Abstract
Soft-tissue segmentation has always been difficult point in the medical research of diagnosis of soft-tissue defects. Especially for Anterior Cruciate Ligament (ACL) rebuilding surgery, ACL segmentation from all soft-tissue inside knee joint, including Posterior Cruciate Ligament (PCL) and meniscus, is a very important task. In this paper, we propose a novel ACL segmentation method: Space Model Contrast Clustering-based (SMC-based) ACL Segmentation. Unlike the widely used processing method, such as segmentation by MRI gray values and Mimics segmentation drawing, the proposed method relies 3D model of knee joint to segment soft tissue by self-adaptive K-means clustering. Extensional experiments demonstrate that the proposed method can be capable of solving the problem of soft-tissue segmentation well and has achieved higher ACL segmentation efficiency.
Please note that the LNCS Editorial assumes that all authors have used the western naming convention, with given names preceding surnames. This determines the structure of the names in the running heads and the author index.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Soft-tissue segmentation
- Anterior cruciate ligament
- SMC-based ACL segmentation
- Self-adaptive K-means clustering
1 Introduction
With some advanced medical devices, like CT, MRI, being widely used, medical 3D reconstruction has been increasingly important research topic in recent years [1, 2]. 3D reconstruction can largely help doctors detect and diagnose some sickness. Nowadays, more and more surgeries are operated under the guide of scheme designed by computer. Besides, 3D reconstruction has many application in kinds of medical sub-topics, such as tissue segmentation [2], tumor detection and location.
Mostly, the first processing steps is to collect data from patients by CT or MRI. Then reconstruct the 3D model of region of interest (ROI), like the knee joint or brain. Among all these tasks, tissue segmentation is the most basic and simplest one. Because independent ROI can provide the most direct object without any other noise and interference. However, tissue data from CT/MRI can hardly be segmented well due to rough source data and the existed techniques. Especially for soft tissue, segmentation would become much harder [3, 4]. Most of soft tissues has the similar scanning values, and CT scan seems to react same to all soft tissue. For example, inside knee joint, there are mainly three kinds soft tissues, Anterior Cruciate Ligament (ACL), Posterior Cruciate Ligament (PCL) and meniscus. And with the interference of fleshy tissue, it would be a large challenge to segment ROI from them.
ACL segmentation is very important because it the key step of injury detection. With the heavy traffic and more sports activities, more and more people suffer from the injury of ACL. Now the accurate way to ACL injury detection is to observe the biological characteristic of ACL under the arthroscope [5], and this way is called as arthroscopy. However arthroscopy is minimally invasive operation after all, it can hardly be accepted by each patient. Then the ideal way is to reconstruct 3D model of ACL and to detect its physical parameters to give the correct diagnosis. Segmentation of ACL from scanning data of knee joint then become so important.
The most widely used method is segmentation by threshold of gray values and Mimics editor. As said before, gray values is not sensitive among soft tissues, so ACL segmentation by gray values threshold would be cause large error. Editing by people is not also a good choice, because it would need much time for people and it is not impersonal, and cannot be applied by each one. In this paper, we propose a novel ACL segmentation method: Space Model Contrast Clustering-based (SMC-based) ACL Segmentation which relies 3D model of knee joint to segment soft tissue by self-adaptive K-means clustering. Extensional experiments demonstrate that the proposed method can be capable of solving the problem of soft-tissue segmentation well and has achieved higher ACL segmentation efficiency.
The remainder of the paper is organized as follow: the proposed method: SMC-based ACL segmentation is introduced in Sect. 2. The experimental settings and results are demonstrated in Sect. 3. Finally, we make a conclusion objectively in Sect. 4.
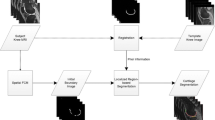
2 The Proposed Method
Knee joint is one of the most important tissue to keep stable for people. Nowadays the most widely used way to segment ACL from knee joint is to edit the scanning data using some software like Mimics. The most important factor which would affect result is the experience of editor. Always, only some clinician who have been familiar with the anatomical structure of knee joint are eligible. From the anatomical data, we find that the main interior structure is always the same, especially for adults, and they have the same structure of knee joint. So we think it is feasible to refer the existed space distribution of interior structure of knee joint to achieve ACL segmentation from scanning data. Then the SMC-based ACL segmentation method is proposed. As shown in Fig. 1, the anatomical structure of knee joint is constant to each man. It has the main tissue, as following: bones, including Thighbone, Patella, Fibula and Tibia; soft tissue, like Anterior cruciate ligament, Posterior cruciate ligament, Lateral collateral ligament, Medial collateral ligament, Lateral meniscus, Medial meniscus. Our study in this paper has not taken collateral ligaments into consideration due to they are not connected with ACL and PCL. So the main task is to segment ACL from bones and soft tissues, including PCL and meniscuses. The 3D model of these tissue can be seen like shown in Fig. 2. The structure model with green color is ACL.
