Abstract
The G-quadruplex DNA formed by the stack of guanines in human telomere sequence is a promising anticancer target. In this study we used the energy landscape theory to elucidate the folding mechanisms for the thrombin aptamer, Form 1 and Form 3 G-quadruplexes. The three G-quadruplexes were simulated with all-atom Gō-model. Results show that, the three G-quadruplexes fold through a two-state mechanism. In the initial stage of the folding process, the compact structures are formed. The G-quadruplexes need to form G-triplex structures on the basis of the compact structures before folding to the native states. The folding free energy barrier of Form 3 G-quadruplex is higher than thrombin aptamer and Form 1, which shows that the structure of Form 3 G-quadruplex has more stability than the other two G-quadruplexes.
Xue Wu, Ting Fu, Peijun Xu and Jinguang Wang have been contributed equally to this paper.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Telomere, a nucleoprotein complex, is located at the ends of linear eukaryotic chromosomes. It is important for maintaining the chromosomal stability and the integrity of the genome [1, 2]. In the process of the replication of eukaryotic chromosomes packed by the DNA, the ends of telomeric DNA cannot be copied by DNA polymerase, which is due to lack of a template strand in the extreme 3′ end of a DNA sequence [3, 4]. As a result, the 3′ end of telomeric DNA is eroded, and the telomeric DNA is shortened without compensation mechanism. Nevertheless, the telomeric DNA in the tumour cell is not shortened during the replication. The telomerase is important for maintaining the stability and integrity of the telomeric DNA in most of proliferating tumour cells [5]. It is interesting to study the telomere and telomerase because of the difference between the somatic and tumour cells for the maintenance of telomere. Human telomeric DNA is composed of thousands of tandem repeats of the guanine-rich sequences, and the 3′-end overhangs 100–200 nt [6]. The G-quadruplex structures can be built from the vertical stacking of planar G · G · G · G tetrads in the G-rich DNA sequences in vitro, and this structures have been found in the telomeric sequences and telomeres [7–11]. The activity of telomerase is inhibited by these structures, so the G-quadruplexes in human telomere sequences are the promising anticancer targets. It is meaningful to study these structures as the promising anticancer targets, and the research about the folding of nucleic acids is useful to understand the biological natures, so here we study the folding dynamics for DNA tetraplex.
To date, there have been reported various G-quadruplex structures [12, 13]. The NMR structure of intramolecular quadruplex formed by four repeats of d(TTAGGG) in the Na(+)-containing solution have been reported [9]. In this structure, three G-quartets are held together by strands in the alternating orientations, and two lateral loops and a central diagonal loop connect these G-quartets. Under approximately physiological ionic conditions, the G-quadruplex has a different conformation in present of the K+ solution comparing with the structure in Na+ solution. In the K+ solution, this crystal structure is consisted of all four parallel strands [12]. The thrombin aptamer sequence d(GGTTGGTGTGGTTGG) could form a G-quadruplex structure in the K+ solution, and this structure is composed of two guanine quartets connected by two T-T loops and a T-G-T loop [14]. The human telomeric DNA in physiologically was observed to form the (3 + 1) G-quadruplex topology (Form 1 and Form 2) in K+ solution, and this structure is consisted of three strands oriented in one direction and the fourth in the opposite direction [15, 16]. Both Form 1 and Form 2 contain one double-chain-reversal and two edgewise T-T-A loops, but the two structures differ in loop arrangement. Furthermore, a novel G-quadruplex fold (Form 3) has been found in K+ solution, and this structure is a basket-type G-quadruplex with two G-tetrad layers [17].
