Abstract
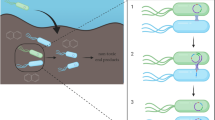
Genes encoding biodegradation of oil hydrocarbons are often localized on large conjugative plasmids. Transmissibility of catabolic plasmids ensures the dissemination of biodegradation features within indigenous microbial populations. Horizontal gene transfer mediated by plasmids plays an important role in the adaptation of bacteria to polluted environments and can be useful for genetic bioaugmentation.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Environmental pollution by petroleum and the resulting ecological damage are a growing worldwide problem. A variety of environmental cleanup systems have been developed and utilized to try to provide environmental remediation and protection from recalcitrant organic pollutants. Bioremediation provides cost-effective and contaminant-specific treatments. Biodegradation is the main process that results in removing environmental pollutants and is carried out by microorganisms, predominantly bacteria. The capability of microorganisms to transform or degrade oil hydrocarbons is well documented and allows them to be used for the bioremediation of polluted environments. Great potential for developing new bioremediation technologies, and improving existing ones, lies in improving our understanding of the metabolism and genetic control of biodegradation processes, especially involvement of catabolic plasmids and ecology of microorganisms capable of degrading oil and oil products.
Mechanisms of bioremediation include biostimulation or bioaugmentation. Biostimulation is the addition of nutrient amendments to soil and/or groundwater to stimulate indigenous microorganisms capable of degrading environmental pollutants and consequently biodegradation. If biodegradative activity of indigenous microflora is not sufficient or absent, bacteria with degradation capacity can be introduced in situ in the soil of the site. It is particularly promising for heavy fractions of oil which are hard to degrade by microbes due to low bioavailability. Bioaugmentation is the addition of specifically selected pre-grown microorganisms to degrade contaminants. The enhancement of the microbial population present at a site subsequently improves contaminant cleanup through ensuring the presence of sufficient quantities of microorganisms in the soil to complete biodegradation and additionally reduces cleanup costs and time.
2 Degrader Microorganisms and Catabolic Plasmids
Crude oil is a complex mixture of hydrocarbons and other chemicals. The composition varies widely depending on where and how the oil was formed. The hydrocarbons in crude oil are mostly alkanes, cycloalkanes, and various aromatic hydrocarbons, while the other organic compounds contain nitrogen, oxygen, and sulfur and trace amounts of metals such as iron, nickel, copper, and vanadium.
A large number of bacteria with oil hydrocarbon-degrading capabilities have been reported; among them are bacteria from different genera such as Pseudomonas, Sphingomonas, Burkholderia, Mycobacterium, Corynebacterium, Aeromonas, Rhodococcus, and Bacillus. Bacteria belonging to the genus Pseudomonas are noted possessing great metabolic potential and are able to acquire new capabilities during adaptation to specific conditions such as polluted environment.
Bacteria can degrade the following petroleum or hydrocarbon products with relative ease: gasoline, diesel, and fuel oil; hazardous crude oil compounds, such as benzene, toluene, and xylene; polycyclic aromatic hydrocarbons, such as naphthalene, phenanthrene, and pyrene; and alkanes.
Some chemicals are only partially degradable, or sometimes wastes are so mixed and variable that they degrade at different rates and may leave some toxic chemicals behind.
Degradation of different pollutants is often controlled by plasmids. Bacterial plasmids are mobile genetic elements able to replicate autonomously. Catabolic (degradative) plasmids confer their bacterial host’s ability to degrade some unusual substrates such as oil hydrocarbons or synthetic compounds. The first reports of catabolic plasmids were made in the 1970s. To date a range of plasmids, catabolic transposons, and gene clusters have been identified which encode the degradation of naturally occurring and synthetic pollutants.
The generally accepted classification of plasmids is based on incompatibility groups. In 1971, Hedges and Datta proposed a plasmid classification scheme based on the instability of identical or related plasmids in one bacterial cell during growth, a phenomenon called incompatibility. Two plasmids are incompatible if they cannot coexist in the same bacterial cell simultaneously or if either is less stable in the presence of the other than it was by itself. Incompatibility group is determined by the genetic information specified by plasmid DNA, more specifically by their replication/partitioning functions. Generally, closely related plasmids are incompatible. Incompatibility grouping had been used to group plasmid of Pseudomonas species (Jacoby 1977) and the Enterobacteriaceae (Couturier et al. 1988). At present there are 14 listed plasmid incompatibility groups in Pseudomonas (Boronin 1992) and about 30 incompatibility groups in Enterobacteriaceae.
One of the fundamental properties of bacterial plasmids is their host range that is primarily determined by the plasmid replication system. A plasmid’s host range is defined as the range of bacterial hosts in which the plasmid can replicate. Usually this range differs from the range of hosts into which plasmid can transfer by conjugation and the range of hosts in which plasmid is able to maintain stability without selective pressure. Most plasmids have a narrow host range (NHR) allowing only intraspecies transfer and replication. These plasmids are sometimes called “specialist.” However, some plasmids have a broad host range (BHR) and can therefore maintain in many species of bacteria. BHR plasmids (“generalist”) may be either self-transmissible (conjugative) or not self-transmissible but mobilizable (Sota and Top 2008). It seems that BHR plasmids are most important for horizontal gene transfer (HGT) between distantly related bacterial hosts. Plasmids can evolve to expand or at least shift their host range. High frequency of conjugal transfer of BHR plasmids belonging to IncP-1 group within an isogenic population accelerates the plasmids’ adaptation and stability in new bacterial hosts which initially were unfavorable for these plasmids (Heuer et al. 2007).
Among the plasmids encoding degradation of naturally occurring organic molecules, IncP-9, IncP-7, and IncP-2 are dominant, whereas IncP-1 plasmids are prevalent among those determining degradation of man-made compounds (Top et al. 2000). There is at least one exception – CAP plasmids which determine degradation of ε-caprolactam and belong to P-2, P-7, and P-9 incompatibility groups. ε-Caprolactam, a man-made compound, is used as raw stuff to produce polymer materials (polycaproamide, nylon 6) for industry, agriculture, medicine, and household activities (Esikova et al. 1990). However, CAP plasmids are beyond the scope of this chapter.
Catabolic plasmids are large and often contain the full set of conjugal transfer genes as well as all genes organized in catabolic operons for biodegradation of various hydrocarbons. Transmissibility of such plasmids ensures the dissemination of biodegradation features within and between microbial populations and influences the process of bacterial microevolution. When hydrocarbon-degrading bacteria, containing appropriate plasmids, are introduced to polluted soils, these microorganisms not only degrade the contaminant but potentially also serve as plasmid donors for indigenous microorganisms. Degradative plasmids encode utilization of aliphatic compounds (octane, decane, hexadecane, etc.) and aromatic (phenol, toluene, xylene) and polycyclic aromatic hydrocarbons (naphthalene, phenanthrene, pyrene, etc.).
