et al. 2015).
Occurrence of divergent CTV isolates in individual citrus farms in many citrus growing regions of India is reported. For instance, an sweet orange farm, IARI, New Delhi has shown at least three CTV variants (e.g., VT and D13 types) and another mandarin farm, IARI-Regional Station of the Darjeeling hills has at least three CTV genotypes (e.g., VT and B165). Similarly, Intra-farm diversity of CTV has been observed in many other individual farms in India suggesting that intra farm diversity of CTV might be common in India (Biswas et al. 2012a; Sharma et al. 2012).
8 Complete Genome Sequence of Indian Isolate of CTV
Based on biological property and host range study, a isolate Kpg3 was identified as a decline inducing CTV strain in the mandarin growing areas of the Darjeeling hills (Biswas et al. 2012a, b). The complete genome, 19,253 nt in length, of isolate Kpg3 was sequenced, analyzed and submitted in NCBI database as accession number HM 573451 (Biswas et al. 2012a). The Kpg3 genome contains all the 12 putative ORFs similar with the other CTV genomes reported earlier. In phylogenetic relationship, the Kpg3 is closely related to Israel severe CTV isolate VT and it is a recombinant strain (Biswas et al. 2012a). The isolates Kpg3 and VT, both might have originated from distantly related ancestors through a complex evolutionary pathway by multiple recombination events exchanging sequences between diverged CTV variants (Biswas et al. 2012a).
Further, 3′ half of the genome (8398 nt) comprising ten genes (ORFs 2–11) of four other CTV isolates, B5 of Bangalore (HQ912023), D1 of Delhi (HQ912022), G28 of Assam (KJ914661) and Kat1 of Vidarbha (KJ914662) of India were sequenced and compared with other Asian and internationally recognized CTV genotypes (Biswas et al. 2016). All the Asian isolates categorized into six genogroups, whereas the Indian isolates fell into four, and other Asian isolates into three genogroups. Indian isolates B5, D1, Kat1 and Kpg3 grouped together (Kpg3Gr) along with Florida isolate T3. However, the isolate B5 was placed distantly from other members of Kpg3Gr. Thus isolate B5 might be a new isolate. The isolate G28 was found to be distinct lineage.
9 Cross Protection
Cross protection is the ability of mild strains or isolates to protect the severe or more virulent strains or isolates of the same virus. The phenomenon of cross protection and use of mild cross protecting strain (MCPS) have been known for a long time for potential management of severe viruses. Cross protection was applied in very few crops and not accepted widely due to lack of pure MCPS. However, this concept of cross protection has been successfully used in management of CTV in many countries like Brazil, Australia, Japan and South Africa (da Graça and van Vuuren 2010; Roistacher et al. 2010). The unique aspect of cross protection in CTV is that the mild strain is easily inoculated to the target plant by grafting scions collecting from mild strain infected mother plants.
Cross protection of CTV was initiated in the years of 1970s and many CTV strains were identified based on vector specificity (Capoor and Rao 1967; Capoor and Chakroborty 1980). The mild and severe CTV strains were identified and effort was made for cross protection of CTV in Tirupathi and Bangalore region (Balaraman and Ramakrishnan 1977). Unfortunately, the experiment was failed and it might be due to appearance of severe strains mixed with the mild strain. Recently, effort was made to identify pure MCPS of CTV with the help of in silico molecular-based codon biasness analysis using CP gene of CTV isolates from Northeast India and their biological evaluation (Biswas et al. 2016). Two Indian isolates Mnp1 from Manipur and MB3 from Meghalaya were identified mild CTV type. This finding will be effective for designing a proof concept of MCPS after biological evaluation challenging with severe strain (Biswas et al. 2016).
10 Conclusion Remarks
Citrus orchards of many citrus growing areas of India, particularly in Northeast India, are being wiped out due decline or slow death of the citrus trees caused by CTV. The genomics of CTV, genetic diversity, evolution of CTV complex population and geographical distribution of virus variants have clearly been understood in India that has helped in understanding of the disease epidemiology. Nucleotide sequence analysis and phylogenetic relationships of large number of CTV isolates determined occurrence of seven to ten CTV variants in India, some of them are new to India or new to the world. These studies would lead to develop an improved diagnostics targeting diiferent virus strains/variants using specific primers, and also to make molecular-based management strategy targeting conserved sequence of the virus through gene silencing. Recombination events, negative selection and gene flow play major role for evolution of in CTV variants.
In cross protection, super infection exclusion occurs between isolates of the same strain but not between isolates of different CTV strain. As it is known that severe CTV isolates of the virus frequently represent a mixture of different virus strains, for practical applications of cross protection in the field, broad-spectrum mild strains are needed against multiple CTV strains. The technologies for detecting CTV in planting material by biological, serological and molecular methods have been perfected and are being utilized to detect virus at early stages. These techniques would help to develop a long term disease management. The distribution of the CTV variants in India has been studied. The genes of interest have been cloned and transgene constructs were made for transformation of citrus to develop virus resistant transgenic plants in India in near future.
