Abstract
The existing directed graph clustering algorithms are born with some problems such as high latency, resource depletion and poor performance of iterative data processing. A distributed parallel algorithm of structure similarity clustering on Spark (SparkSCAN) is proposed to solve these problems: considering the interaction between nodes in the network, the similar structure of nodes are clustered together; Aiming at the large-scale characteristics of directed graphs, a data structure suitable for distributed graph computing is designed, and a distributed parallel clustering algorithm is proposed based on Spark framework, which improves the processing performance on the premise of the accuracy of clustering results. The experimental results show that the SparkSCAN have a good performance, and can effectively deal with the problem of clustering algorithm for large-scale directed graph.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
With the wide application of network data, such as gene regulatory network, social network and other network data in various fields, the scale of directed graph is growing explosively. How to manage and use the massive data has become a hot research topic in recent years [1]. Directed graph contains a wealth of data relationships, such as the behavior of users in social networks. In order to discover the hidden cluster structure in the network, traditional clustering methods are based on the link density, such as Newman [2] algorithms and Kernighan-Lin [3] algorithms, which make the distance between the nodes in the cluster closer, and make the distance between the cluster nodes is far away to achieve the effect of clustering. However, the algorithms above ignore the directed interaction and different functions that nodes may have in the graph data. Based on the link density, Xu Xiao-wei [4] proposed the SCAN algorithm, which is based on the structural similarity. However, the algorithm is only useful to the undirected network of clustering and it doesn’t consider the variety of data relationships in the real environment. Zhou Deng-yong [5] proposed a way making the directed edges convert to the undirected edges, but the way ignore the structure information of directed graph. Literature [6] transformed the network clustering problem into the optimization problem of weighted cutting of directed graph for further study. However, literatures [5, 6] did not distinguish the different functions of the nodes.
Chen Jia-jun [7] proposed a directed graph clustering algorithm DirSCAN based on SCAN. Chen Ji-meng [8] proposed a parallel clustering algorithm PDirSCAN which used MapReduce based on literature [7]. Zhao W [9] proposed a clustering algorithm based on MapReduce by looking for connected components, however, there are some problems such as high delay, high I/O operation of HDFS file system, and the poor performance of iterative data processing. In this paper, we propose a structure similarity clustering algorithm based on Spark framework with the advantages of iterative computation. We calculate the similarity of the vertices and build the initial cluster with the distributed framework; we perform cluster label expansion and synchronize operation in parallel to achieve the clustering of the vertices in the graph to reduce the running time and computational cost. The experimental results show that the algorithm can efficiently clustered in the big data environment for directed graph clustering.
The rest of the paper is organized as follows. In Sect. 2, we give a brief introduction to the concept of graph clustering and Spark. In Sect. 3, we introduce our SparkSCAN algorithm in detail. Section 4 presents the experimental results and analysis. Finally we provide our conclusions in Sect. 5.
2 Preliminary
2.1 Spark
Spark [10] is a common parallel framework for the Berkeley AMP Lab UC. Spark has the advantages of MapReduce Job, and intermediate output results can be saved in memory, Job no longer need to read and write HDFS. Thus, Spark can be better applied to data mining and machine learning.
2.2 Rdd
RDD [11] (Resilient Distributed Datasets), is an abstract concept of distributed memory. RDD provides a highly constrained shared memory model, which is a read-only collection of records, and can only be created by performing a set of transformations (such as, join, and map) in the other RDD. These constraints make the cost of achieving fault tolerance very low.
2.3 PDirSCAN Algorithm
A PDirSCAN algorithm based on directed graph is proposed in the paper [8].
Definition 1
(Neighborhood). Given a directed graph G = {V, E}. The directed edge which from v to u is signed as < v, u >, v, u ∈ V. The Neighborhood is a set of nodes and itself which starting from the one step of v, denoted by Γ (v).
Definition 2
(Structural Similarity). For two nodes, the more coincident nodes can be reached, the more likely to belong to the same cluster. The definition of structural similarity, denoted by σ, is given by:
Definition 3
(ε Neighborhood-Nodes). The definition of ε Neighborhood-Nodes, denoted by Nε (u), is given by:
The ε is used to divide the ε neighbor node and non-neighbor node threshold.
Definition 4
(CORE). If a node has enough ε neighborhood-nodes, we called it core. Node u is core if |Nε (u)| ≥ μ, u∈V. μ is a threshold.
Definition 5
(Directly Structure Reachable). If u is one of the v’s ε neighborhood-nodes, where v is a core. Then u must belongs to the same cluster with v.