2.1 The OCT Plasmids
n-Octane is a common component of gasoline and other petroleum products. Many microorganisms, including genus Pseudomonas, are able to use linear alkanes as their sole source of carbon and energy (van Beilen et al. 1994).
The genetics and biochemical pathway of alkane metabolism have been well investigated for Pseudomonas putida (oleovorans), which is able to oxidize C5–C12 n-alkanes. Genes encoding biodegradation of octane are localized on the OCT plasmid and called alk genes. The naturally occurring OCT plasmid (IncP-2 group) consists of three distinct plasmids, namely, OCT plasmid responsible for octane utilization, MER plasmid encoding mercury resistance, and transfer plasmid (factor K). Initial OCT plasmid is conjugative; however, after dissociation into three distinct plasmids, OCT is non-conjugative but is mobilizable.
The OCT plasmid of Pseudomonas oleovorans contains two operons, alkBFGHJKL and alkST, which encode all proteins necessary for the degradation of n-octane and other 5- to 12-carbon linear alkanes to the corresponding acyl-CoA derivatives. Both clusters of the genes (alkBFGHJKL and alkST) encoding the P. putida GPo1 alkane degradation pathway are regulated by the AlkS protein. When no alkanes are present, alkS is expressed at low levels. The first catabolic operon of OCT plasmid encodes seven proteins, of which at least three are involved in alkane hydroxylation and alkanol dehydrogenation; the final product of this pathway, octanoyl-CoA, enters the beta-oxidation cycle and can be utilized as carbon and energy sources (van Beilen et al. 1994). Growth on alkanes requires a functional chromosomally encoded fatty acid degradation system in addition to the plasmid-borne alk system; such a system usually is active in P. putida. The nucleotide composition of the alk genes (47 % G + C) differs considerably from the G + C content of the P. oleovorans genome, suggesting that the alk regulon may originate from an unrelated organism. It has been shown that OCT plasmid encodes, among others, a methyl-accepting transducer protein (AlkN) that may be involved in chemotaxis to alkanes (van Beilen et al. 2001).
Two other plasmid-located genetic systems for alkane utilization have been partially characterized. Pseudomonas maltophilia N246-1 bears large IncP-2 plasmid allowing the host strain to grow on C8–C14 of n-alkanes. The OCT plasmid can transfer to Rhodopseudomonas sphaeroides with a low frequency (Lee et al. 1993). The ability of Pseudomonas sp. strain C12B to utilize medium-chain-length n-alkanes (C9–C12) is encoded by plasmid pDEC. The enzyme system encoded by the putative dec genes present on plasmid pDEC differs from the system coded by the alk genes of plasmid OCT in the size range of hydrocarbons preferentially used (Kostal et al. 1998).
2.2 Plasmids Involved in Phenol Biodegradation (PHE Plasmids)
A common pathway for metabolizing an aromatic compound like phenol is to dihydroxylate the benzene ring to form a catechol. Catechol is oxidized via ortho pathway (intradiol cleavage) by catechol 1,2-dioxygenase or via meta pathway (extradiol cleavage) by catechol 2,3-dioxygenase. Further degradation is realized by distinct sets of enzymes. The final products of both the pathways are molecules that can enter the tricarboxylic acid cycle. Dihydroxylation is carried out by multi- or single-component phenol hydroxylases (Kivisaar et al. 1990).
The catabolic plasmid pVI150 of Pseudomonas sp. strain CF600 encodes all the genetic information required for the metabolism of phenol and some of its methyl-substituted derivatives as sole carbon and energy sources. The plasmid is very large and conjugative and belongs to the incompatibility group P-2. Biochemical pathway for the dissimilation of phenolic compounds involves a multicomponent phenol hydroxylase and subsequent meta cleavage pathway. These enzymes are encoded by 15 dmp structural genes in a single operon of pVI150. The operon is regulated by dmpR gene product belonging to the NtrC family of transcriptional activators that regulate transcription from -24/-12 promoters (Shingler et al. 1993).
The naturally occurring plasmid pPGH1 from Pseudomonas putida H also encodes inducible degradation of phenol and some of its methylated derivatives via the meta cleavage pathway (Herrmann et al. 1995). The plasmid DNA region about 16 kb contains all genetic information necessary for inducible degradation of phenolic compounds. Degradative genes are organized into a single operon encoding for a multicomponent phenol hydroxylase and meta cleavage pathway enzymes. Catabolic operon is subject to positive control by the product of regulatory gene.
Plasmids carrying genes for phenol degradation (phe genes) through catechol ortho cleavage pathway obtained as a result of long-term cultivation of the Pseudomonas putida multiplasmid strain EST1020 on phenol have been characterized (Kivisaar et al. 1990). PHE plasmids determine phenol monooxygenase (gene pheA) and catechol 1,2-dioxygenase (gene pheB), the key enzyme for ortho pathway. Sequencing of pheB has shown the relationship between the pheB gene and other C12O-encoding genes. The comparison of the pheB sequence with sequences of catA of Alcaligenes calcoaceticus, tfdC of A. eutrophus, and clcA of P. putida demonstrated that there are conserved residues in all the four protein products of these genes (Kivisaar et al. 1991).
Pseudomonas putida PaW85, carrying recombinant plasmid pAT1140 that contains the inducible pheBA operon, is able to degrade phenol using hybrid plasmid–chromosome-encoded pathway. The synthesis of the plasmid-encoded phenol monooxygenase and catechol 1,2-dioxygenase is induced by cis,cis-muconate. The pheBA operon is positively controlled by the regulatory protein CatR that is chromosomally encoded in P. putida (Kasak et al. 1993).
The bacterial strain Pseudomonas putida PhCN resistant to heavy metals has genetic systems for biodegradation of phenol. The strain contains two plasmids, a 120 kb catabolic plasmid that encodes the degradation of phenol (pPhCN1) and pPhCN2 plasmid (100 kb) that codes for cadmium and copper resistance (El-Deeb 2009).
The analysis of nine oil-degrading strains isolated from contaminated soils in Western Siberia able to grow on phenol as sole carbon and energy sources reveled that at least for two strains, degradation of phenol is encoded by conjugative plasmids about 100 kb in size (Makarenko et al. 2002).