The old citrus orchards, particularly in Northeast India, are severely infected by CTV. Thus, sanitation and replanting with virus free propagative materials are foremost needed to reduce the economic losses of citrus. The random distribution of CTV infected citrus planting materials as means of virus dissemination is common in India. Thus legislation/notification is needed from the Government sector to develop budwood certification programmes and provide disease-free budwood or seedlings to the growers. As the disease is horizontally spread through planting material and vertically through aphids, establishment of new orchards using disease free planting material and keeping the orchards free from aphids with regular inspection are essential to maintain the citrus industry viable and profitable. In this regard a strategy for production of CTV-free planting material for the Darjeeling hill developed earlier (Biswas et al. 2009) could be followed. Efforts have been made to produce disease free citrus planting materials through seed certification program and shoot tip grafting successfully practiced in new plantations in many citrus growing areas in India.
References
Ahlawat YS (1997) Viruses, greening bacterium and viroids associated with citrus (Citrus species) decline in India. Indian J Agr Sci 67:51–57
Anonymous (2012) In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy: classification and nomenclature of viruses. Ninth Report of the International Committee on Taxonomy of Viruses. International Union of Microbiological Societies, Virology Division, Amsterdam, pp 871–879. 11 March
Balaraman K, Ramakrishan K (1977) Studies on strains and strain interaction in Citrus tristeza virus, Technical series bulletin. University of Agricultural Sciences, Bangalore, p 62
Bar-Joseph M, Marcus R, Lee RF, (1989) The continuous challenge of citrus tristeza virus control. Annu Rev Phytopathol 27(1):291–316
Bhagabati KN, Ahlawat YS, Chakroborty NK, Borthakur BC (1989) Distribution of greening, tristeza amd mosaic disease of citrus in North eastern staes of India. Indian Phytopath 42:552–555
Biswas KK (2008) Molecular diagnosis of Citrus tristeza virus in mandarin (C. reticulata) orchards of Darjeeling hills of West Bengal. Indian J Virol 19:26–31
Biswas KK (2010) Molecular characterization of Citrus tristeza virus isolates from the Northeastern Himalayan region of India. Arch Virol 155:959–963
Biswas KK, Manjunath KL, Marais LJ, Lee RF (2004) Single aphids transmit multiple genotypes of citrus tristeza virus, but often with changed population dynamics. Phytopathology 94:S8
Biswas KK, Tarafdar A, Jayakumar BK, Pun KB (2009) Strategy for the production of Citrus tristeza virus-free mandarin (Citrus reticulata) planting materials in the Darjeeling hills of India. Indian Phytopathol 62:376–380
Biswas KK, Tarafdar A, Diwedi S, Lee RF (2012a) Distribution, genetic diversity and recombination analysis of Citrus tristeza virus of India. Virus Genes 45:139–148
Biswas KK, Tarafdar A, Sharma SK (2012b) Complete genome of mandarin decline Citrus tristeza virus of Northeastern Himalayan hill region of India: comparative analyses determine recombinant. Arch Virol 157:579–583
Biswas KK, Godara S, Nayak D (2014a) Distribution of Citrus tristeza virus in the Darjeeling hills and their biological symptoms in mandarin orchards. Indan J Hort 71:408–411
Biswas KK, Tarafdar A, Sharma SK, Singh JK, Dwivedi S, Biswas K, Jayakumar BK (2014b) Current status of Citrus tristeza virus incidence and its spatial distribution in citrus growing geographical zones of India. Indian J Agric Sci 84:8–13
Biswas KK, Pal Choudhury S, Godara S (2016) Decline of mandarin orange caused by Citrus tristeza virus in Northeast India: conventional and biotechnological management approaches. J Agric Eng Food Technol 3(3):236–241
Borah M, Nath PD, Saikia AK (2012) Serological detection of Citrus tristeza virus affecting citrus tree species in Assam. Indian Phytopath 65:289–293
Brown WR (1920) The Orange: a trial of stock at Peshawar. In: Bulletin 93, Indian Agricultural Research Institute, Pusa, p 7
Capoor SP (1961) Kagzi lime: an indicator plant of citrus ‘decline’ virus in India. Indian Phytopathol 14:109–112
