Abstract
Most proteins need to avoid the complex topologies when folding into the native structures, but some proteins with nontrivial topologies have been found in nature. Here we used protein unfolding simulations under high temperature and all-atom Gō-model to investigate the folding mechanisms for two trefoil knot proteins. Results show that, the contacts in β-sheet are important to the formation of knot protein, and if these contacts disappeared, the knot protein would be easy to untie. In the Gō-model simulations, the folding processes of the two knot proteins are similar. The compact structures of the two knot proteins with the native contacts in β-sheet are formed in transition state, and the intermediate state has loose C-terminal. This model also reveals the detailed folding mechanisms for the two proteins.
Xue Wu, Ting Fu, Peijun Xu and Jinguang Wang have been contributed equally to this paper.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
The protein molecule performs the biological function through folding into the compact structure. In the folding process, most proteins avoid complex topologies, but some proteins are able to fold into nontrivial topologies, especially the main chain fold into a knotted conformation [1–3], which is an evolutionary curiosity. If pulling the knotted protein from both the two ends, this structure can’t be disengaged. So far, most of the discovered knotted proteins are belong to the 31 knot, and the others are belong to the 41, 52 or 61 knots [4–6]. Though the knotted proteins are existent, but how the knotted proteins overcoming the energy barrier fold into the complicated and intact topologies from the disordered linear polypeptide is still a mystery. The shape of the protein and the chain connectivity of its backbone may determine the folding routes of a well-designed protein sequence [7]. So the structure based protein models can capture the essential features of protein folding through separating from the effects of topology and eliminating all non-native energetic traps [8–10]. From the unfolded state to the native state of protein, the energy landscape directs this folding route of protein, and the diverse sizes and shapes of the free energy barriers are directed by the pattern of contacts especially the native contacts [11–13]. The knot protein with the complicated topology may not fold easily because of the emerging unlikely configurations in the folding process [14–16]. The folding of knot protein needs right crossing of polypeptide, otherwise may have an unknotted protein or a wrong chirality. Based on the structure-based model, the information of protein folding pathway is contained in the folded configuration, so it is a good model for studying the folding process of knot protein. The time scale of protein folding is incompatible with the time scale of molecular dynamics simulations, so studying the protein unfolding process under high temperature is another meaningful method for studying the folding process of protein. At high temperature, it is easy to cross the energy barrier of knot protein, so this knot could be untied in this condition, and through studying the unfolding progress of knot protein to get the information of the folding process. Here we used the high-temperature unfolding method, all-atom and Ca structure-based model to research the folding pathways of knot proteins. The all-atom model can supply more accurate thermodynamic information for the folding of knotted protein than the Ca structure-based model, which is good for uncovering the folding mechanisms for the simple knot proteins.
For studying the formation of knot protein, various biochemical and biophysical techniques have been employed, like chemical denaturants, single-molecule atomic force microscopy (AFM) measurements [17]. The studies for the folding of protein YibK from H. influenzae and YbeA from E. coli in experiment were through using the denaturant urea to get the unfolded structure reversibly which lacked secondary or tertiary structure, and then gave a detailed folding study for protein [18, 19]. The folding pathways of protein YibK have been extensively studied. The double-jump refolding experiment has been used to investigate the presence of multiple unfolded states of protein YibK [20]. The folding mechanism of YibK has been probed by using single-site mutants, this folding process of protein YibK was from the denatured state, but this structure was not unfold completely [21]. The theoretical investigations of knot protein can be started from the wholly unfolded structure, which does not contain a knot, so the theoretical investigations can make up the defects of the experimental studies and give more information for the folding of knot protein. The atomistic simulations have been used for studying the unfolding of bovine carbonic anhydrase II [22]. On the coarse-grained level, the simulations also could be used for the studies of protein folding. The simulations of Gō-model for knot protein was applied to some studies, and this model could make the protein fold from the unfolding structure to the topologically frustrated, knotted structure. Generally, the Gō-model reduces the protein to its Ca-backbone. Wallin undertook