For better understanding the structure and biological nature, the investigation of the folding dynamics for G-quadruplex structures is needed. The stopped-flow mixing coupled with rapid wavelength scanning method was used to study the folding dynamics for G-quadruplexes [18]. The folding of G-quadruplex structures from the G-rich oligonucleotides may be via intermediates. Some reports about the G-quadruplexes were shown a triplex structure which was important for the formation of G-quadruplex [19, 20]. The theoretical research has been used to study the folding dynamics of G-quadruplexes. A thrombin-bind DNA aptamer was investigated by replica exchange molecular dynamics simulation method at the all-atom level for giving more insight into its fold in atomic detail [21]. Though the structure and the folding of G-quadruplexes have been investigated, how this molecule overcoming the energy barrier folds from the G-rich oligonucleotide to the correct G-quadruplex topology still needs to be investigated. Here we used a different approach from these studies to discuss the folding mechanisms of G-quadruplexes. The folding routes of a well-designed sequence with the reduced effect of the local trap may be determined by the shape of this molecule and the chain connectivity of its backbone [22]. So the structure-based model can capture the essential folding features through isolating the effect of topology and removing all non-native energetic trap [23–25]. In the folding process, the energy landscape directs the folding of protein from unfolded state to the native state, the pattern of contacts directs the diverse sizes and shapes of the free energy barriers, and the native contacts may be more favorable than the nonnative contacts [26–28]. The DNA molecules have been studied widely [29–31], and this structure-based model has been used for the folding of nucleic acid in theory [32–34]. So here we used the all-atom structure-based model to study the folding pathways of thrombin-bind DNA aptamer, Form 1 and Form 3 G-quadruplexes. The PDB entries for thrombin-bind DNA aptamer, Form 1 and Form 3 G-quadruplexes are 148d, 2jsm and 2kf8, respectively. The study for the folding of the three G-quadruplexes demonstrated that all the three G-quadruplexes had a two-state folding behavior. In the folding process, the thrombin aptamer formed the two T-T loops firstly, and then the two G-quartets stacked together by the native contacts between the two ends. The Form 1 and Form 3 G-quadruplexes needed to form the compact structures first, and then through forming the G-triplex structures to fold into the native states. The energy barrier of Form 3 was higher than the other two G-quadruplexes, which may explain the reason that the stability of Form 3 G-quadruplex is higher than the other two G-quadruplexes.
2 Results and Discussion
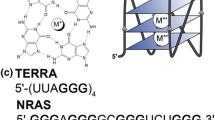
The thrombin aptamer is consisted of two stacked G-quartets (Fig. 10.1a). Two T-T loops at the two ends and one T-G-T loop link the two G-quartets. The guanines in the two G-quartets have the alternative glycoside orientations. The two G-quartets compose of G1syn · G6anti · G10syn · G15anti and G2anti · G5syn · G11anti · G14syn [14]. The Form 1, a (3 + 1) quadruplex,is consisted of one anti · syn · syn · syn and two syn · anti · anti · anti G-tetrads (Fig. 10.1b). This G-quadruplex has one narrow, one wide and two medium grooves, and the double-chain-reversal loop is located in a medium groove [16]. The Form 3 G-quadruplex structure has two layers of G-tetrads like the thrombin aptamer, but forms antiparallel-stranded basket-type structure (Fig. 10.1c). The glycosidic conformations of guanines in the two G-tetrads are G1syn · G14syn · G20anti · G8anti and G2anti · G15anti · G19syn · G7syn. Form 3 has one diagonal and two edgewise loops.
The secondary structures and sequences alignment for thrombin aptamer, Form 1 and Form 3 G-quadruplexes. a Thrombin aptamer ribbon diagram. b The ribbon diagram for Form 1 G-quadruplex. c Form 3 G-quadruplex ribbon diagram. d Sequences alignment for thrombin aptamer, Form 1 and Form 3 G-quadruplexes. These guanines form the G-quartets in the three G-quadruplexes marked in red
2.1 Transition States in All-Atom Gō-Model
Thrombin aptamer. The free energy as a function of the number of native contact at folding temperature is shown in Fig. 10.3a. There are two basins corresponding to the folded and denatured states. The two states are separated by a free energy barrier as the transition state near the number of native contact of ~0.5. The transition state is defined as the state that near the maximum free energy as a function of the number of native contact [35]. The temperature wasn’t defined as the usual Boltzmann constant but an arbitrary chosen constant in this study. The native contact maps for thrombin aptamer is shown in Fig. 10.2a. Comparing with the native state, the structures in the transition state have been formed some of the native contacts. In the transition state, the native contacts between G2 and G5, G1 and G6, G11 and G14, and G10 and G15 at the two ends were formed. Between G5 · G6 and G10 · G11, the native contacts G5-G11 and G6-G10 were appeared. In this state the native contacts in the T7-G8-T9 loop were formed. Some structures formed the native contacts G2-G14, G1-G15, and T4-T13 between the loops of the two ends, but these native contacts had low probability than the other native contacts in the two G-tetrads. So in the transition state part of structures formed most of the native contacts, but most of the structures only presented the native contacts G2-G5, G1-G6, G11-G14, and G10-G15 at the two ends. Before reaching to the native state, the G-triplex structures were found, which were formed by the native contacts G5-G11 and G6-G10 and the native contacts at the two ends.