2.3 The TOL Plasmids
The ability to degrade toluene or BTEX compounds (benzene, toluene, ethylbenzene, xylene) is common among soil microorganisms such as Pseudomonas. Therefore, biochemical pathways and the genetic control of toluene degradation are well characterized. At least five different pathways are known for aerobic catabolism of toluene. The pathway for the catabolism of toluene and some substituted toluenes via meta cleavage of catechol utilization is very often encoded by plasmids collectively called TOL plasmids. The archetype TOL plasmid pWW0 was first described in 1974 (Williams and Murray 1974). Plasmid pWW0 was assigned to the P-9 incompatibility group. The xyl genes of Pseudomonas putida TOL plasmid encoding degradation of toluenes and xylenes are organized in four transcriptional units: the upper operon xylUWCAMBN for conversion of toluenes/xylenes into benzoates/alkylbenzoates; the meta operon xylXYZLTEGFJQKIH, which encodes the enzymes for further conversion of these compounds into Krebs cycle intermediates; and xylS and xylR, which are involved in transcriptional control. The XylS and XylR proteins are members of the XylS/AraC and NtrC families, respectively, of transcriptional regulators. The xylS gene is constitutively expressed at a low level from the Ps2 promoter. The XylS protein is activated by interaction with alkylbenzoates, and this active form stimulates transcription from Pm. The xylR gene is also expressed constitutively. The XylR protein, which in the absence of effectors binds in a non-active form to target DNA sequences, is activated by aromatic hydrocarbons and ATP; it subsequently undergoes multimerization and structural changes that result in stimulation of transcription from Pu of the upper operon. Once activated, the XylR protein also stimulates transcription from the Ps1 promoter of xylS without interfering with expression from Ps2 (Ramos et al. 1997). The pWW0 plasmid’s degradative genes are located within class II (Tn3-like) catabolic transposons Tn4651 (56 kb) and Tn4653 (70 kb), and the latter transposon includes the former (Tsuda et al. 1989). The analysis of the complete pWW0 nucleotide sequence revealed 148 putative open reading frames. Of these, 77 showed similarity to predicting functions for plasmid replication, stable maintenance, conjugal transfer, catabolic determinants, and transposition (Greated et al. 2002). All identifiable transposition functions are located within the boundaries of transposon Tn4653, leaving a 46 kb region containing the IncP-9 plasmid’s core functions. Thus, the sequence of pWW0 and its comparison to others suggest that the current structure can be derived from a single original insertion of the Tn4563 transposon into IncP-9 ancestor plasmid. To the author’s opinion, the complexity and size of pWW0 are largely the result of the mosaic organization of the transposable elements that it carries, rather than the backbone functions of IncP-9 plasmids.
Investigations of many environmental Pseudomonas isolates capable of growing on toluene, m-xylene, and m-toluate have shown that TOL-like plasmids are widely distributed in the environment. A number of TOL plasmids which carry xyl genes that are strongly homologous to those in pWW0 have been described. The plasmids differ in size, incompatibility, and ability to conjugal transfer. The plasmid pWW53 isolated from Pseudomonas putida MT53 is a 107 kb nontransmissible plasmid that does not belong to the P-9 incompatibility group. This plasmid carries a single upper pathway operon, two highly homologous but distinguishable meta operons, a single xylR gene, and three xylS-homologous genes (xylS1, xylS2, and xylS3) (Keil et al. 1985). Thus, the naturally occurring TOL plasmid may carry genes for two meta cleavage dioxygenases. The second homologous gene for catechol 2,3-dioxygenase also is present on naturally occurring TOL plasmids pWW5, pWW74, and pWW88. Other plasmids, pWW14 and pWW84, carried the second but nonhomologous C23O gene C23OII (Chatfield and Williams 1986).
The TOL plasmid pDK1 (129 kb in size) isolated from Pseudomonas putida HS1 and belonging to IncP-7 group carries two homologous but nonidentical copies of xylS regulatory genes. The comparison of the organization of the xyl catabolic operons on pDK1 and pWW53 indicates that the catabolic region on pDK1 was derived from a replicon on which the xyl genes are organized similarly to pWW53 and that a genetic rearrangement has taken place involving a reciprocal recombination internal to two of its xylS homologues (Assinder et al. 1993). Complete nucleotide sequence of pDK1 and comparative analysis revealed that toluene catabolic gene clusters of this plasmid were derived through homologous recombination, transposition, and site-specific recombination from the xyl gene clusters homologous to another TOL plasmid, pWW53. Moreover, recipient host range of conjugal transfer of pDK1is limited to only two Pseudomonas strains in spite of the fact that the mini-replicon of pDK1 is maintained in at least six Pseudomonas strains. These results indicate that IncP-7 plasmids have narrow host range and that IncP-7-specified conjugal transfer was narrower than that of its replication (Yano et al. 2010).
The majority of pSVS plasmids in P. putida strains isolated from different contaminated sites in Belarus carry xyl genes homologous to those of pWW53 and are organized in a similar manner (Sentchilo et al. 2000). These plasmids contain two distinguishable meta operons, one upper pathway operon and three xylS-homologous regions. Two other pSVS plasmids carry one upper operon, one meta operon, and one copy of each regulatory gene.
Pseudomonas putida O2C2, isolated from oil-contaminated soil in the Netherlands, is able to grow on toluenes and methyl-substituted toluenes and contains the large plasmid pWW102 (the size is between 220 and 270 kb). The relative location of xyl operons differs from the organization of those in archetype plasmid pWW0: the upper pathway operon xylUWCAMBN being located downstream of the meta operon xylXYZLTEGFJQKIH, the two regulatory genes xylS and xylR are found immediately downstream of the meta pathway operon (Aemprapa and Williams 1998).
2.4 The NAH Plasmids
Naphthalene and its substituted derivatives are commonly found in crude oil and oil products. Naphthalene-degrading microorganisms are widely distributed in nature and are easy to isolate from different coal tar- and oil-contaminated soils. Among taxonomic groups of microorganisms able to degrade polycyclic aromatic hydrocarbons (PAH), a high proportion of the isolates belong to Pseudomonas, Shingomonas, and Burkholderia strains. The key intermediate of naphthalene biodegradation is salicylate, which is further catabolized via catechol or gentisate by catechol 2,3-dioxygenase (meta pathway) or catechol 1,2-dioxygenase (ortho pathway) and gentisate 1,2-dioxygenase, respectively.