the coarse-grained model on the knot protein YibK for studying the folding kinetics, and through introducing the attractive nonnative interactions on the knot protein, this protein could take the knotted mechanism of plug motion to form native structure [23]. The coarse-grained model has been used for probing the folding processes of protein YibK and YbeA, and succeeded in forming a native knot structure in 1–2 % of the simulations with native interactions through using this model [24]. In the folding processes of protein YibK and YbeA, an intermediate configuration with a slipknot was involved, and the appearance of this configuration was aimed at reducing the topological bottlenecks. The researches about slipknots of proteins also revealed that these slipknots could give contribution to the thermal stability for the slipknot feature [25]. The molecular dynamics simulations have been used widely in the studies of proteins [26–34], so here molecular dynamics simulation methods were used to study the folding of knot proteins. Two different approaches comparing with these previous studies have been used for probing the folding mechanisms of knot proteins. The method of protein unfolding under high temperature and all-atom Gō-model were applied to two 31 knot proteins for studying the folding mechanisms and thermodynamics of the two proteins.@@@@@
When folding to the correct native structure, the knot protein has to avoid the topological traps and kinetic traps on the landscape. In the folding process, the tying of knot protein is refer to a problem about the chain crossing, and a topological constraint may solve this problem, otherwise this process is not allowed. The geometric constraint of the native structure may dominate the knot protein through a subset of possible folding pathways. In theory, the protein with the minimally frustrated structure is supposed to have the energy landscape of funnel shape. In the folding process of protein, the shape of the landscape is dominated by the strong energetic bias, which could reduce traps caused by non-native interactions. Thus, this geometric constraint model is better for determining the folding mechanisms of proteins. Using this geometric constrain model also is necessary for guiding the chain to form knot, and the final folded structure of protein plays a major role in determining its foldability, so this model may make the protein have more chances to fold into the native state. Under high temperature, the knot protein has more probability to cross the energy barrier, so this protein has more chances to unfold. The all-atom model can make up the gap between coarse-grained models and all-atom empirical forcefields. Hence, here we used the method of high-temperature unfolding and all-atom model to research two knot proteins in order to have more information about the folding mechanisms and topological constraint effects of these proteins.
In this study, the knot proteins are the smallest knot protein MJ0366, from Methanocaldococcus jannaschii [5, 19], and protein VirC2, the border-specific endonuclease, from Agrobacterium tumefaciens [25]. The two proteins have trefoil knot structures (Fig. 8.1). At high temperature, the protein MJ0366 could unfold. The conformational clustering method was used to find the transition state, and this state has the native contacts in β-sheet. The unfolding process has relation ship with the stability of this β-sheet. The all-atom model for the smallest knot protein shows the intermediate state has native contacts in β-sheet, and slipknot and plug knotting routes are found at folding temperature. The protein VirC2 is prone to have traps in the folding process and through backtracking to fold into the native state.
2 Results and Discussion
In this paper, we study two trefoil knot proteins which are two simple examples of nontrivial knots. The protein MJ0366 with 82 residues belongs to α/β protein. The trefoil knot is one end of the chain through into a loop. The C-terminal of protein MJ0366 threads into the loop consists of α1, α2 and their linkers, and the N-terminal threads into the loop which is comprised of β2, α3 and their linkers. The protein VirC2 with 121 residues has ribbon-helix-helix (RHH) fold. This protein has two β-strands, and four α-helices like protein MJ0366. The C-terminal of protein VirC2 threads into the loop which is created by α1, α2 and the linkers between α2 and β2, and the N-terminal threads into the loop formed by β2, α3 and their linkers.
2.1 Protein MJ0366 Unfolding Pathway
Here we used molecular dynamics simulations under high temperature to study the protein unfolding process. We selected 530 K for studying the protein unfolding process, and the molecular dynamics simulation of native state was in 298 K as a comparison. The Ca root-mean-square deviation (Ca RMSD) cluster method was used to find out the transition state. We took nine unfolding simulation trajectories for protein MJ0366, knot_1-knot_9. The transition states were identified at 8.175 ns in knot_1, 23.42 ns in knot_2, 14.076 ns in knot_3, 4.345 ns in knot_4, 14.073 ns in knot_5, 17.831 ns in knot_6, 8.431 ns in knot_7, 9.511 ns in knot_8, and the transition state in the last trajectory knot_9 was not found.