2.2 Form 1 G-quadruplex
The free energy as a function of the number of native contact at folding temperature is shown in Fig. 10.4a. Two basins corresponding to the folded and denatured states are presented like the thrombin aptamer. The denatured states have the number of native contact of ~0.15, and the number of native contact in the folded state is near 0.65. The transition state with the number of native contact of ~0.35 separates the folded state and denatured state. The native contact maps of the structures in the native and transition states are shown in Fig. 10.2b. The contact map of the transition state had only two regions, so some of the native contacts weren’t formed. One of the regions in the contact map had the native contacts G3-G21 and G4-G22 at the two ends of Form 1 G-quadruplex. At one end of the G-quadruplex, the native contacts G17-G21, G16-G22, and G15-G23 were appeared at the transition state. In the transitions state, the Form 1 G-quadruplex folded into the compact structure through forming the native contacts between the two ends. The native contacts A2-T19 and A2-A20 stabilized this compact structure. The native contact T18-A20 also stabilized this structure. The G9-G10-G11 strand did not stack with the other three strands. So the G-triplex structures were formed in the transition state, and these structures included the formed native contacts G3-G21, G4-G22, G17-G21, G16-G22, and G15-G23.
2.3 Form 3 G-quadruplex
The free energy as a function of native contact at the folding temperature is shown in Fig. 10.5a. There are two basins corresponding to the folded and denatured states like thrombin aptamer and form 1 G-quadruplex. The folded state has the number of native contact of ~0.7, and the number of native contact in the denatured state is around 0.15. The transition state with the number of native contact of ~0.3 separates the folded and denatured states. The native contact maps of the folded and transition states for Form 3 G-quadruplex are shown in Fig. 10.2c. Comparing with the native state, the some of the native contacts were formed at the transition state. The native contacts G1-G14 and G2-G15 were appeared at the transition state. Few structures in the transition state had native contacts G1-G8 and G2-G7. The native contact between G3 and T5 was appeared in the transition state. This native contact may stabilize the loop at one end, and promote the formation of the native contacts G1-G14 and G2-G15. The native contact G3-A18 was formed in the loop at one end, but the high probability of this native contact did not promote the formation of native contacts G14-G20 and G15-G19. The structures were compact at transition state, and the native contacts T5-T17 and T5-A18 were good for the formation of these compact structures. The native contact between G9 and T11 was appeared, and this contact was benefit to the formation of native contacts G1-G14, G2-G15, G1-G8 and G2-G7. All the above, few structures in the transition state showed the G-triplex conformations formed by the native contacts G1-G14, G2-G15, G1-G8 and G2-G7, but most of the structures had native contacts G1-G14 and G2-G15 between the loops of the two ends.
2.4 The Folding Pathway of G-quadruplexes
Thrombin aptamer. The constant temperature simulations of thrombin aptamer were performed at folding temperature. The landscape for free energy as a function of the number of native contact and the Ca root-mean-square deviation (RMSD) is shown in Fig. 10.3b. In the folding process, the RMSD decreased as an increasing number of native contact, and the RMSD didn’t have the sharp change. The transition state with the native contact of ~0.5 separates the two folded and denatured regions. The free energy landscape as a function of the number of native contact and the radius of gyration is shown in Fig. 10.3c. The L-shaped landscape indicated that the radius of gyration decreased sharply in the initial stage of the folding process, when the structure reached the transition state with the number of native contact of ~0.5, the radius of gyration decreased little further. So in the initial stage of the folding process, the compact structures were formed. The Fig. 10.3d is shown for the free energy landscape as a function of the number of native contact and the distance between 5′-end and 3′-end. In the initial stage of the folding process, the distance between 5′-end and 3′-end decreased sharply, so the 5′-end and the 3′-end was in close distance, and this G-rich oligonucleotide has formed a compact structure. After transition state the number of native contact increased rapidly as the slowly decreasing distance between the 5′-end and the 3′-end, and the two ends was in short-distance as the native state. So the unfolded state were formed a compact structure first through the stack between the 5′ and 3′ ends. More details on the folding mechanism can be derived from the free energy landscape as a function of the number of native contact and the relative contact order (RCO) parameter (Fig. 10.3e). The relative contact order was defined as a function of the distance between two residues that formed native contact [36]. Two regions as the denatured and folded states were shown in this plot. The unfolded region had the RCO value of ~0.3, the number of native contact of the unfolded region was inferior to 0.4, and the folded region with the RCO of ~0.5 presented the number of native contact higher than 0.6. In the folding process, the RCO increased as the increasing number of native contact, so the local native contacts were important for the initial stage of the folding process. With the increasing number of native contact the non-local native contacts were increased, so the local native contacts may promote the stack of the local structures, and then stabilized the whole structure with non-local native contacts.