Capoor SP (1963) Decline of citrus trees in India. Bul Nat Inst Sci India 24:48–64
Capoor SP, Chakraborty NK (1980) Further studies of citrus seedling yellow yellows in Deccan trap country. Indian J Hort 37:97–100
Capoor SP, Rao DG (1967) Tristeza virus infection of Citrus in India. In: Proceedings of the international symposium on subtropical and tropical horticulture. Horticulture Society of India, Bangalore, pp 723–736
Chakroborty NK, Ahlawat YS, Varma A, Chandra KJ, Ramapandu S, Kapur SP (1992) Serological reactivity in Citrus tristeza virus strains in India. In: Proceedings of the 12th conference on IOCV, pp 108–112
Chander V, Godara S, Kumar A, Saha B, Biswas KK (2015) Nucleotide sequence of ORF 11-p23 suppressor gene of Citrus tristeza virus isolate of different citrus growing states of India and determination of extensive genetic diversity. J Mycopathol Res 53(1):25–29
da Graça JV, van Vuuren SP (2010) Managing Citrus tristeza virus losses using cross protection. In: Karasev AV, Hilf ME (eds) Citrus tristeza virus complex and tristeza diseases. APSPress, Eagan, pp 247–260
Dawsona WO, Folimonova SY (2013) Virus-based transient expression vectors for woody crops: a new frontier for vector design and use. Annu Rev Phytopathol 51:321–337
Febres VJ, Ashoulin L, Mawasaki M, Frank A, Barjoseph M, Manjunath KL, Lee RF, Niblet CL (1996) The p27 protein is present at one end of citrus tristeza virus particles. Phytopathology 86:1331–1335
Folimonova SY (2012) Superinfection exclusion is an active virus-controlled function that requires a specific viral protein. J Virol 86:5554–5561
Garnsey SM, Civerolo EL, Gumf DJ, Yokomi RK, Lee RF (1987) Toward a standardized evolution of the biological properties of Citrus tristeza virus. Phytophylactica 19:151–157
Ghosh SP (2007) Citrus fruits. ICAR, New Delhi
Ghosh DK, Das AK, Singh S (2003) Individual and mixed infection of CTV and greening Liberobacter and their association with sweet orange decline in the state of Maharashtra. J Mycol Plant Pathol 33(1):69–72
Ghosh DK, Aglave B, Roy A, Ahlawat YS (2009a) Molecular cloning, sequencing and phylogenetic analysis of coat protein gene of a biologically distinct CTV isolate occurring in Central India. J Plant Biochem Biotechnol 18:105–108
Ghosh DK, Das AK, Shivankar VJ (2009b) Implementing citrus budwood certification program. Indian Farming 59(8):41–45
Harper SJ (2013) Citrus tristeza virus: evolution of complex and varied genotypic groups. Front Microbiol 4:1–18
Karasev AV, Boyko VP, Gowda S, Nikolaeva OV, Hilf ME, Koonin EV, Niblet CL, Cline K, Gumpf DJ, Lee RF (1995) Complete sequence of the citrus tristeza virus RNA genome. Virology 208:511–520
Kumar S, Singh L, Ferretti L, Barba M, Zaidi AA, Hallan V (2013) Evidence of Grapevine leafroll associated virus-1–3, Grapevine fleckvirus and Grapevine virus B occurring in Himachal Pradesh. India Indian J Virol 24:66–69
Lu R, Folimonov A, Shintaku M, Li WX, Falk BW, Dawson WO, Ding SW (2004) Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc Natl Acad Sci U S A 101:15742–15747
Manjunath KL, Pappu HR, Lee RF, Niblett CL, Civerolo E (1993) Studies on coat prtein genes of four Indiam isolates of Citrus tristeza closterovirus: cloning sequencing and expression. In: Proceedings of the 12th conference IOCV, Riverside, CA, pp 20–27
Martin S, Sambade A, Rubio L, Vives MC, Moya P, Guerri J, Elena SF, Moreno P (2009) Contribution of recombination and selection to molecular evolution of Citrus tristeza virus. J Gen Virol 90:527–1538
Melzer MJ, Borth WB, Sether DM, Ferreira S, Gonsalves D, Hu JS (2010) Genetic diversity and evidence for recent modular recombination in Hawaiian Citrus tristeza virus. Virus Genes 40:111–118
Moreno P, Ambros S, Albiach-Marti MR, Guerri J, Pena L (2008) Citrus tristeza virus: a pathogen that changed the course of the citrus industry. Mol Plant Pathol 9:251–268
Nagpal RL (1959) Tristeza found in Bombay state. Calif Citrograph 44:402–405
Nariani TK, Sahambi HS, Chona BL (1965) Occurrence of tristeza virus in citrus in Northern India. Indian Phytopath 18:220