Naphthalene biodegradation by Pseudomonas spp. is often determined by large conjugative plasmids. Plasmids carrying naphthalene catabolic genes represent a class of well-characterized degradation plasmids, known collectively as NAH plasmids. The plasmids NAH7 from Pseudomonas putida G7 and pDTG1 from P. putida NCIB 9816 are the best-studied naphthalene catabolic plasmids. The nah genes for naphthalene catabolism are organized in the upper pathway operon (nahAaAbAcAdBFCQED) which controls initial oxidation and subsequent degradation to salicylate (nah operon) and the lower operon (nahGTHINLOMKJ) for salicylate oxidation and further catechol meta cleavage pathway to tricarboxylic acid cycle intermediates (sal operon). Downstream of the nahJ gene of P. putida strain G7, two additional genes coding for an unknown function (nahX) and chemotaxis toward naphthalene transducer protein (nahY) were found (Habe and Omori 2003). A methyl-accepting chemotaxis protein, NahY, is cotranscribed with the degradation genes. In both strains a second operon encodes the enzymes for catabolism of salicylate to central metabolites via catechol and extradiol cleavage by catechol 2,3-dioxygenase. Both nah and sal operons are activated by a trans-acting positive regulator encoded by nahR gene and commonly are induced by a naphthalene intermediate – salicylate. The product of nahR gene, NahR is a member of the LysR family of transcriptional activators that are widely distributed in bacteria. Positive regulatory protein NahR binds to the promoters of nah and sal operons and activated their transcription after interaction with salicylate (Shell 1990). NahR proteins from different naphthalene-degrading P. putida strains exhibit a highly conserved helix–turn–helix motif and a putative enhancer-binding region in the N-terminal domain. Gene activation by NahR is consistent with the general transcriptional mechanism of class I transcription factors, by protein–protein interactions between alphaRNAP and NahR (Park et al. 2002).
Plasmids NAH7 and pDTG1 are conjugative and 83 kb in size and belong to incompatibility group P-9. Naphthalene catabolic genes on the NAH7 plasmid are located within a 37.5 kb region, which is the defective class II transposon, Tn4655.
The complete sequence of naphthalene-degrading 83 kb plasmid pDTG1 from P. putida strain NCIB 9816-4 has revealed that the upper and lower naphthalene-degrading operons occupy 9.5 kb and 13.4 kb, respectively (Dennis and Zylstra 2004).
The complete nucleotide sequence of 102 kb plasmid pND6-1 isolated from naphthalene-degrading Pseudomonas sp. strain ND6 was also determined. Among the 23 naphthalene catabolic genes in pND6-1, almost all have 99–100 % identity in amino acid sequences homologous to their nearest counterparts found in plasmids pDTG1 and NAH7 and in a chromosome region in Pseudomonas stutzeri AN10 except for two duplicated genes, ND013 and ND016 (Li et al. 2004). Interestingly, that plasmid pND6-1 contains two duplicate naphthalene catabolic genes. ND013 and ND016 were duplicate genes of ND058 (nahF) and ND091 (nahG), respectively. Later ND016 was identified as nahU (isofunctional gene of the classical salicylate hydroxylase gene), encoding a new salicylate hydroxylase, NahU, which possesses a higher binding ability to salicylate and cofactors and catalytic efficiency in comparison with NahG (Zhao et al. 2005).
In NAH7, the regulatory gene nahR is located between nah and sal operons and the direction of transcription of the two operons is identical. In pDTG1 and pND6-1, gene nahR is located upstream of nah2 operon, and both operons are transcribed in opposite direction toward each other. The intervening region between the upper and lower nah operons of NAH7 does not show any homology with the corresponding regions of pDTG1 and pND6-1 plasmids. These data suggest that the nah operons on NAH7 have been acquired differently from those of pDTG1 and pND6-1 (Sota et al. 2006). The comparison of NAH7 with two other completely sequenced IncP-9 catabolic plasmids, pDTG1 and pWW0, revealed that the three plasmids share very high nucleotide similarities in a 39 kb region encoding the basic plasmid functions (the IncP-9 backbone). The backbone of NAH7 is phylogenetically more related to that of pDTG1 than that of pWW0 (Sevastsyanovich et al. 2008). The IncP-9 naphthalene-degrading plasmid pNAH20 (83 kb in size) from multiplasmid Pseudomonas fluorescens strain PC20 exhibits a similarity to another naphthalene plasmid, pDTG1. However, the positions of insertion sequence (IS) elements significantly alter both catabolic and backbone functions provided by the two plasmids (Heinaru et al. 2009). The P. fluorescens strain PC20 harbors two large plasmids – self-transmissible pNAH20 and mobilizable pPHE20. These plasmids enable the host strain to degrade naphthalene and phenol simultaneously via catechol meta and ortho degradation pathways, respectively. The transfer frequency of pNAH20 is 100 times higher than that of pDTG1 probably due to the insertion of the pCAR1 ISPre2-like element into the mpfR gene coding for a putative repressor of the mpf operon responsible for mating pilus formation. The plasmid pNAH20 can mobilize pPHE20, and the IncQ broad-host-range plasmid, the RSF1010-based PHE plasmid pEST1412 (Heinaru et al. 2009).
Naphthalene catabolic plasmids differ in size, restriction patterns, and incompatibility groups and revealed a high level of DNA homology among nah genes, and the gene order within either upper or meta pathway operons seems to be invariable. However, the relative position of both the operons and the regulatory genes varies. Naphthalene operons can be located in trans as it has been shown for P. putida strains bearing NPL-1, pBS1141, and pBS1191. In the first two cases, nah operon is located on the plasmids, and sal operon on the host bacterial chromosome; in the third case, the gene encoding key enzyme of sal operon (salG) is located on another plasmid – pBS1192. However, plasmids NPL-1, pBS1141, and pBS1191 contain nonfunctioning salG gene (Kosheleva et al. 2003). The plasmid NPL-1 contains at least two class II transposons of the Tn3 family; one of them is located between two catabolic operons near regulatory nahR gene. These transposons are involved in intraplasmid rearrangements, such as deletions and inversions, and can influence the expression of the catabolic and regulatory genes borne by biodegradation plasmids. The formation of a strong NahR-independent constitutive promoter by the inversion of a DNA fragment may be responsible for changing the character of naphthalene dioxygenase synthesis from inducible (in the case of plasmid NPL-1) to constitutive (in the case of plasmid NPL-41).
Most of the naphthalene biodegradation plasmids contain the genes for salicylate metabolism via meta pathway of catechol oxidation. However, genetic control of naphthalene biodegradation through salicylate and gentisate (“gentisate” pathway) also has been studied.