The typical structures for the folding process are shown in Fig. 10.6a. The two ends of the unfolded structure folded to the native state firstly, but the native contacts G5-G11 and G6-G10 were not formed. The native contacts in the region of loop T7-G8-T9 were formed in the denatured state, and were stable in the whole folding process. The native contacts in the two ends were not formed, but the compact structures were appeared through forming the native contacts in the T7-G8-T9 loop. So the local native contacts had the main contribution for the compact structures. After the native contacts at the two ends formed, the native contacts G5-G11 and G6-G10 were appeared, at this time the G-triplex was formed by the native contacts G1-G6, G2-G5, G6-G10, G5-G11, G11-G14, and G10-G15. The native contacts G2-G14 and G1-G15 were the last formed in the G-tetrads, and these native contacts made the two ends stack together. In the experimental study, two-state folding process was found, the two ends folded firstly, and G-triplex structures were needed for the formation of the native state [37]. This folding process studied by the all-atom Gō-model is consistent with the conclusion of the experimental research. In the folding process, the native contacts in the loops contributed to the formation of the G-tetrads. The free energy landscape as the function of the number of native contact in the two G-triplexes is shown in Fig. 10.3(F). The G1 and G2 represent the native contacts of the G-quartets G1 · G6 · G10 · G15 and G2 · G5 · G11 · G14, respectively. The number of native contact in G1 increased rapidly as an increasing number of native contact of G2 before folding to the native state, so the native contacts in G1 may be formed prior to G2. The local native contacts formed in these loops also promoted the formation of the compact structure, and the native contact T4-T13 between the loops at the two ends stabilized the compact structure. After the formation of G-triplex structure the two ends stacked together and folded to the native state.
2.5 Form 1 G-quadruplex
We used constant temperature simulations for Form 1 G-quadruplex at folding temperature. The free energy landscape as a function of the number of native contact and the RMSD is shown in Fig. 10.4b. The RMSD decreased sharply with slowly increasing the number of native contacts, and after the number of native contact reached ~0.4, the RMSD decreased little further. The transition area with the number of native contact of ~0.3 separates the two regions of folded and denatured states. The Fig. 10.4c is shown for the free energy as a function of the number of native contact and the radius of gyration. The L-shaped landscape showed the radius of gyration for the integral G-quadruplex was decreased rapidly with an increasing number of native contact, but once the number of native contact reached ~0.3 the radius of gyration decreased little further. In the initial stage of the folding process, the sharply decreased radius of gyration implied the unfolded G-quadruplex formed a compact structure, which was regarded as a basis for folding to the native state. The landscape of the free energy as a function of the number of native contact and the distance between 5′-end and 3′-end is shown in Fig. 10.4d. In this plot, the distance between 5′-end and 3′-end was decreased sharply in the initial stage of the folding process, and the closeness between the two ends made the unfolded state have a compact structure. After the formation of the compact structure with the number of native contact ~0.3 the distance between the two ends decreased little further. The transition state with the distance between 5′-end and 3′-end of ~2 nm separates the two regions of the folded and denatured states. The distance between the two ends in the folded state was lower than 2 nm, and the distance in denatured state was higher than 2.5 nm. More detailed about the folding mechanism can be derived from the free energy landscape of a function of the number of native contact and RCO parameter (Fig. 10.4e). In the folding process, the RCO increased with an increasing number of native contact, but this plot didn’t show the sharply increasing RCO. The transition state with the RCO of ~0.1 separates the folded and denatured states. The RCO in the folded state was higher than 0.2, and the number of native contact was higher than 0.6. The denatured state had the RCO of ~0.01 and the number of native contact ~0.2. In the denatured state, the RCO increased with an increasing the number of native contact, so the local native contacts had the main contribution for the formation of the compact structure in the initial stage of the folding process. After the compact structure formed, the RCO was increased because of the increasing non-local native contacts. Hence the local native contacts promoted the formation of the compact structure like thrombin aptamer G-quadruplex, and the non-local native contacts further contributed to the formation of the native structure.