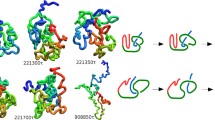
The number of native contact for protein MJ0366 as a function of time in a typical trajectory knot_6 is shown in Fig. 8.2. Under high temperature, the number of native contact was obviously changed as the time growth, and the change trend of the number of native contact for the whole protein was the same as the number of native contact in the β-sheet. The native contacts in β-sheet decreased along with the decreasing number of native contact of the whole protein, so the β-sheet unfolding may have significant impact on the whole system. After the β-sheet untied, the whole system may have low stability, so the knot would be easier to unfold. The unfolding process of protein MJ0366 in the typical trajectory knot_6 is shown in Fig. 8.3. Under high temperature, the α-helices especially the α2 unfold firstly. The native contacts between α2 and the other secondary structures were few (Fig. 8.5b), which may effect the stability of this helix. In the trajectory knot_6, the α-helices were almost disappeared after 1 ns, and the native contacts in β-sheet were still existent. The α-helices disappeared after 2 ns, at this time the β-sheet still was stable, and the protein MJ0366 formed a compact structure. The β-sheet disappeared after 5 ns, and the new β-sheet between the position of α1 and α3 was appeared. Though the β-sheet was disappeared, the two β-strands were in close distance, and the loop controlled by this β-sheet was enlarged, so the C-terminal may have the chance to unfold. The untied protein was appeared for the first time at ~9.71 ns, in this time scale the β-sheet was reformed, and the two terminals formed a new β-sheet. This new β-sheet made the protein fluctuate around N-terminal, so the C-terminal could have the chance to unfold in short time. From this time, the knot protein entered a fluctuant stage lasting for ~7 ns, the knot protein was varied between the untied state and the knot state. In the fluctuant stage, the contacts in β-sheet were diminished, which made the protein change to a loose structure, and then made protein easier to untie. In this stage, the β-sheet was prone to form loops to make the knot untie. Though the C-terminal has formed loop, and it seems to be excluded from the loop formed by α1, α2 and their linkers, but the contacts in the β-sheet were still existent, which effected the unfolding of the C-terminal of knot protein. From the above, the β-sheet is important for the stability of knot protein. The β-sheet disappeared completely after ~11 ns. At 16.09 ns, the protein was untied, and did not form the knot again. The β-sheet between the two terminals was disappeared, and the terminal of β1 was prone to form a loop, which made the β1 exclude from the loop formed by β2, α3 and their linkers. At ~17 ns, the knot protein got to the transition state. After the transition state the contacts between β-strands were disappeared, the N-terminal excluded from the loop formed by β2, α3 and their linkers, and then the C-terminal had the chance to exclude from the loop formed by α1, α2 and their linkers. Under high temperature, the β-sheet is prone to be destroyed first, and then the C-terminal may have the chance to exclude from the loop formed by α1, α2 and their linkers. The unfolding trajectories are considered in reverse as a description of the folding pathway. The α-helixes of protein MJ0366 have been disappeared in the early stage of the folding process, and then the β-sheet disappeared, so the β-sheet may be formed earlier than α-helixes. The process that C-terminal unfolds firstly may consume more energy for the knot protein, so this protein chooses the pathway that the N-terminal unfolds firstly. Hence, this protein may choose a pathway that a structure with the β-sheet is formed firstly, and then the C-terminal thread into this loop controlled by the β-sheet for folding into the native state.
2.2 Transition States for Protein MJ0366 Under High Temperature
In the protein unfolding process, the transition state was decided by the Ca-RMSD cluster method. The Ca-RMSD has been used as a crucial criterion for the convergence measure of the protein systems [35, 36]. The Ca-RMSD for protein MJ0366 in a typical trajectory knot_6 is shown in Fig. 8.4a. Before performing the unfolding simulations under high temperature, the dynamic behavior of protein MJ0366 was investigated at room temperature. Under room temperature, the knot protein was stable, and the Ca-RMSD for this protein was remained at ~2.0 Å during the 40 ns simulation. Under the temperature of 530 K, the Ca-RMSD of protein MJ0366 had a rapid structural deviation comparing with the crystal structure in the native state at ~17 ns in the typical trajectory, and the transition state was found through the method of Ca-RMSD cluster at ~17 ns. The knot position can be characterized by its depth, the distance along the sequence from N-terminal and C-terminal of the knot [24]. Here we used the residues that form the knot to monitor this protein. The knot protein server was used for the detection of knot proteins [37]. The size of knot protein as a function of time under temperature of 530 K is shown in Fig. 8.4a. At transition state, the knot of protein MJ0366 was untied, and before reaching the transition state the protein fluctuated between folded and untied states. After transition state, the protein was untied and no longer formed a knot. The contact map for the knot protein in native state is shown in Fig. 8.5a. In transition state, some of the native contacts in β-sheet were existent, which implied the two β strands fluctuated in the close distance