Palchoudhury S, Sharma SK, Biswas MK, Biswas KK (2015) Diversified Citrus tristeza virus causing decline disease in Khasi mandarin in Manipur State of Northeast India. J Mycol Plant Pathol 45:317–323
Palchoudhury S, Ghimiray P, Biswas MK, Biswas KK (2017) Citrus tristeza virus variants and their distribution in mandarin orchards in Northeastern Himalayan hill region of India. Int J Curr Microbiol App Sci 6(5):1680–1690, https://doi.org/10.20546/ijcmas.2017.606.196
Peremyslov VV, Andreev IA, Prokhnevsky AI, Duncan GH, Taliansky ME, Dolja VV (2004) Complex molecular architecture of beet yellows virus particles. Proc Natl Acad Sci U S A 101, 5030l
Raychaudhuri SP, Nariani TK, Ahlawat YS (1977) Dieback of citrus in India. Proc Int Soc Citriculture 3:914–918
Roistacher CN, daGraça JV, Muller GW (2010) Cross protection against Citrustristezavirus-a review. In: Hilf ME, Timmer LW, Milne RG, da Graça JV (eds) Proceedings of conference of the international organization of Citrus Virology, 17th (Riverside, CA: IOCV), pp 1–27
Roy A, Brlansky RH (2010) Genome analysis of an orange stem pitting Citrus tristeza virus isolate reveals a novel recombinant genotype. Virus Res 151:118–130
Roy A, Manjunath KL, Brlansky RH (2005) Assessment of sequence diversity in the 5-terminal region of Citrus tristeza virus from India. Virus Res 113:132–142
Rubio L, Ayllon MA, Kong P, Fernandez A, Polek M, Guerri J, Moreno P, Falk BW (2001) Genetic variation of isolates from California and Spain: evidence for mixed infections and recombination. J Virol 75:8054–8062
Satyanarayana T, Gowda S, Mawassi M, Albiach-Marti MR, Ayllon MA, Robertson C, Garnsey SM, Dawson WO (2000) Closterovirus encoded HSP70 homolog and p61 in addition to both coat proteins function in efficient virion assembly. Virology 278:253–265
Satyanarayana T, Gowda S, Ayllon MA, Albiach-Marti MR, Rabindram R, Dawson WO (2002) The p23 protein of Citrus tristeza virus controls asymmetrical RNA accumulation. J Virol 76:473–483
Schneider H (1959) The anatomy of tristeza-virus-infected citrus. In: Wallace JM (ed) Citrus virus diseases. University of California, Division of Agriculture Sciences, Berkeley, pp 73–84
Sharma SK, Tarafdar A, Khatun D, Sumita K, Biswas KK (2012) Intra-farm diversity and evidence of genetic recombination of Citrus tristeza virus isolates in Delhi region of India. J Plant Biochem Biotechnol 21:38–43
Singh JK, Tarafdar A, Sharma SK, Biswas KK (2013) Evidence of recombinant Citrus tristeza virus isolate occurring in acid lime cv. pant lemon orchard in Uttarakhand Terai Region of Northern Himalaya in India. Indian J Virol 24:35–41
Tanaka H, Yamada S, Nakanishi (1971) Approach to eliminating tristeza virus from citrus trees by using trifoliate orange seedlings. Bull Hort Res Station Japan Sec 11:157–163
Tarafdar A, Ghosh PD, Biswas KK (2012) In planta distribution, accumulation, movement and persistence of Citrus tristeza virus in citrus host. Indian Phytopathol 65:184–188
Tarafdar A, Godara S, Dwivedi S, Biswas KK (2013) Characterization of Citrus tristeza virus and determination of genetic variability in North-east and South India. Indian Phytopathol 66:302–307
Tatineni S, Robertson C, Garnsey SM, Dawson WO (2011) A plant virus evolved by acquiring multiple nonconserved genes to extend its host range. Proc Natl Acad Sci U S A 108:17366–17371
Vasudeva RS, Capoor SP (1968) Citrus decline in Bombay state. FAO Plant Prot Bul 6:91
Vasudeva RS, Varma PM, Rao DG (1959) Transmission of Citrus decline virus by Toxoptera citricudus (Kirk) in India. Curr Sci 28:418–419
Verma PM, Rao DG, Capoor SP (1965) Transmission of tristeza virus by Aphis craccivora Koch and Dactynotus jaceae (L.) India J Entomol 27:67–71
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Biswas, K.K., Palchoudhury, S., Ghosh, D.K. (2017). Closterovirus in India: Distribution, Genomics and Genetic Diversity of Citrus Tristeza Virus. In: Mandal, B., Rao, G., Baranwal, V., Jain, R. (eds) A Century of Plant Virology in India. Springer, Singapore. https://doi.org/10.1007/978-981-10-5672-7_8
Download citation
DOI: https://doi.org/10.1007/978-981-10-5672-7_8
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-5671-0
Online ISBN: 978-981-10-5672-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)