The folding process with the typical structures is shown in Fig. 10.6b. The starting structure was unfolded. In the initial stage of the folding process a compact structure was formed. The native contacts in the 3′-end were formed firstly, and the native contacts G17-G21, G16-G20, and G15-G23 were appeared. The loop T18-T19-A20 had fewer native contacts in the denatured state, and the loops in the regions T6-T7-A8 and T12-T13-A14 were formed in the denatured state and stable in the folding process. The native contacts in the loop T18-T19-A20 were increased with the formation of the G-triplex at 3′-end. After forming the native contacts at the 3′-end, the native contacts between 5′-end and 3′-end were formed, and the compact structure was appeared. In the process of forming the compact structure, the local native contacts were important, which implied the native contacts in loop regions may have the main contribution for this structure. The formation of the native contacts in these loops drew the 5′-end and the 3′-end toward each other, so the two ends could have the chance to form the native contacts. The G1, G2 and G3 represent the three G-quartets G3 · G9 · G17 · G21, G4 · G10 · G16 · G22 and G5 · G11 · G15 · G23, respectively. The landscape of the number of native contact of G1 and G2 is shown in Fig. 10.4f, and the landscape for the number of native contact of G1 and G3 is shown in Fig. 10.4g. The number of native contact of G1 increased as the increasing number of native contact of G2, and this change trend is the same as the landscape of the number of native contact of G1 and G3. Hence, the three G-quartets may be formed in the same time. The compact structure had the native contacts at the two ends, and the strand of G9-G10-G11 didn’t stack with the other strands. The native contacts G3-G21, G4-G22, G17-G21, G16-G22, and G15-G23 formed a G-triplex structure, and then this structure folded to the native state through stacking the strand G9-G10-G11 with the other three strands.
2.6 Form 3 G-quadruplex
The constant temperature simulations at the folding temperature were used for Form 3 G-quadruplex. The free energy landscape as a function of the number of native contact and RMSD is shown in Fig. 10.5b. In the initial stage of the folding process, the RMSD decreased sharply as an increasing number of native contact, but once the number of native contact of ~0.3 was fromed the RMSD decreased little further. The transition state with the number of native contact of ~0.3 separates two regions of the folded and denatured states, and the folded state had the number of native contact of ~0.8. In Fig. 10.5c, the free energy landscape as a function of the number of native contact and radius of gyration of the whole molecule for Form 3 G-quadruplex is shown. The L-shaped landscape implied that the radius of gyration decreased rapidly with an increasing number of native contact, after the number of native contact reached ~0.3, the radius of gyration decreased little further. The transition state with the radius of gyration of ~1.2 nm separates the folded and denatured states in this landscape. Hence, the unfolded state of this G-quadruplex formed a compact structure in the initial stage of the folding process, and then through adjusting this conformation to fold into the native state. The landscape as a function of the number of native contact and the distance between 5′-end and 3′-end at the folding temperature is presented in Fig. 10.5d. In this L-shaped landscape, the distance between the two ends decreased sharply with an increasing number of native contact, after the number of native contact got up to ~0.3 the distance decreased little further. The transition state with the distance between the two ends of ~2 nm separates folded and denatured states in this L-shaped landscape. In the initial stage of the folding process, the 5′-end was closed to 3′-end for the formation of a compact structure. The Fig. 10.5e presents the free energy landscape as a function of the number of native contact and RCO parameter at the folding temperature. The RCO increased sharply with an increasing number of native contact until the number of native contact reached ~0.3 in the initial stage of the folding process. The transition state with the RCO of ~0.25 divided the two regions of folded and denatured states in this landscape. In the initial stage of the folding process, the compact structure was formed, so the local native contacts had the main contribution for this process. The RCO increased sharply in the denatured state, so the contribution for the formation of the compact structure can’t eliminate the non-local native contacts. These non-local native contacts may be mainly come from the native contacts between the 5′-end and one strand.