The Pseudomonas putida АК5 that was isolated from slime pit of Nizhnekamskneftekhim chemical factory can metabolize naphthalene via salicylate and gentisate. Naphthalene and salicylate catabolic genes are localized on non-conjugative IncP-7 plasmid pAK5, about 115 kb in size. The “classical” nah operon and the novel sgp operon (salicylate–gentisate pathway) are involved in naphthalene-degrading by P. putida АК5 (Izmalkova et al. 2013). The sgp operon contains six open reading frames (sgpAIKGHB). The four ORFs code for the entire salicylate 5-hydroxylase-oxidoreductase component (sgpA), large and small subunits of oxygenase component (sgpG and sgpH), and 2Fe–2S ferredoxin (sgpB). Genes for gentisate 1,2-dioxygenase (sgpI) and fumarylpyruvate hydrolase (sgpK) incorporate in salicylate 5-hydroxylase gene cluster between sgpA and sgpG. The putative positive regulator for sgp operon gene sgpR is located upstream from the sgpA gene and oriented in the opposite direction from sgpA. Putative maleylacetoacetate isomerase gene locates apart, directly downstream from the sgp operon. The sgp operon organization and phylogenetic analysis of deduced amino acid sequences indicate that this operon has a mosaic structure.
All NAH catabolic plasmids investigated to date encode a single upper pathway for the conversion of naphthalene to salicylate. High level of homology between NAH genes on different plasmids isolated from Pseudomonas sp. suggests that these catabolic plasmids are related.
Most of the naphthalene catabolic plasmids belong to incompatibility groups P-9 and P-7. Environmental studies have suggested a wide distribution of IncP-9-like replicons in nature and their involvement in natural horizontal gene transfer of the naphthalene-degrading trait. It should be noted that IncP-9 plasmids have a moderate range of host bacteria, whereas IncP-7 plasmids are those with a narrow host range. Naphthalene-degrading plasmids of IncP-7 group can be isolated more rarely than IncP-9.
A large collection of naphthalene-degrading fluorescent Pseudomonas strains isolated from sites contaminated with coal tar and crude oil were screened for the presence of degradative plasmids. About 50 % of strains were found to carry naphthalene catabolic plasmids of IncP-9 group ranging in size from 83 to 120 kb. The analysis of several strains bearing IncP-7 naphthalene catabolic plasmids revealed that the structure of IncP-7 plasmids is more various than the structure of IncP-9 plasmids. All IncP-7 plasmid-bearing strains were isolated from hardly contaminated soils, while IncP-9 plasmid-bearing strains were more widely spread and can be isolated from the relatively pristine environments too. Prevalence and diversity of IncP-7 and IncP-9 plasmids in polluted and pristine environments have been also determined by using primers specific for IncP-9 and IncP-7 backbone segments. While a pool of IncP-7 plasmids were detected in contaminated soil samples, IncP-9 plasmids seem to be ubiquitous. About 30 % of NAH plasmids have not been assigned to a particular group. It may be that these plasmids do belong to known groups, but at present they cannot be assigned to one. The absence of DNA probes or DNA primers for PCR amplification for all the known groups makes it difficult to perform the classification easily. New incompatibility groups of catabolic plasmids could be established when the relation between unclassified plasmids and the defined groups will be studied in detail.
3 Genetic Bioaugmentation
3.1 Horizontal Transfer of Catabolic Plasmids
Horizontal gene transfer (HGT) is defined to be the movement of genetic material between bacteria other than by descent in which information travels through the generations as the cell divides. Plasmids as mobile genetic elements play an important role in HGT resulting in microbial adaptation to environmental changes and in the spread of existing catabolic pathways. Moreover horizontal exchange of degradative genes among bacteria in microbial communities plays a significant role in the evolution of catabolic pathways. It has been suggested that the complete catabolic pathways for degradation of at least aromatic hydrocarbons have evolved through modular fusion (Bosch et al. 1999). Catabolic operons are often located within transposons and bordered by insertion sequences. Transposons and IS elements play a significant role in recruitment of catabolic clusters by the replicon and also may increase further DNA rearrangements and exchange of the genes between different bacterial hosts and different replicons. The suggestion is made that the genes nahAc and nahG evolved independently and occur in Pseudomonas sp. strains in different combinations. Thus, nah-like genetic systems, despite conservative organization and high level of homology, demonstrate significant variability. Various configurations of the plasmids in different Pseudomonas sp. strains have arisen over time, and catabolic pathways are continuously subjected to selective pressure. Coexistence of duplicated genes or even operons within one plasmid as in the abovementioned TOL and NAH plasmids provides opportunities for recombination between them. Genetic plasticity of catabolic operons has appeared to play a significant role in adaptation of microorganisms to a wide variety of environmental conditions. In order to assess the prevalence of different hydrocarbon catabolic genotypes in nature and to monitor the evolution of modern catabolic pathways, there is a need for more complete nucleotide sequences for plasmids and degradative operons found in different hydrocarbon-degrading microorganisms. Bacterial plasmid genome sequence comparisons indicate that plasmids have complex genetic histories resulting from transposition, homologous recombination, and illegitimate recombinational events.
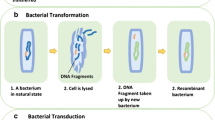
It is well documented that bacteria can readily exchange genetic information under artificial conditions typically used in most laboratory studies as well as to some extent in nature. The three mechanisms by which such genetic exchange can occur are transformation, transduction, and conjugation. Bacterial conjugation is the most widespread mechanism for horizontal DNA transfer in nature. As it has been mentioned above in addition to their degradative genes, most of the catabolic plasmids are transmissible via conjugation and contain clusters of transfer (tra) genes. Ecologically, plasmid-encoded pathways are advantageous because they provide genetically flexible systems and can be transferred between bacterial species.
When the in situ degradation potential is very low or absent, application of effective oil-degrading microorganisms is necessary. Introduced active microorganisms can themselves accelerate biodegradation of target pollutants; however, they have to compete with indigenous microbial flora which is usually more adapted to specific conditions of site. If the survival of used bacteria or bacterial consortium is poor, the introduction of degradative genes and operons, located on catabolic plasmids, into well-established, competitive indigenous microbial populations may play an important role in bioaugmentation. This approach is termed “plasmid-mediated” or “genetic” bioaugmentation. In genetic bioaugmentation, introduced microorganisms appear to be not only degrader strains but also donors of the catabolic genes for horizontal transfer to well-establish and competitive indigenous bacterial populations of a site. Spreading of catabolic genes increases the diversity of microorganisms capable of degrading target hydrocarbons and therefore accelerates the bioremediation process.