The cluster is generated from the core in this algorithm. If the v is one of the core u’s ε neighborhood-nodes, v is assigned to the same cluster with u. The cluster continued to grow until all the clusters could not be further increased.
Definition 6
(Hub and Outlier). Assume node u does not belong to any cluster. Node u is hub just has node v and w exist in Γ (u), which v and w do not belong to the same cluster. Otherwise u is outlier.
3 SparkSCAN
In order to adapt to the large-scale clustering of directed graph, this section we design a parallel algorithm SparkSCAN on Spark.
Definition 7
(Structure Reachable). Given by a directed graph G = {V, E}. Given a series of vertices v1, v2, …,vn, v = v1, u = vn. We called v and u is structure reachable, which vi and vi-1 is Directly Structure Reachable. If v and u is structure reachable, then u should also belong to the same cluster with v. Any pair node of the same cluster is structure reachable.
In this paper, the operation of the SparkSCAN algorithm is mainly divided into three steps:
-
1.
Parallel recognize ε neighbors nodes and core node, then build the initial clusters;
-
2.
Execute cluster expansion through synchronizing cluster label in parallel, and then, achieve clustering merge;
-
3.
Analysis of clustering results and recognize the hub and outlier node.
In the first step, each node can independently calculate the structural similarity between the other nodes; In the second step, each sub-cluster can be independently calculate the label, cluster labels of vertices can be synchronized according to the intermediate result of the algorithm; In the third step, algorithm can be used to analyze the clustering results of each cluster label. In the process of identifying the hubs and the outliers, each node can do it by itself. SparkSCAN algorithm with the fault tolerance of Spark, so that the whole task will not collapse because of one processing node’s paralysis, so as to achieve the purpose of parallel processing.
The overall flow chart of the algorithm is shown in Fig. 1:
3.1 Data Structure of SparkSCAN
Data Structure of Graph Storage.
There are two kinds of storage methods of giant graph, Edge-Cut and Vertex-Cut. Because per vertex just be stored once by Edge-Cut, so it can save storage space. But if the calculation involves two vertices on the edge are divided into different machines, it communicate and transfer data by crossing machine and cost the network communication traffic. So we use Vertex-Cut way, which each edge only stores one time, and only appears on a machine. This way increases the storage overhead, but it can significantly reduce the amount of network traffic. This method has the advantages especially in large data.
In order to store the graph data effectively, this paper design the data structures to store the nodes and edges with RDD as follows:
Vertex (ID: Long, Arr: String)
Where ID represents the ID of vertex, Arr is a String which represent the properties of vertex, just like “property 1, property 2”;
Edge (srcID: Long, dstID: Long, Arr: String)
Where srcID represents ID of the begin node of the edge, dstID represents ID of the end node of the edge, Arr is a String which represents the properties of vertex, just like “property 1, property2”;
With the characteristics of Spark, this paper design the data structures to represent the collection of storage points and edges as follows:
VertexRDD = RDD [Vertex (Long, String)];
EdgeRDD = RDD [Edge (Long, Long, String)];
Data Structure of Algorithm.
This algorithm involves some important intermediate variables. In order to realize the parallel of the algorithm, we design the data structures as follows:
The data structure of the storage vertices and their son nodes is shown as follows:
neighborRDD = RDD[(Long, Array[Long])]
Each element of the data structure is a key-value pair, denoted by (Long, Array [Long]) where the first Long value represents the ID of vertex, the second Array represents an array of son nodes of vertex.
The data structure of the storage vertices and their ε neighborhood-nodes shown as follows:
eNeighborRDD = RDD[(Long, Array[Long])]
Each element of the data structure is a key-value pair, denoted by (Long, Array [Long]) where the first Long value represents the ID of vertex, the second Array represents an array of the ε neighborhood-nodes of vertex.
The data structure of the storage cores and their ε neighborhood-nodes shown as follows:
uNeighborRDD = RDD[(Long, Array[Long])]
Each element of the data structure is a key-value pair, denoted by (Long, Array [Long]), where the first Long value represents the ID of core, the second Array represents an array of ε neighborhood-nodes of core.
The data structure of the storage the sub-cluster and their cluster label shown as follows:
uAllNeiRDD = RDD[Array[(Long, Long)]]
Each element of the structure is an array, denoted by Array [(vid, label)], storage the relationship between all the vertices of a sub cluster and the cluster labels. Each element of the array is a key-value pair, denoted by (Long, Long), where the first Long value represents ID of vertex, the second Long represents the cluster label of vertex.