The snapshots for the folding process are shown in Fig. 10.6c. The starting structure was unfolded. The compact structure was formed in the initial stage of the folding process like Form 1 G-quadruplex. The free energy landscape as a function of the number of native contact of G1 (G1 · G8 · G20 · G14) and G2 (G2 · G7 · G19 · G15) is shown in Fig. 10.5f. The number of native contact of G1 increased as an increasing number of native contact of G2. So the two G-quartets may be formed in the same time. The native contacts in loop G9-T10-T11-A12-G13 were formed in the denatured state with high probability. The loops G3-T4-T5-A6 and T16-T17-A18 formed fewer native contacts comparing with the loop G9-T10-T11-A12-G13. The formed native contacts especially the native contacts in loop G9-T10-T11-A12-G13 contributed to the formation of the compact structure in the initial stage of the folding process. The native contacts G1-G14 and G2-G15 constituted the compact structure, and the native contacts in loop G9-T10-T11-A12-G13 could stabilize this structure. After forming the compact structure, the native contacts G1-G8 and G2-G7 were formed at the 5’-end. Hence, in this stage the G-triplex was formed by native contacts G1-G8, G2-G7, G1-G14 and G2-G15. The 3′-end of the Form 3 G-quadruplex was loose comparing with the other three strands. After the 3′-end stacking with the other three strands, the native structure was formed.
2.7 Comparing the Folding Mechanisms of Thrombin Aptamer, Form 1 and Form 3 G-quadruplexes
The thrombin aptamer has two G-quartets like the Form 3 G-quadruplex. The Form 1 G-quadruplex has three G-quartets and folds to the (3 + 1) G-quadruplex. The Form 3 G-quadruplex only has three different bases in the 5′ and 3′ ends comparing with Form 1 G-quadruplex but folds to the basket form. Though the structures of the three G-quadruplexes are obviously different, the folding processes for the three G-quadruplexes are similar. A compact structure formed firstly, and then the G-triplex structure was appeared for folding into the native state. The compact structure of the thrombin aptamer was formed by the native contacts in the two ends and the loop between the two ends. The compact structure was formed by the native contacts between the two ends for Form 1 G-quadruplex. The Form 3 G-quadruplex had the compact structure formed by the native contacts between one strand and the 5′-end. The sequences alignment for the three G-quadruplexes is shown in Fig. 10.1d. In the folding process of thrombin aptamer, the native contact between T4 and T13 stabilized the G-triplex, and comparing with the Form 3 G-quadruplex the native contact T5-T17 stabilized the loops at the two ends. The native contact G3-T5 in the G-triplex of Form 3 determined the loop, and the native contact between G9 and T11 gave the contribution for the formation of the loop. The native contacts G1-G14 and G2-G15 were formed in the G-triplex of Form 3, but these native contacts were formed at the last stage in the folding process of thrombin aptamer. The native contacts in loop T7-G8-T9 were formed and stable in denatured state, and the native contacts in the loops of the two ends were existent, so these loops were determined in the initial stage of the folding process of thrombin aptamer for defining the stacking way of G-quartets, which was different from Form 3. Some of the native contacts in loops G9-T10-T11-A12-G13 and T16-T17-A18 of Form 3 were formed in the denatured state, and most of the native contacts in these loops were formed in folded state. These native contacts made the 3′-end and one strand in a close distance, so the compact structure of Form 3 could be formed. Hence, the local native contacts in these loops have made a difference between thrombin aptamer and Form 3 for the stacking ways of G-quartets in the denature state. The native contacts in loop T18-T19-A20 of Form 1 G-quadruplex were appeared, and then these local native contacts promoted the formation of the native contacts in the 3′-end. The native contacts in loop G9-T10-T11-A12-G13 of Form 3 for stabilizing this region were higher than the number of the native contact in the corresponding loop T12-T13-A14 of Form 1, and these native contacts in Form 3 were formed and stable in the denatured state. The formed native contacts in the loops of Form 1 made the two ends stack together, however the stable native contacts in the loop G9-T10-T11-A12-G13 of Form 3 promoted the stack between the 3’-end and one strand of G-triplex. So the local native contacts formed in the denatured state may impact the folding pathways of G-quadruplexes. The native contacts T5-T17, G3-A18, G3-A6, and G9-G13 in Form 3 stabilized the G-triplex structure, and the Form 1 had the native contacts A2-T19 and A2-A20 to stabilize this G-triplex structure. The folding free energy barrier of Form 1 G-quadruplex was ~0.83 kbT, but the Form 3 G-quadruplex had the free energy barrier higher than 1.15 kbT. Hence, the Form 3 needs more free energy than the Form 1 for folding into the native state, and the Form 3 is more stable than the Form 1 G-quadruplex. This result is consistent with experimental studies. In experiment, the Form 3 G-quadruplex with basket-type fold had high structural stability, because of the base pairing and the stacking in the loops such as G21 · G9 · G13, T21 · T11, A6 · G3 · A18, and T5 · T17 [17]. Here we found the thrombin aptamer with free energy barrier of ~0.82 kbT as the Form 1 G-quadruplex may have lower stability than the Form 3.