between each other. This state effected the excluding of C-terminal from the loop formed by α1, α2 and their linkers. The number of native contact of residues A8-I53, R7-S57 and T8-L60 were higher than 30 % in the transition state. Some of the native contacts between the loop of N-terminal and β2 were maintained at high level. The number of native contact K5-E60 was higher than 40 %. The residues K3-E57, K3-E65 had the number of native contact higher than 30 %. In the transition state, the non-native contacts for this knot protein were increased, especially the contacts in the β-sheet and between C-terminal and N-terminal. The non-native contacts between N-terminal and β2 were increased, which implied the β1 was prone to exclude from the loop formed by β2, α3 and their linkers, and the contacts in the β-sheet effected the unfolding of knot protein. The non-native contacts between C-terminal and the region around β1 were appeared. The decreasing native contacts in β-sheet made the surrounding secondary structures of C-terminal become loose, so the C-terminal had more chances to have contacts with N-terminal. All the above, the contacts in the β-sheet is important for the protein stability, if breaking these contacts may promote protein untie. The molecular dynamics simulations for this knot protein were in water, and in transition state the solvent accessible surface area (SASA) was changed (Fig. 8.4b). The SASA values of C-terminal and N-terminal were decreased. The emerging non-native contacts between the C-terminal and N-terminal made the two regions eliminate the surrounding water molecules. The changes in the surrounding loop of C-terminal made the contacts among α-helices decreased, which may impact the SASA value of the C-terminal in α1, and this region had SASA decreased. In transition state, the whole system did not unfold, so the SASA of the knot protein was not changed very much.
The average native contact maps for protein MJ0366. a The contact map of the trajectory at 298 K. b The native contact map for the transition states of the nine simulation trajectories at 530 K. The upper triangular presents the nonnative contacts, and the lower triangular presents the native contacts
2.3 Protein MJ0366 Folding Pathway in Gō-model
We performed constant temperature molecular dynamics simulations to obtain the free energy landscape for the monomer structure of knot protein at folding temperature. Each simulation of all-atom model included the folded/knotted state and unfolded/unknotted state. The folding process for protein MJ0366 was monitored by reaction coordinates. The free energy as a function of the number of native contact is shown in Fig. 8.6a. In the folding process, the knot protein had three states. The unfolded state was near the number of native contact 0.15, and then this protein folded into the intermediate state. This result is consistent with the investigation by Jeffrey K. Noel et al., and they made use of Gaussian-type contact potential to study knot protein [38]. After crossing the free energy barrier with the number of native contact ~0.4, the protein folded into the native state. The free energy as a function of two reaction coordinates, the number of native contact and RMSD, is shown in Fig. 8.6b. In the folding process, the RMSD of knot protein were changed with the increasing number of native contacts. The RMSD of the unfolded state for knot protein was near 20 Å. When the RMSD value decreased to ~3 Å, the knot protein folded into the native state. The two-dimensional free energy landscape as a function of the number of native contact and radius of gyration (Fig. 8.6c) was not shown an obvious L-shaped landscape, which indicated the whole system not aggregated rapidly. The radius of gyration of knot protein decreased with an increasing number of native contact. When the value of radius of gyration decreased to ~3 Å, the protein folded into the native state. The landscape as a function of the number of native contact and the number of native contact formed in the β-sheet was shown three states (Fig. 8.6d). The number of native contact in β-sheet increased rapidly with an increasing number of native contact of the whole protein, but stayed low and increased little further once the number of native contact in β-sheet of ~0.7 was formed. In the folding process, the intermediate state was appeared, which near the number of native contact of 0.7. After intermediate state the protein needed to cross the energy barrier to form a knot. In the folding process, protein must overcome an energy barrier to form the β-sheet, and this state could form a loop, which is necessary for the formation of the knot. This loop needs to twist correctly, otherwise the protein may form the topological trap structures like the results of the investigation by Jeffrey K. Noel et al. The C-terminal needed to thread into this loop for the formation of native structure, and this step required to cross the high energy barrier. The C-terminal may thread into this loop through plug or slipknot motion [38]. Here we found when the loop formed by α1, α2 and their linkers was loose, the C-terminal was prone to thread into this loop with plug motion, otherwise the C-terminal tended to adopt slipknot motion. From the above, the native contacts between C-terminal and the loop formed by α1, α2 and their linkers are stable, so more energy is needed to destroy these contacts than the native contacts in β-sheet. Under high temperature, the protein chooses to untie the β-sheet firstly, which is because of the unstability of this region comparing with C-terminal of the knot protein.