The data structure of the storage vertex and the minimum cluster label in all the sub clusters is shown as follows:
minRDD [(Long, Long)]
Each element of the structure is a key-value pair, denoted by (Long, Long), where the first Long value represents the ID of vertex, the second Long represents the minimum cluster label in all the sub clusters of vertex.
3.2 Parallel Recognition ε Neighbors and Core Nodes
Parallel recognize ε neighbor-nodes and cores in three stages:
Stage I, we can get the relationship between node and their son nodes through the calculation of the relationship between the vertices of the graph. Then transform the relationships to key-value pairs and put them into a collection, denoted by neighborRDD. For the convenience of parallel computing, we transform neighborRDD to an array, denoted by neighborArr, and broadcasts the array in all machine.
Stage II, computing structure similarity between vertices in parallel to get all vertices and their ε neighborhood-nodes. Assuming that exist an element, denoted by (vidi, Arrayi), we can get Γ (vidi) = {vidi}∪Arrayi.. We can get the structural similarity between vertices through the calculation of the element with each other.
The second stage can be described as follows process:
Step.1 Get the current calculation of the element, denoted by (vidi,Arrayi);
Step.2 Assuming that vidi’s has an array to storage the ε neighborhood-nodes, denoted by eArrayi. Computing the structure similarity between elements in neighborArr and (vidi, Arrayi). We put the element’s ID into eArrayi if the structure similarity is greater than ε.
Step.3 Remove the element from eArrayi which equals vidi. Then constitute a new element (vidi, eArrayi) as the result to return.
The second stage of each element is executed independently in parallel. And then all the calculations results of each element are merged to a collection. The collection include all key-value pairs which storage vertex and its ε neighborhood-nodes.
Stage III, filter elements in eNeighborRDD to find the elements, which the size of ε neighborhood-nodes is greater than μ. Merge them to the core collection, denoted by uNeighborRDD.
The process can be described by the following example:
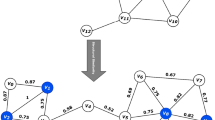
Assuming that exist a directed graph G = {V, E}, its structure is shown in Fig. 2:
We can get a set of vertex and its son nodes which has the out-degree, as show in column “Son Set” in Table 1.
For illustration of purposes, we set the adjustable parameter ε = 0.4, μ = 1;
Take the ID of vertex is 1 as the example, denoted by Vertex 1, its structure similarity as show in Table 2.
According to ε = 0.4 we can get the node’s Nε (Vertex 1) is {6}.
Similarly we can get all vertices and their Nε (Vertex 1) as show in column “ε neighborhood-nodes” in Table 1.
According to μ = 1, we can get set of core is {1,6,7,9,10,13,14,15,16}.
3.3 Extension and Synchronization of Cluster Label in Parallel
Extension and synchronization of cluster label in parallel is in two stages:
Stage I. Build and initialize the sub-clusters. Each core and their Nε (u) are transformed to a sub-cluster. The sub-cluster is an array of the key-value pairs with the ID and cluster label of vertex, which in Nε (u) ∪ the ID. The label is initialized to the ID of vertex. The first stage can be described by the following process:
Each element of the uNeighborRDD (core and its ε neighbor-nodes) calculate by the following steps:
Step.1 Get the current calculation of the element, denoted by (vidi, Arrayi);
Step.2 Put vidi into Arrayi;
Step.3 Set labelj = vidj;
Step.4 Transform Arrayi to an array which includes all vertices and their label, denoted by Array[(vidj,labelj)], as the result to return.
The stage I of each element of the stage is executed independently in parallel. And then all the calculation results of element are merged a collection, denoted by uAllNeiRDD, after calculation. The algorithm steps can be described by the following example:
According to the example given by Sect. 4.2, we can build the sub-clusters, results of calculation as show in column “vertices of sub-cluster (initial stage)” in Table 3. Among them, each line represents a sub-cluster, where the key-value pair (id, label) is represents the ID and cluster label of the vertex. Next step, parallel synchronizing the only cluster label of the vertices to all vertices in different sub-cluster.
According to Definition 7, if there exists directly structure reachable of any two vertices in the same cluster, then there exists the structure reachable of any two vertices in the sub-clusters if the sub-clusters has same vertices. The vertices of structure reachable should belong to the same cluster. In this paper, vertices are in same cluster if their have same cluster label. Execute cluster expansion through synchronization cluster label in parallel, and then, achieve clustering merge.