For these point clouds, following parameters would have great impact on segmentation result: barycenter (G), angles across with X, Y, Z plane respectively \( (\alpha ,\beta ,\delta ) \), and the distance between the point and origin (d). The barycenter G can be computed by these equations:
where \( G_{x} ,G_{y} \,{\text{and}}\,G_{z} \) are three dimensional value respectively, \( v \) is the volume of point cloud model, and \( \rho (x,yz) \) denotes the density of the model structure. For us human body, each organ is made by some certain materials, so in medical measurement, the density of organ is constant which would greatly help solve the equations above. Then barycenter G of each model can be computed by another simple method, Eq. (4) as follow:
where N is the total number of points in each model, and \( G^{i} \) denotes the ith point in point cloud. Compared to the former equations, Eq. (4) could reduce the time consumption dramatically. The distance d can be computed by Eq. (5):
Then the angles can be solved with distance d using the following equations:
All of these data from model can be used as arguments into segmentation method. In our study, we choose K-means for the consideration of its simplicity [6–8]. K-means is an unsupervised classification algorithm based on clustering, the basic theory is described in Eqs. (9)–(10).
Assuming that there are N data points in total, they are divided into K cluster. The K-means algorithm is to minimize J, \( \gamma_{ik} \) is 1 or 0. When we get the smallest \( J,u_{k} \) should be met Eq. (10). The iteration cannot be stopped until its convergence. K-means is a high-efficiency method to clustering, however there are two problems hindering on the way. The two existed problems are how to initialize and how to judge convergence well. And the difference in our study is that we use the model data to set initialization and convergence condition.
As the input into K-means, the scanning data of knee joint has the following structure that the algorithm need to segment: Thighbone, Patella, Fibula and Tibia; Anterior cruciate ligament, Posterior cruciate ligament, Lateral collateral ligament, Medial collateral ligament, Lateral meniscus, Medial meniscus. To reduce the time consumption of this task, the algorithm can remove the collateral ligaments directly due to they are outside the knee bones gap. So the number of clusters (K) is 7, 4 kinds of bones and 3 kinds of soft tissues including ACL. Then the barycenter G of these 7 model structures, which are computed by Eqs. (1)–(3), would be used as centriods in initialization processing. And the most important is convergence condition. Experimental results demonstrate that the best segmentation can hardly be extracted. And the algorithm always miss the and the global optimization between two iterations. In this paper, to reduce the time consumption and the risk of local optimization, we take the variance of angles and distance into consideration of convergence. The convergence condition is shown in Eq. (11).
where C denotes the convergence conditional value. The evaluation setting is also important to the segmentation. No matter large or small, it would cause the bad results we cannot stand. We will talk about it in experiments part.
The proposed segmentation method in this paper need the other two traditional segmentation method: threshold segmentation and software editing to remove some other noise data, such as skin and fat. And the proposed processing algorithm is concluded in Algorithm 1 as follow:
Algorithm 1: SMC-based ACL Segmentation
3 Experiments and Analysis
In this paper, the experimental setting is as follow, hardware: Thunderobot with Inter(R) Core(TM) i7-4710 CPU@2.5 GHz, 16GRAM, NVIDIA GTX 870; software: Windows 8, Mimics 16.0, Geomagic Studio 12.0, MatlabR2012b, SPSS19.0, Geomagic Studio 2012. And the MRI scanning data all are provided by Chongqing Xinqiao Hospital.
Threshold segmentation and erasing editor are two necessary steps in the SMC-based ACL Segmentation method. The basic processing flow of threshold segmentation is shown in Fig. 3. Mimics provides the analysis tool to extract threshold which is different in different patient because it can be affected by some other factors, such as weather, machine. The threshold range of soft tissue of the patient in Fig. 3 is from 1100 to 1800. Then these extracted thresholds can be used to produce segmentation masks. Figure 4a shows us the mask we set in which the green is bone mask while the yellow is soft tissue mask. This figure demonstrates threshold segmentation causes so much noise. Figure 4b shows the results after erasing editor.