The folding routes of knot protein MJ0366 from all-atom Gō-model at folding temperature T = 111. a The free energy as a function of the number of native contact. b The free energy as a function of the number of native contact and Ca-RMSD. c Two-dimensional free energy landscape as a function of the number of native contact and radius of gyration. d The free energy as a function of the number of native contact of the whole protein and the number of native contact in β-sheet
2.4 Intermediate and Transition States for Protein MJ0366 in Gō-model
The native contact maps in intermediate state and transition state for protein MJ0366 are shown in Fig. 8.7. The intermediate and transition states were defined according to the free energy as a function of the number of native contact. The intermediate state had the number of native contact of ~0.2, and the transition state was located in the maximum free energy as the function of the number of native contact. In intermediate state, all the native contacts in β-sheet were almost appeared, but the C-terminal was loose. After the intermediate state the protein entered the transition state with high energy barrier. The transition state appeared some native contacts, such as the native contacts between C-terminal and the surrounding region of β1, and the C-terminal and α1. So the C-terminal was ready to thread into the loop formed by α1, α2 and their linkers in transition state. In addition, some structures in the transition state formed the native contacts between α2 and α4, which means the main native contacts needed by the formation knot protein were appeared, so some structures have formed knot and the formation knot protein maybe at the late transition state.
2.5 Protein VirC2 Folding Pathway in Gō-model
We used constant temperature molecular dynamics simulations of Ca Gō-model to get the free energy landscape of the structure of protein VirC2 at folding temperature for better understanding the folding mechanism of trefoil protein. The free energy as a function of the number of native contact and radius of gyration is shown in Fig. 8.8b. The L-shaped landscape indicated the radius of gyration decreased rapidly with an increasing number of native contact, but once the number of native contact of ~0.4 was formed, the radius of gyration decreased little further. The unfolded state had the radius of gyration of ~0.1. After the radius of gyration reached ~0.6, the protein folded to the nature state. The two states were separates by the area of transition state. The sharply decreased radius of gyration implied the system of knot protein had the initial collapse. The landscape of the free energy as a function of the number of native contact for the whole protein and the number of native contact in β-sheet (Fig. 8.8c) was showed the β-sheet formed first, subsequently the number of native contact increased until the protein folded into the native state. So the native contacts in β-sheet may promote the formation of compact structure. The free energy landscape was plotted as a function of the number of native contact and the relative contact order (RCO) parameter (Fig. 8.8d), which can be used to investigate more detail about the folding mechanism of this knot protein [39]. In the folding process of protein VirC2, the RCO increased with an increasing number of native contact. The change trend of RCO value coincided with the number of native contact. This implied that the local native contacts were formed in the initial stage of the folding process, and then the long-range native contacts were formed as an increasing number of native contact. In the Ca Gō-model, the β-sheet formed firstly, which promoted the compaction between N-terminal and the other parts of this knot protein. In this process, the local native contacts were important for the formation of β-sheet. After forming the native contacts in β-sheet, the knot protein needed to cross the transition state to fold into the native state. A typical folding process for this protein with all-atom Gō-model at T = 103 is shown in Fig. 8.9. In the folding process, the structure of knot protein formed the native contacts in β-sheet at the number of native contact of ~0.2. When the number of native contact reached ~0.5, the protein entered a state with a compact structure, but the C-terminal can not thread into the loop formed by α1, α2 and their linkers, it is likely that this process needed to adjust the conformation of this loop. When the number of native contact decreased to ~0.2 again, this loop was readjusted, and the orientation of this loop was changed. After this process the C-terminal could thread into this loop. The folding process for this trefoil knot protein was similar to the protein MJ0366. The formation of the native contacts in β-sheet was important for the whole protein, after forming the native contacts in β-sheet, the N-terminal could have chances to thread into the loop formed by α1, α2 and their linkers. At the last stage of the folding process, the C-terminal was prone to adopt slipknot motion to thread into this loop.