The occurrence of HGT in natural microbial communities has been well documented (Top and Springael 2003). These data were obtained by either retrospective analysis or by direct experimental studies. The retrospective approach is the investigation of distribution of very similar or highly homologous plasmids and genes among different microorganisms. For example, the presence of highly conserved nahAc allele among phylogenetically diverse bacteria bearing naphthalene catabolic plasmids (pDTG1-like) and isolated from geographically distant regions provides evidence for in situ horizontal transfer at coal tar-contaminated sites (Herrick et al. 1997). Data that horizontal transfer of naphthalene catabolic plasmids has indeed occurred between microorganisms in the soil bacterial community was also obtained in different experiments with soil microcosms using conjugative plasmids labeled by additional selectable marker. Conjugation transfer of catabolic plasmids was demonstrated in naphthalene-contaminated laboratory soil microcosms (Akhmetov, et al. 2008). The soil used in this work contained indigenous bacterial degraders of naphthalene which were isolated and identified as Pseudomonas putida and Pseudomonas fluorescens. In these experiments, both indigenous microorganisms and the introduced laboratory strain Pseudomonas putida BS394 (pNF142::TnMod-OTc) served as donors of naphthalene biodegradation plasmids. As a recipient for indigenous catabolic plasmids, well-characterized P. putida strain KT2442 was used. Transconjugant strains harboring indigenous catabolic plasmids possessed high salicylate hydroxylase and low catechol 2,3-dioxygenase activities, in contrast to indigenous degraders, which had a high level of catechol 2,3-dioxygenase activity and a low level of salicylate hydroxylase. The transfer of the labeled plasmid pNF142::TnMod-OTc to the introduced plasmid-free recipient P. putida KT2442 and to indigenous soil microorganisms of the genus Pseudomonas was shown both under selective pressure (in the presence of naphthalene) and in its absence. The isolated transconjugant strains belonged to several species of the genus Pseudomonas (Filonov et al. 2010). This work has demonstrated that the plasmid-bearing P. putida strains KT2442 (pNF142::TnMod-OTc) and BS394(pNF142::TnMod-OTc) could dominate over indigenous naphthalene destructors under selective pressure (soil contamination) and were quickly eliminated in the absence of the pollutant. The transfer of naphthalene biodegradation plasmids in soil microbial populations seems to enhance the efficiency of hydrocarbon biodegradation under field conditions due to the increase of microbial degradative potential. The occurrence of new bacterium-plasmid combinations resulting from horizontal transfer can lead to appearance of more efficient and competitive degrader strains that can be used for a successful bioremediation. However, authors could not present direct evidence for plasmid conjugal transfer-mediated biodegradation of naphthalene, since introduced strains themselves could survive, compete with indigenous microorganisms, and effectively degrade aromatic hydrocarbon.
Another example of HGT was demonstrated in a field bioaugmentation experiment with the abovementioned multiplasmid P. fluorescens strain PC20 (pNAH20, pPHE20) which is an effective degrader of pollutants (Heinaru et al. 2009). Occurrence of a natural transconjugant of pNAH20 was observed in birch rhizosphere. During in vitro transfer experiments, transconjugants of both plasmids pNAH20 and pPHE20 were obtained. It has been proposed that mobilization of the pPHE20 plasmid by the helper plasmid pNAH20 into the indigenous bacteria of the genus Pseudomonas might take place under natural conditions as well.
The best-studied catabolic plasmid pWW0 also has been used as a model for development of bioremediation strategies involving environmental release of plasmid-containing strains. Horizontal transfer of pWW0 in microbial biofilm communities has been investigated. The results demonstrated that plasmid transfer does occur when a donor interacts with a growing or established biofilm but that transfer to endogenous bacteria only occurs at the interface between the donor and recipient cultures and does not spread throughout the recipient population (Christensen et al. 1998). The mycorrhizosphere Pseudomonas fluorescens strain OS81 supplied with the TOL plasmid pWW0::Km was inoculated in microcosms with and without pine seedlings mycorrhized with Suillus bovinus (Sarand et al. 2000). After 3 months of regular treatment with m-toluate, the introduced catabolic plasmid was disseminated in the indigenous populations in both mycorrhizosphere and soil uncolonized by the fungus. Transconjugants were represented by the genera Pseudomonas and Burkholderia. It has been shown that inoculation of P. fluorescens OS81 (pWW0::Km) into microcosms influences the development of plants in contaminated soil protecting them from m-toluate. Since the inoculated strain was not detected after 3 months, protective effect was attributed to transconjugants receiving TOL plasmid and capable of degrading m-toluate.
3.2 Factors Affecting Horizontal Transfer and Expression of Catabolic Genes
Horizontal gene transfer is affected by many factors (environmental conditions) such as temperature, pH, soil textural type, moisture, and nutrient availability. A major factor was shown to be the level of nutrients as far as they stimulate plasmid transfer by enhancing the numbers and activities of donor and recipient bacteria (van Elsas et al. 2000). The presence of plant roots providing additional organic substrates allows higher frequency of catabolic gene transfer as well as higher metabolic activity compared to bulk soils. Exudates derived from the plant can help to stimulate the survival and action of bacteria, which subsequently results in a more efficient degradation of pollutants. Plant roots have nutrient-rich surfaces which provide conditions for microbial colonization and activities, e.g., plasmid conjugal transfer. The very important factor which dramatically affects the plasmid transfer rate in soil is selective pressure. Top et al. proposed that positive effect of the presence of the pollutant on the number of transconjugants is almost certainly due to the proliferation of the transconjugants, but not to a direct effect of the pollutant on the conjugation efficiency (Top et al. 2002). For successful bioaugmentation, it is desirable that effectively degrading pollutant transconjugants would be the numerically dominant population in the bacterial community. In order to grow to high density, the transconjugants need to have a selective advantage over the other indigenous bacteria which are usually more competitive without pollutants. Moisture, temperature, and soil pH influence on cell physiology in whole, and obviously these factors also effect on plasmid transfer. Indeed, plasmid transfer rates were highest at near-neutral pH values, whereas under acid conditions transfer was not detected and it was only detected at a favorable temperature for bacterial cells (van Elsas et al. 2000).