Stage II, parallel computing the results of Stage I to synchronize the cluster label in every sub-cluster. Then computing the minimum of the cluster labels of the vertices, which have the same ID. The minimum as the only cluster label of the vertex. Iterative above process until the label of vertices not change any more. Finally we can output the result set. The algorithm process can be described as follows:
Step.1 In each sub-cluster, sort the vertices by their cluster label. We get the minimum as the cluster label of the sub-cluster, and set all vertices’s cluster label to this minimum. Then execute Step.2;
Step.2 Merge the elements to a new collection, denoted by allRDD [(vid, label)]. Then execute Step.3;
Step.3 Get the minimum cluster label of vertices, which have the same ID, set the minimum as the only cluster label. Then merge the key-value pair which made up by the ID and only cluster label of vertex to a new collection, denoted by minRDD[(vid, label)]. Then execute Step.4;
Step.4 According to minRDD [(vid, label)], synchronization the only cluster label to all vertices in different sub-cluster in parallel.
Step.5 If the minRDD is the same with the last iterative result, then the iterative is completed, minRDD as the result and return; otherwise execute Step.1.
The process can be described by the following example:
According to the example given by Sect. 4.3, we simulate an iterative process. First, synchronize the cluster label in the sub-cluster, the result as show in column “vertices of sub-cluster (after synchronize in sub-cluster)” in Table 3. Then get every element of all sub-cluster, merge to allRDD as show in column “cluster labels of vertex” in Table 4:
Then computing the minimum cluster label of the vertices, which have the same ID. The minimum as their only cluster label. Next set the cluster label to minimum. The result as show in column “only cluster labels of vertex” in Table 4.
According to the result, we synchronize the cluster label in every sub-cluster. The result as show in Table 5:
Now we finish an iteration.
In the above example, the final result as show in column “only cluster labels of vertex (final)” in Table 4.
3.4 Clustering Result Analysis
In the result of the parallel clustering algorithm, the vertices of the same cluster label should belong to same cluster.
In this stage, we transform (id, label), which is element in result, to (label, id). Then merge the id of vertices by the same cluster label as the clustering result.
According to example of Sect. 4.3 we get three clusters. They are {1, 6, 7, 16},{9, 10},{13, 14, 15}.
According to Definition 6, the vertex is hub or outlier, if it is not in any cluster. According to example, hubs are {8, 3}, outliers are {2, 4, 11, 12}.
4 Evaluation
4.1 Data-sets
-
1.
We test and verify the accuracy of algorithm by using binary_networks, which is a tool for generated social network randomly. Datasets generated as follows:
-
(1)
Dataset binary_networks1K, which includes 1,000 vertices, edges is randomly generated.
-
(2)
Dataset binary_networks10K, which includes 10,000 vertices, and edges are randomly generated.
-
(3)
Dataset binary_networks100K, which includes 100,000 vertices, and edges are randomly generated.
-
(1)
-
2.
We test and verify parallel efficiency of algorithm by:
-
(1)
Random dataset built by binary_networks tool includes 100,000 vertices and 1,532,964 edges.
-
(2)
Dataset soc-sign-slashdot090216 of Slashdot Zoo signed social network from February 21 2009 includes 82,144 vertices and 549,202 edges.
-
(3)
Dataset amazon0302 of Amazon product co-purchasing network from March 2 2003 includes 262,111 vertices and 1,234,877 edges.
-
(4)
Dataset Wiki-vote of Wikipedia who-votes-on-whom network includes 7,115 vertices and 103,689 edges.
-
(1)
Among the above dataset, dataset 1 is simulation and the others are from Stanford University big data network.
4.2 Algorithm Evaluation Index
We used the Precision (P), Recall (R), F1 and Rand Index (RI) to verify the accuracy of algorithm. It is a correct result if two vertices of same real cluster is belong to the same cluster. The greater the value of the four evaluation index, more similar to the real world and better clustering.
We used the speedup verify the parallel efficiency of algorithm, which is the ratio of serial and parallel processing with the shortest time. The greater of speedup, the shorter of parallel time.
4.3 Environment of Experiment
See Table 6.
4.4 Parameters of Cluster
This paper select the value of ε and μ by the method of SCAN in literature [4]. This involves making a k-nearest neighbor query for a sample of vertices and noting the nearest structural similarity. The query vertices are then sorted in ascending order of nearest structural similarity. The knee indicated by a vertical line represents a separation of vertices belonging to clusters to the right from hubs and outliers to the left. We recommend a value for μ, of 2.
4.5 Results and Analysis of Experiment
We test and verify the accuracy of algorithm by using binary_networks, which is a tool for generating network randomly.
In binary_networks1K Case, if μ = 2, the result on different values of ε as show in Table 7:
From experiments on different values of ε, we can see that value of ε has a significant effect on the accuracy of the clustering results in SparkSCAN. When value of ε is too small, it easily divide different clusters vertices into one cluster; however, when value of ε is too big easily create too much hubs and outlier.