The folding routes of knot protein VirC2 from Ca Gō-model at folding temperature T = 146. a The free energy as a function of the number of native contact. b The free energy as a function of the number of native contact and radius of gyration. c Two-dimensional free energy landscape as a function of the number of native contact for the whole protein and the native contacts in β-sheet. d The free energy as a function of the number of native contact and RCO parameter. e Two-dimensional free energy landscape as a function of Ca-RMSD and the number of native contact in β-sheet
2.6 Transition State for Protein VirC2 in Gō-model
The free energy as a function of the number of native contact is shown in Fig. 8.8a. The transition state has the number of native contact of ~0.4. The native contact map of transition state for protein VirC2 in Ca Gō-model is shown in Fig. 8.10. In transition state, the native contacts in β-sheet were formed, some of the native contacts were formed between N-terminal and C-terminal, and between C-terminal and α2. Comparing with the native state, most of the native contacts have been formed for some structures in the transition state, which implied this knot may be formed at this stage. Hence, the protein VirC2 may be formed in the late transition state like the trefoil knot MJ0366. The free energy as a function of Ca-RMSD and the number of native contact in β-sheet (Fig. 8.8e) was presented a state with the number of native contacts of ~0.3 in the β-sheet. This state formed the native contacts in the β-sheet, and the C-terminal was loose like the intermediate state of protein MJ0366. So the knot protein VirC2 may have the intermediate state, in this state the loop was controlled by the β-sheet which needed to be readjusted, and then the protein could fold into the native structure.
3 Conclusions
We simulated two trefoil proteins with Gō-model, and high-temperature unfolding simulations was used for the study of trefoil protein MJ0366. The unfolding process of protein MJ0366 showed the contacts in β-sheet decreased firstly, and then the C-terminal of knot MJ0366 could thread out of the loop controlled by the contacts in β-sheet. In all-atom Gō-model, the native contacts in β-sheet promote the formation of a loop, and then the C-terminal threads into this loop to form the native state. The folding processes of the two trefoil knots were similar, and the formation of β-sheet was important for the two knot proteins. The C-terminal was prone to thread into the loop formed by secondary structures in correct size with slipknot motion, but when the loop was loose, the C-terminal was probably to thread into the loop with plug motion. In the intermediate state, the compact structure with the native contacts in the β-sheet was formed, but the C-terminal was loose. In transition state, the native contacts in β-sheet were formed, and the C-terminal was prone to thread into the loop.
4 Materials and Methods
High-temperature unfolding. The molecular dynamics simulations for protein MJ0366 were performed through using the software package GROMACS 4.0.7 with GROMOS force field [40]. The starting structure of protein MJ0366 was taken from the NMR structure of the Protein Data Bank. This protein had nine simulation trajectories at 530 K for 40 ns, and a molecular dynamics simulation in native state was performed under 298 K at neutral PH. For preparing the molecular dynamics simulations, the starting structure was solvated with SPC216 water, and then subjected to 20,000 steps of steepest descent minimization. The nearest distance between solute and box was 1.2 nm. Following the minimization, the whole system was subjected to 500,000 steps molecular dynamics simulations under NVT canonical ensemble and NPT constant pressure and constant temperature ensemble, respectively. The initial velocities were assigned from the Maxwellian distribution. The time step for these molecular dynamics simulations was 2 fs, and the neighboring list was updated every 5 steps. The transition states in the high-temperature unfolding process were determined by the conformational cluster method which was based on the Ca root-mean-square deviation (Ca RMSD) among the structures taken from the molecular dynamics simulation trajectories. For the nine simulation trajectories, the Ca RMSD values of the whole trajectory were used to generate positive definite matrix. The Michael Levitt’s projecting co-ordinate spaces method was used to project this positive definite matrix onto the best plane [41]. The structures in the last 5 ps of the first obvious cluster were regarded as the transition state.
All-Atom Model. The all-atom model has been described [42] and has an available web server [43]. In the all-atom model of protein, only the heavy atoms were included. The single bead with unit mass was used to represent each atom. The harmonic potentials were used to restrain the bond lengths, bond angles, improper dihedrals, and planar dihedrals. The attractive 6–12 interactions were used for the nonbonded atom pairs which formed the native contacts, and the repulsive interactions were given to the other nonlocal interactions. For the Ca coarse-grained protein model [44], the single bead was centered in the Ca position to represent each residue. The contact map was constructed by including all residue pairs that at least had one atom-atom contact between them. Here we used GROMACS 4.0.7 software package to perform the molecular dynamics simulations [40]. The constant temperature molecular dynamics simulations at folding temperature were used to get thermodynamics datas, and these datas were compiled through using weighted histogram analysis method [45].