Even if transfer of degradative plasmid into some indigenous recipients was successful, expression of catabolic genes in new transconjugants can be limited. Alternative substrate availability may significantly change expression of catabolic genes transferred in a new recipient strain. The presence of glucose concurrently with toluene has been shown to trigger catabolite repression in Pseudomonas putida cells harboring the TOL plasmid (del Castillo and Ramos 2007). E. coli RP-1 cells containing the same TOL plasmid exhibited very low degradative functions when toluene is a sole carbon source. Large enhancement of toluene degradation rate and enzyme activities was observed when cells were exposed simultaneously on toluene and glucose or toluene and Luria-Bertani broth as easily degradable carbon source (Ikuma and Gunsch 2012). In spite of bacteria belonging to the Enterobacteriaceae family being very unlikely to use in bioremediation technology, these results indicated that additional carbon source may play a significant role in enhancement of catabolic abilities of degrader strains.
4 Conclusions
The role of plasmids and their horizontal transfer in evolution of bacterial genomes and adaptation of microbial populations to specific environmental changes is generally accepted nowadays. Despite that only a few reports have clearly demonstrated a direct effect of gene transfer on accelerated biodegradation, genetic bioaugmentation seems to be perspective for environmental cleanup biotechnology. For successful genetic bioaugmentation, knowledge about microbial communities of bioremediated sites is necessary to predict possible changes in their biodegradative potential. Changes in bacterial community structure in response to the contamination as well as the addition of nonindigenous microorganisms should be taken into account in the development of cleanup strategy. To access the fate and behavior of introduced plasmid in an open environment, preliminary studies are necessary concerning incompatibility group, host range, stability, and expression of catabolic genes in a new bacterial host’s background. Catabolic plasmids are of great environmental significance. The type of catabolic plasmids and their horizontal transfer should be monitored for estimation of biodegradative potential of oil-contaminated soil.
References
Aemprapa, S., Williams, P.A.: Implication of the xylQ gene of TOL plasmid pWW102 for the evolution of aromatic catabolic pathways. SGM Microbiol. 144, 1387–1396 (1998)
Akhmetov, L.I., Filonov, A.E., Puntus, I.F., Kosheleva, I.A., Nechaeva, I.A., Yonge, D.R., Petersen, J.N., Boronin, A.M.: Horizontal transfer of catabolic plasmids in the process of naphthalene biodegradation in model soil systems. Microbiology 77, 29–39 (2008)
Assinder, S.J., De Marco, P., Osborne, D.J., Poh, C.L., Shaw, L.E., Winson, M.K., Williams, P.A.: A comparison of the multiple alleles of xylS carried by TOL plasmids pWW53 and pDK1 and its implications for their evolutionary relationship. J. Gen. Microbiol. 139, 557–568 (1993)
Boronin, A.M.: Diversity of Pseudomonas plasmids: to what extent? FEMS Microbiol. Lett. 79, 461–467 (1992)
Bosch, R., Garcia-Valdes, E., Moore, E.R.B.: Genetic characterization and evolutionary implications of a chromosomally encoded naphthalene-degradation upper pathway from Pseudomonas stutzeri AN10. Gene 236, 149–157 (1999)
Chatfield, L.K., Williams, P.A.: Naturally occurring TOL plasmids in Pseudomonas strains carry either two homologous or two nonhomologous catechol 2,3-oxygenase genes. J. Bacteriol. 168, 878–885 (1986)
Christensen, B.B., Sternberg, C., Andersen, J.B., Eberl, L., Moller, S., Givskov, M., Molin, S.: Establishment of new genetic traits in a microbial biofilm community. Appl. Environ. Microbiol. 64, 2247–2255 (1998)
Couturier, M., Bex, F., Bergquist, P.L., Maas, W.K.: Identification and classification of bacterial plasmids. Microbiol. Rev. 52, 375–395 (1988)
del Castillo, T., Ramos, J.L.: Simultaneous catabolite repression between glucose and toluene metabolism in Pseudomonas putida is channeled through different signaling pathways. J. Bacteriol. 189, 6602–6610 (2007)
Dennis, J.J., Zylstra, G.J.: Complete sequence and genetic organization of pDTG1, the 83 kilobase naphthalene degradation plasmid from Pseudomonas putida strain NCIB 9816–4. J. Mol. Biol. 341, 753–768 (2004)
El-Deeb, B.: Natural combination of genetic systems for degradation of phenol and resistance to heavy metals in phenol and cyanide assimilating bacteria. Malays. J. Microbiol. 5(2), 94–103 (2009)
Esikova, T.Z., Grishchenkov, V.G., Kulakov, L.A., Morenkova, M.A., Boronin, A.M.: Incompatibility groups of caprolactam degradative plasmids of Pseudomonas sp. Mol. Genet. Microbiol. Virusol. 4, 25–28 (1990)
Filonov, A.E., Akhmetov, L.I., Puntus, I.F., Esikova, T.Z., Gafarov, A.B., Kosheleva, I.A., Boronin, A.M.: Horizontal transfer of catabolic plasmids and naphthalene biodegradation in open soil. Microbiology 79, 184–190 (2010)
Greated, A., Lambertsen, L., Williams, P.A., Thomas, C.M.: Complete sequence of the IncP-9 plasmid pWW0 from Pseudomonas putida. Environ. Microbiol. 4, 856–871 (2002)
Habe, H., Omori, T.: Genetics of polycyclic aromatic hydrocarbon metabolism in diverse aerobic bacteria. Biosci. Biotechnol. Biochem. 67, 225–243 (2003)
Heinaru, E., Vedler, E., Jutkina, J., Merit, A., Heinaru, A.: Conjugal transfer and mobilization capacity of the completely sequenced naphthalene plasmid pNAH20 from multiplasmid strain Pseudomonas fluorescens PC20. FEMS Microbiol. Ecol. 70, 563–574 (2009)
Herrick, J.B., Stuart-Keil, K.G., Ghiorse, W.C., Madsen, E.L.: Natural horizontal transfer of a naphthalene dioxygenase gene between bacteria native to a coal tar-contaminated field site. Appl. Environ. Microbiol. 63, 2330–2337 (1997)