If ε = 0.5, μ = 2, the result on different values of ε as show in Table 8:
The experimental results show that SparkSCAN algorithm with good accuracy performance by selecting P, R, F1, RI reasonably.
Conducting experiments using PDirSCAN and SparkSCAN to verify the parallel efficiency of algorithm:
The speedup of binary_networks100K as show in Fig. 3:
The speedup of soc-sign-slashdot090216 as show in Fig. 4:
The speedup of amazon0302 as show in Fig. 5:
The speedup of Wiki-vote as show in Fig. 6.
Compared with PDirSCAN algorithm, and SparkSCAN has better performance on the above datasets, especially on large datasets. When the number of computer increases, the time algorithm spend is reducing. The result in real world is better than simulation dataset. This is due to the data relationships between simulated data sets is too complex. Synchronizing cluster label increases the network overhead and leading to decrease the effect of parallel. The parallel effect on Wiki-vote dataset is not obvious because of the size of dataset is too small, at this time consuming by Spark framework itself is more apparent.
In conclusion, SparkSCAN improves the processing speed on the premise of the accuracy of clustering results. SparkSCAN has good performance in large data environment and high practical value. However, the accuracy of the algorithm is more dependent on ε and μ parameters.
5 Conlusion
This paper proposes a structure similarity clustering algorithm based on Spark for directed graph. The experimental results show that SparkSCAN can effectively improve the efficiency and the speed of the directed graph clustering and has a greater practical value in the large-scale environment data. However, the selection of parameter values has great influence on accuracy and computational efficiency. In the future, we will study further the reasonable allocation scheme of the related parameters to achieve better results.
References
Ding, Y., Zhang, Y., Li, Z.-H., Wang, Y.: Researach and advances on graph data mining. J. Comput. Appl. 32(1), 182–190 (2012)
Lancichinetti, A., Fortunato, S., Kertész, J.: Detecting the overlapping and hierarchical community structure in complex networks. New J. Phys. 11(3), 033015-1–033015-18 (2009)
Fallani, F.D.V., Nicosia, V., Latora, V., et al.: Nonparametric resampling of random walks for spectral network clustering. Phys. Rev. E 89(1), 012802-1–012802-5 (2014)
Xu, X.-W., Yuruk, N., Feng, Z.-D., et al.: SCAN: a structural clustering algorithm for networks. In: Proceedings of the 13th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Jose, pp. 824–833 (2007)
Zhou, D.-Y., Huang, J.-Y., Schölkopf, B.: Learning from labeled and unlabeled data on a directed graph. In: Proceedings of the 22nd International Conference on Machine Learning, Bonn, pp. 1036–1043 (2005)
Meila, M., Pentney, W.: Clustering by weighted cuts in directed graphs. In: Proceedings of the 7th SIAM International Conference on Data Mining, Minneapolis, pp. 135–144 (2007)
Chen, J.-J.: Research on Clustering Algorithms for Large—Scale Social Networks based on Structural Similarity. Nankai University (2013)
Chen, J.-M., Chen, J.-J., Liu, J., Huang, Y.-L., Wang, Y., Feng, X.: Clustering algorithms for large-scale social networks based on structural similarity. J. Electron. Inf. Technol. 02, 449–454 (2015)
Zhao, W., Martha, V., Xu, X.: Pscan: a parallel structural clustering algorithm for big networks in mapreduce. In: 2013 IEEE 27th International Conference on Advanced Information Networking and Applications (AINA), pp. 862–869. IEEE (2013)
Zaharia, M.A.: An Architecture for Fast and General Data Processing on Large Clusters. University of California, Berkeley (2013)
Zaharia, M., Chowdhury, M., Das, T., et al.: Resilient distributed datasets: a fault-tolerant abstraction for in-memory cluster computing. In: Proceedings of the 9th USENIX Conference on Networked Systems Design and Implementation, p. 2. USENIX Association (2012)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media Singapore
About this paper
Cite this paper
Zhou, Q., Wang, J. (2016). SparkSCAN: A Structure Similarity Clustering Algorithm on Spark. In: Chen, W., et al. Big Data Technology and Applications. BDTA 2015. Communications in Computer and Information Science, vol 590. Springer, Singapore. https://doi.org/10.1007/978-981-10-0457-5_16
Download citation
DOI: https://doi.org/10.1007/978-981-10-0457-5_16
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-0456-8
Online ISBN: 978-981-10-0457-5
eBook Packages: Computer ScienceComputer Science (R0)