Reaction Cordinates. We used QAA and QCA as the reaction coordinates. QAA is the fraction of native contact which is the probability of interactional atoms comparing with the native state. If any atom-atom interaction between two residues within 1.2 times the native distance σij are considered as the native contact. QCA is the fraction of native contact for the Ca coarse-grained model which includes the residue pairs whose Ca atoms within 1.2 times their native distance.
References
Virnau P, Mirny LA, Kardar M (2006) Intricate knots in proteins: function and evolution. PLoS Comput Biol 2:1074–1079
Taylor WR (2000) A deeply knotted protein structure and how it might fold. Nature 406:916–919
Taylor WR (2007) Protein knots and fold complexity: some new twists. Comput Biol Chem 31:151–162
Mansfield ML (1994) Are there knots in proteins? Nat Struct Biol 1:213–214
Bölinger D, Sułkowska JI, Hsu HP, Mirny LA, Kardar M, Onuchic JN, Virnau P (2010) A Stevedore’s protein knot. PLoS Comput Biol 6:e1000731
King NP, Yeates EO, Yeates TO (2007) Identification of rare slipknots in proteins and their implications for stability and folding. J Mol Biol 373:153–166
Gosavi S, Chavez LL, Jennings PA, Onuchic JN (2006) Topological frustration and the folding of interleukin-1 beta. J Mol Biol 357:986–996
Go N (1983) Theoretical studies of protein folding. Annu Rev Biophys Bioeng 12:183–210
Nymeyer H, Garcia AE, Onuchic JN (1998) Folding funnels and frustration in off-lattice minimalist protein landscapes. Proc Natl Acad Sci USA 95:5921–5928
Koga N, Takada S (2001) Roles of native topology and chain-length scaling in protein folding: a simulation study with a go-like model. J Mol Biol 313:171–180
Bryngelson JD, Wolynes PG (1987) Spin glasses and the statistical mechanics of protein folding. Proc Natl Acad Sci USA 84:7524–7528
Leopold PE, Montal M, Onuchic JN (1992) Protein folding funnels—a kinetic approach to the sequence structure relationship. Proc Natl Acad Sci USA 89:8721–8725
Onuchic JN, Wolynes PG (2004) Theory of protein folding. Curr Opin Struct Biol 14:70–75
Bayro MJ, Mukhopadhyay J, Swapna GV, Huang JY, Ma LC, Sineva E. Dawson PE, Montelione GT, Ebright RH (2003) Structure of antibacterial peptide microcin J25: a 21-residue lariat protoknot. J Am Chem Soc 125:12382–12383
Duff AP, Cohen AE, Ellis PJ, Kuchar JA, Langley DB, Shepard EM, Dooley DM, Freeman HC, Guss JM (2003) The crystal structure of Pichia pastoris lysyl oxidase. Biochemistry 42:15148–15157
Wagner JR, Brunzelle JS, Forest KT, Vierstra RD (2005) A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438:325–331
Vimau P, Mallam A, Jackson S (2011) Structures and folding pathways of topologically knotted proteins. J Phys: Condens Matter 23:1–17
Mallam AL, Jackson SE (2005) Folding studies on a knotted protein. J Mol Biol 346:1409–1421
Mallam AL, Jackson SE (2007) A comparison of the folding of two knotted proteins: YbeA and YibK. J Mol Biol 366:650–665
Mallam AL, Jackson SE (2006) Probing nature’s knots: the folding pathway of a knotted homodimeric protein. J Mol Biol 359:1420–1436
Mallam AL, Morris ER, Jackson SE (2008) Exploring knotting mechanisms in protein folding. Proc Natl Acad Sci USA 105:18740–18745
Ohta S, Alam MT, Arakawa H, Ikai A (2004) Origin of mechanical strength of bovine carbonic anhydrase studied by molecular dynamics simulation. Biophys J 87:4007–4020
Wallin S, Zeldovich KB, Shakhnovich EI (2007) The folding mechanics of a knotted protein. J Mol Biol 368:884–893
Sulkowska JI, Sulkowski P, Onuchic J (2009) Dodging the crisis of folding proteins with knots. Proc Natl Acad Sci USA 106:3119–3124
Lu J, den Dulk-Ras A, Hooykaas PJ, Glover JN (2009) Agrobacterium tumefaciens VirC2 enhances T-DNA transfer and virulence through its C-terminal ribbon-helix-helix DNA-binding fold. Proc Natl Acad Sci U S A 106:9643–9648
Li J, Wei DQ, Wang JF, Yu ZT, Chou KC (2012) Molecular dynamics simulations of CYP2E1. Med Chem 8:208–221