Threshold and editing segmentation are the most widely used method in medical processing, and it can segment bones from soft tissue well. As shown in Fig. 5a is the 3D objects by threshold masks while (b) is soft tissue 3D object. But the structure of ACL and other soft tissue cannot be observed by any users. The proposed SMC method can segment different soft tissue, like shown in Fig. 5c in which the green is ACL, the red one is PCL while the violet one is meniscus.
In this paper, we also use Geomagic Studio 2012 to optimize the ACL 3D object. Nurbs curve surface is made by some non-uniform ration B-splines [9] which is shown in Eq. (12). And GS2012 can optimize the B splines to remove some corners and to make ACL more smooth, like the comparison shown in Fig. 6 in which (a) is the results from SMC segmentation method, while (b) is the optimization result.
where \( P_{i,j} \) is corner point, \( N_{j,k} (u) \), \( N_{j,l} (v) \) are B-splines in u, v directions respectively, \( W_{i,j} \) is weight factor.
Figure 7 shows us the segmentation results of 3D model of ACL. In this figure, the segmented ACL has unambiguous edges which would help doctors diagnose. In our study, we use model parameters to initialize K-mean clustering. Experimental results demonstrate that this initialization greatly reduce the iterations and time consumption of algorithm processing. Tables 1 and 2 show the iterations and the time consumption. Of the tables, the data in the parentheses are the results from random initialization. From the comparison, parameters initialization reduces at least half of iterations of random initialization. For ACL segmentation task, iterations are cut down to 1.4 from 7.5, and the time consumption is also reduced into 138 ms from 682 ms.
4 Conclusion
In this paper, we propose a new segmentation method: SMC-based ACL Segmentation which can segment ACL from knee joint well. This new method solves the traditional medical problem, segmentation of soft tissue. The ACL 3D objects from SMC has unambiguous edges which would help doctors diagnose. And model parameters initialization greatly reduce the iterations and time consumption of the proposed algorithm processing.
References
Koehler C, Wischgoll T, Golshani F (2010) Reconstructing the human ribcage in 3d with x-rays and geometric models. Multimedia 17(3):46–53
Bouchet A, Pastore JI, Di Meglio L, Robuschi L, Ballariin V (2013) Segmentation and 3D Reconstruction of Microbial Biofilms. Latin Am Trans 11(1):324–328
Malik OA, Arosha SMN, Zaheer D (2015) An intelligent recovery progress evaluation system for ACL reconstructed subjects using integrated 3-D kinematics and EMG featrues. J Biomed Health Inf 19(2):453–463
Saha PK et al (2011) A new Osteophyte segmentation algorithm using the partial shape model and its applications to rabbit femur anterior cruciate ligament transection via Micro-CT imaging. IEEE Trans Biomed Eng 58(8):2212–2227
Spillmann J, Tuchschmid S, Harders M (2013) Adaptive space warping to enhance passive haptics in an arthroscopy surgical simulator. IEEE Trans Visual Comput Gr 19(4):626–633
Vora P, Oza B (2013) A survey on K-mean clustering and particle swarm optimization. Int J Sci Mod Eng 1(3):24–26
Agrawal A, Gupta H (2013) Global K-means clustering algorithm: a survey. Int J Comput Appl 79(2):20–24
Rajput GG, Patil PN (2014) Detection and classification of exudates using K-means clustering in color retinal images. In: International conference on signal and image processing, pp 126–130
Rouhani M, Sappa AD, Boyer E (2015) Implicit B-spline surface reconstruction. IEEE Trans Image Process 24(1):32–33
Acknowledgment
The authors would like to thank the anonymous reviewers for their helpful comments. This work is partly supported by the National Natural Science Foundation of China (Grant No. 61300092), the Fundamental Research Funds for the Central Universities of China, (Grant No. ZYGX2013J068), Natural Science Foundation of Fujian Province, China (Grant No 2015J05132).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media Singapore
About this paper
Cite this paper
Rao, Y., Ding, X., Gou, J., Ma, Y. (2016). An Efficient ACL Segmentation Method. In: Hung, J., Yen, N., Li, KC. (eds) Frontier Computing. Lecture Notes in Electrical Engineering, vol 375. Springer, Singapore. https://doi.org/10.1007/978-981-10-0539-8_6
Download citation
DOI: https://doi.org/10.1007/978-981-10-0539-8_6
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-0538-1
Online ISBN: 978-981-10-0539-8
eBook Packages: Computer ScienceComputer Science (R0)