3 Conclusions
We have simulated thrombin aptamer, form 1 and form 3 G-quadruplexes with all-atom Gō-model for studying the folding mechanisms for the three G-quadruplexes. The folding processes of the three G-quadruplexes are similar. The compact structures were formed in the initial stage of the folding process. The thrombin aptamer had the compact structure through forming the native contacts in the two ends and the medium loop. The native contacts in the loops of Form 1 had main contribution for the formation of the compact structure. The Form 3 had the native contacts formed between the 5′-end and one strand in order to obtain the compact structure. The G-triplex structures were formed before folding to the native states of the three G-quadruplexes. The G-triplex of thrombin aptamer is consisted of the native contacts G5-G11, G6-G10, G2-G5, G1-G6, G11-G14, and G10-G15. The G-triplex of Form 1 is composed of the native contacts G3-G21, G4-G22, G17-G21, G16-G22, and G15-G23. The G-triplex comprises the native contacts G1-G14, G2-G15, G1-G8 and G2-G7 in Form 3 G-quadruplexes. The Form 3 has higher free energy barrier than the other two G-quadruplex structures, and this structure has more structural stability.
4 Materials and Methods
All-Atom Gō-model. The all-atom Gō-model was described previously [34]. This model is available on a web server [38]. In the all-atom Gō-model, all heavy (non-hydrogen) atoms are explicitly included. A single bead of unit mass represents each atom. The harmonic potentials were used for restraining the bond length and angles, and planar dihedrals. The non-bonded atom pairs that are in contact in the native state, are given attractive 6–12 interactions. Nevertheless, all the other non-local interactions are repulsive. Gromacs 4.0.7 software package was used for all simulations [39]. The simulations were started from the unfolded structures. For obtaining the thermodynamic sampling, more than 30 simulation trajectories were performed for the three G-quadruplexes at folding temperature. The Weighted Histogram Analysis Method was used for the calculation of the thermodynamic quantities [40].
Reaction Coordinates. We used the fraction of native residues in contact as the reaction coordinate Q. A native contact is defined as any two atoms in different residues that are within 4 Å of each other and separated by at least 3 bonds [34]. A contact between two atoms is formed if this pair distance is within the 1.2 times their native distance.
References
De Cian A, Lacroix L, Douarre C, Temime-Smaali N, Trentesaux C, Riou JF, Mergny JL (2008) Targeting telomeres and telomerase. Biochimie 90:131–155
Neidle S, Parkinson GN (2003) The structure of telomeric DNA. Curr Opin Struct Biol 13:275–283
Bryan TM, Cech TR (1999) Telomerase and the maintenance of chromosome ends. Curr Opin Cell Biol 11:318–324
Lian P, Liu LA, Shi Y, Bu Y, Wei D (2010) Tethered-hopping model for protein-DNA binding and unbinding based on Sox2-Oct1-Hoxb1 ternary complex simulations. Biophys J 98:1285–1293
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100:57–70
Makarov VL, Hirose Y, Langmore JP (1997) Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88:657–666
Gellert MN, Lipsett MN, Davies DR (1962) Helix formation by guanylic acid. Proc Natl Acad Sci USA 48:2013–2018
Davis JT (2004) G-quartets 40 years later: from 50-GMP to molecular biology and supramolecular chemistry. Angew Chem Int Ed Engl 43:668–698
Wang Y, Patel DJ (1993) Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure 1:263–282
Schaffitzel DL, Berger I, Postberg J, Hanes J, Lipps HJ, Plückthun A (2001) In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci USA 98:8572–8577
Maizels N (2006) Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat Struct Mol Biol 13:1055–1059
Parkinson GN, Lee MP, Neidle S (2002) Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 417:876–880
Phillips K, Dauter Z, Murchie AI, Lilley DM, Luisi B (1997) The crystal structure of a parallel-stranded guanine tetraplex at 0.95 Å resolution. J Mol Biol 273:171–182
Schultze P, Macaya RF, Feigon J (1994) Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG). J Mol Biol 235:1532–1547
Phan AT, Kuryavyi V, Luu KN, Patel DJ (2007) Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res 35:6517–6525
Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ (2006) Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J Am Chem Soc 128:9963–9970
Lim KW, Amrane S, Bouaziz S, Xu W, Mu Y, Patel DJ, Luu KN, Phan AT (2009) Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J Am Chem Soc 131:4301–4309
Gray RD, Chaires JB (2008) Kinetics and mechanism of K+- and Na+-induced folding of models of human telomeric DNA into G-quadruplex structures. Nucleic Acids Res 36:4191–4203
Xu Y, Sato H, Sannohe Y, Shinohara K, Sugiyama H (2008) Stable lariat formation based on a G-quadruplex scaffold. J Am Chem Soc 130:16470–16471
Mashimo T, Yagi H, Sannohe Y, Rajendran A, Sugiyama H (2010) Folding pathways of human telomeric type-1 and type-2 G-quadruplex structures. J Am Chem Soc 132:14910–14918
Kim E, Yang C, Pak Y (2012) Free-energy landscape of a thrombin-binding DNA aptamer in aqueous environment. J Chem Theory Comput 8:4845–4851
Gosavi S, Chavez LL, Jennings PA, Onuchic JN (2006) Topological frustration and the folding of interleukin-1 beta. J Mol Biol 357:986–996
Go N (1983) Theoretical studies of protein folding. Annu Rev Biophys Bioeng 12:183–210
Nymeyer H, Garcia AE, Onuchic JN (1998) Folding funnels and frustration in off-lattice minimalist protein landscapes. Proc Natl Acad Sci USA 95:5921–5928
Koga N, Takada S (2001) Roles of native topology and chain-length scaling in protein folding: a simulation study with a go-like model. J Mol Biol 313:171–180
Bryngelson JD, Wolynes PG (1987) Spin glasses and the statistical mechanics of protein folding. Proc Natl Acad Sci USA 84:7524–7528
Leopold PE, Montal M, Onuchic JN (1992) Protein folding funnels—a kinetic approach to the sequence structure relationship. Proc Natl Acad Sci USA 89:8721–8725
Onuchic JN, Wolynes PG (2004) Theory of protein folding. Curr Opin Struct Biol 14:70–75
Li L, Chen Q, Wei DQ (2012) Prediction and functional analysis of single nucleotide polymorphisms. Curr Drug Metab 13:1012–1023
Wei DQ (2012) New drug design based on multi-targets and system biology approach in light of real time DNA sequencing technologies. Curr Top Med Chem 12:1309
Xiong Y, Liu J, Wei DQ (2011) An accurate feature-based method for identifying DNA-binding residues on protein surfaces. Proteins 79:509–517
Sorin EJ, Nakatani BJ, Rhee YM, Jayachandran G, Vishal V, Pande VS (2004) Does native state topology determine the RNA folding mechanism? J Mol Biol 337:789–797
Hyeon C, Dima RI, Thirumalai D (2006) Pathways and kinetic barriers in mechanical unfolding and refolding of RNA and proteins. Structure 14:1633–1645
Whitford PC, Schug A, Saunders J, Hennelly SP, Onuchic JN, Sanbonmatsu KY (2009) Nonlocal helix formation is key to understanding S-adenosylmethionine-1 riboswitch function. Biophysical J 96:L7–L9
Kouza M, Hansmann UH (2012) Folding simulations of the A and B domains of protein G. J Phys Chem B 116:6645–6653
Plaxco KW, Simon KT, Baker D (1998) Contact order, transition state placement and the refolding rates of single domain proteins. J Mol Biol 277:985–994
Mao XA, Gmeiner WH (2005) NMR study of the folding-unfolding mechanism for the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG). Biophys Chem 113:155–160
Noel JK, Whitford PC, Sanbonmatsu KY, Onuchic JN (2010) SMOG@ctbp: simplified deployment of structure-based models in GROMACS. Nucleic Acids Res doi:10.1093/nar/gkq498
Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447
Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM (1992) The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J Comput Chem 13:1011–1021
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Shanghai Jiaotong University Press, Shanghai and Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Wu, X. et al. (2015). Theoretical Studies on the Folding Mechanisms for Different DNA G-quadruplexes. In: Wei, D., Xu, Q., Zhao, T., Dai, H. (eds) Advance in Structural Bioinformatics. Advances in Experimental Medicine and Biology, vol 827. Springer, Dordrecht. https://doi.org/10.1007/978-94-017-9245-5_10
Download citation
DOI: https://doi.org/10.1007/978-94-017-9245-5_10
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-017-9244-8
Online ISBN: 978-94-017-9245-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)