Gu RX, Liu LA, Wei DQ, Du JG, Liu L, Liu H (2011) Free energy calculations on the two drug binding sites in the M2 proton channel. J Am Chem Soc 133:10817–10825
Wang Y, Wei DQ, Wang JF (2010) Molecular dynamics studies on T1 lipase: insight into a double-flap mechanism. J Chem Inf Model 50:875–878
Wang JF, Yan JY, Wei DQ, Chou KC (2009) Binding of CYP2C9 with diverse drugs and its implications for metabolic mechanism. Med Chem 5:263–270
Gong K, Li L, Wang JF, Cheng F, Wei DQ, Chou KC (2009) Binding mechanism of H5N1 influenza virus neuraminidase with ligands and its implication for drug design. Med Chem 5:242–249
Li L, Wei DQ, Wang JF, Chou KC (2007) Computational studies of the binding mechanism of calmodulin with chrysin. Biochem Biophys Res Commun 358:1102–1107
Arias HR, Gu RX, Feuerbach D, Guo BB, Ye Y, Wei DQ (2011) Novel positive allosteric modulators of the human α7 nicotinic acetylcholine receptor. Biochemistry 50:5263–5278
Zhang T, Liu LA, Lewis DF, Wei DQ (2011) Long-range effects of a peripheral mutation on the enzymatic activity of cytochrome P450 1A2. J Chem Inf Model 51:1336–1346
Wang JF, Gong K, Wei DQ, Li YX, Chou KC (2009) Molecular dynamics studies on the interactions of PTP1B with inhibitors: from the first phosphate-binding site to the second one. Protein Eng Des Sel 22:349–355
Lian P, Wei DQ, Wang JF, Chou KC (2011) An allosteric mechanism inferred from molecular dynamics simulations on phospholamban pentamer in lipid membranes. PLoS ONE 6:e18587
Li J, Wei DQ, Wang JF, Li YX (2011) A negative cooperativity mechanism of human CYP2E1 inferred from molecular dynamics simulations and free energy calculations. J Chem Inf Model 51:3217–3225
Kolesov G, Virnau P, Kardar M, Mirny LA (2007) Protein knot server: detection of knots in protein structures. Nucleic Acids Res 35:W425–W428
Noel JK, Sułkowska JI, Onuchic JN (2010) Slipknotting upon native-like loop formation in a trefoil knot protein. Proc Natl Acad Sci USA 107:15403–15408
Wallin S, Chan HS (2006) Conformational entropic barriers in topology-dependent protein folding: perspectives from a simple nativecentric polymer model. J Phys: Condens Matter 18:S307–S328
Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447
Levitt M (1983) Molecular dynamics of native protein. II. Analysis and nature of motion. J Mol Biol 168:621–657
Whitford PC, Noel JK, Gosavi S, Schug A, Sanbonmatsu KY, Onuchic JN (2009) An all-atom structure-based potential for proteins: bridging minimal models with all-atom empirical forcefields. Proteins 75:430–441
Noel JK, Whitford PC, Sanbonmatsu KY, Onuchic JN (2010) SMOG@ctbp: simplified deployment of structure-based models in GROMACS. Nucleic Acids Res. doi:10.1093/nar/gkq498
Clementi C, Nymeyer H, Onuchic JN (2000) Topological and energetic factors: what determines the structural details of the transition state ensemble and “on-route” intermediates for protein folding? An investigation for small globular proteins. J Mol Biol 298:937–953
Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM (1992) The weighted histogram analysis method for free-energy calculations on biomolecules I. The method. J Comput Chem 13:1011–1021
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Shanghai Jiaotong University Press, Shanghai and Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Wu, X. et al. (2015). Folding Mechanisms of Trefoil Knot Proteins Studied by Molecular Dynamics Simulations and Go-model. In: Wei, D., Xu, Q., Zhao, T., Dai, H. (eds) Advance in Structural Bioinformatics. Advances in Experimental Medicine and Biology, vol 827. Springer, Dordrecht. https://doi.org/10.1007/978-94-017-9245-5_8
Download citation
DOI: https://doi.org/10.1007/978-94-017-9245-5_8
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-017-9244-8
Online ISBN: 978-94-017-9245-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)