Herrmann, H., Muller, C., Schmidt, I., Mahnke, J., Petruschka, L., Hahnke, K.: Localization and organization of phenol degradation genes of Pseudomonas putida strain H. Mol. Gen. Genet. 247, 240–246 (1995)
Heuer, H., Fox, R.E., Top, E.M.: Frequent conjugal transfer accelerates adaptation of broad-host-range plasmid to an unfavorable Pseudomonas putida host. FEMS Microbiol. Ecol. 59, 738–748 (2007)
Ikuma, K., Gunsch, C.K.: Genetic bioaugmentation as an effective method for in situ bioremediation. Bioengineered 3(4), 236–241 (2012)
Izmalkova, T.Y., Sazonova, O.I., Nagornih, M.O., Sokolov, S.L., Kosheleva, I.A., Boronin, A.M.: The organization of naphthalene degradation genes in Pseudomonas putida strain AK5. Res. Microbiol. 164, 244–253 (2013)
Jacoby, G.A.: Classification of plasmid Pseudomonas aeruginosa. In: Schlessinger, D. (ed.) Microbiology-1977, pp. 119–126. American Society of Microbiology, Washington, DC (1977)
Kasak, L., Hôrak, R., Nurk, A., Talvik, K., Kivisaar, M.: Regulation of the catechol 1,2-dioxygenase- and phenol monooxygenase-encoding pheBA operon in Pseudomonas putida PaW85. J. Bacteriol. 175, 8038–8042 (1993)
Keil, H., Keil, S., Pickup, R.W., Williams, P.A.: Evolutionary conservation of genes coding for meta pathway enzymes within TOL plasmids pWW0 and pWW53. J. Bacteriol. 164, 887–895 (1985)
Kivisaar, M., Hõrak, R., Kasak, L., Heinaru, A., Habicht, J.: Selection of independent plasmids determining phenol degradation in Pseudomonas putida and the cloning and expression of genes encoding phenol monooxygenase and catechol 1,2-dioxygenase. Plasmid 24(1), 25–36 (1990)
Kivisaar, M., Kasak, L., Nurk, A.: Sequence of the plasmid-encoded catechol 1,2-dioxygenase-expressing gene, pheB, of phenol-degrading Pseudomonas sp. strain EST1001. Gene 98, 15–20 (1991)
Kosheleva, I.A., Izmalkova, T.Y., Sokolov, S.L., Sazonova, O.I., Boronin, A.M.: Structural and functional variability of genetic systems for catabolizing polycyclic aromatic hydrocarbons in Pseudomonas putida strains. Genetika 39, 1185–1192 (2003)
Kostal, J., Suchanek, M., Klierova, H., Demnerova, K., Kralova, B., McBeth, D.L.: Pseudomonas C12B, an SDS degrading strain, harbors a plasmid coding for degradation of medium chain length n-alkanes. Int. Biodeter. Biodegr. 42, 221–228 (1998)
Lee, M.-Y., Hwang, M.-O., Choi, S.-Y., Min, K.-H.: Min n-Alkane dissimilation by Rhodopseudomonas sphaeroides transferred OCT plasmid. Microb. Ecol. 26, 219–226 (1993)
Li, W., Shi, J., Wang, X., Han, Y., Tong, W., Ma, L., Liu, B., Cai, B.: Complete nucleotide sequence and organization of the naphthalene catabolic plasmid pND6-1 from Pseudomonas sp. strain ND6. Gene 336, 231–240 (2004)
Makarenko, A.A., Bezverbnaya, I.P., Kosheleva, I.A., Kuvichkina, T.N., Il’yasov, P.V., Reshetilov, A.N.: Development of biosensors for phenol determination from bacteria found in petroleum fields of West Siberia. Appl. Biochem. Microbiol. 38, 29–34 (2002)
Park, W., Jeon, C.O., Madsen, E.L.: Interaction of NahR, a LysR-type transcriptional regulator, with the alpha subunit of RNA polymerase in the naphthalene degrading bacterium, Pseudomonas putida NCIB 9816-4. FEMS Microbiol. Lett. 213, 159–165 (2002)
Ramos, J.L., Marqués, S., Timmis, K.N.: Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators. Annu. Rev. Microbiol. 51, 341–373 (1997)
Sarand, I., Haario, H., Jorgensen, K., Romantschuk, M.: Effect of inoculation of a TOL plasmid containing mycorrhizosphere bacterium on development of Scots pine seedlings, their mycorrhizosphere and the microbial flora in m-toluate-amended soil. FEMS Microbiol. Ecol. 31, 127–141 (2000)
Sentchilo, V.S., Perebituk, A.N., Zehnder, A.J.B., van der Meer, J.R.: Molecular diversity of plasmid bearing genes that encode toluene and xylene metabolism in Pseudomonas strains isolated from different contaminated sites in Belarus. J. Bacteriol. 66, 2842–2852 (2000)
Sevastsyanovich, Y.R., Krasowiak, R., Bingle, L.E.H., Haines, A.S., Sokolov, S.L., Kosheleva, I.A., Leuchuk, A.A., Titok, M.A., Smalla, K., Christopher, M., Thomas, C.M.: Diversity of IncP-9 plasmids of Pseudomonas. SGM Microbiol. 154, 2929–2941 (2008)
Shell, M.A.: Regulation of the naphthalene degradation genes of plasmid NAH7: example of a generalized positive control system in Pseudomonas and related bacteria. In: Pseudomonas. Biotransformation, Pathogenesis, and Evolving Biotechnology, pp. 165–176. American Society for Microbiology, Washington, D.C (1990)
Shingler, V., Bartilson, M., Moore, T.: Cloning and nucleotide sequence of the gene encoding the positive regulator (DmpR) of the phenol catabolic pathway encoded by pVI150 and identification of DmpR as a member of the NtrC family of transcriptional activators. J. Bacteriol. 175, 1596–1604 (1993)
Sota, M., Top, E.M.: Horizontal gene transfer mediated by plasmids. In: Lipps, G. (ed.) Plasmids: Current Research and Future Trends, p. 263. Horizon Scientific Press, Norfolk (2008)
Sota, M., Yano, H., Ono, A., Miyazaki, R., Ishii, H., Genka, H., Top, E., Tsuda, M.: Genomic and functional analysis of the IncP-9 naphthalene-catabolic plasmid NAH7 and its transposon Tn4655 suggests catabolic gene spread by a tyrosine recombinase. J. Bacteriol. 188, 4057–4067 (2006)
Top, E.M., Springael, D.: The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr. Opin. Biotechnol. 14, 262–269 (2003)
Top, E.M., Moenne-Loccoz, Y., Pembroke, T., Thomas, C.M.: Phenotypic traits conferred by plasmids. In: Thomas, C.M. (ed.) The Horizontal Gene Pool. Bacterial Plasmids and Gene Spread, p. 419. Harwood Academic Publishers, Amsterdam (2000)
Top, E.M., Springael, D., Boon, N.: Catabolic mobile elements and their potential use in bioaugmentation of polluted soils and waters. FEMS Microbiol. Ecol. 42, 199–208 (2002)
Tsuda, M., Minegishi, K.-I., Iino, T.: Toluene transposons Tn4651 and Tn4653 are class II transposons. J. Bacteriol. 171, 1386–1393 (1989)
van Beilen, J.B., Wubbolts, M.G., Witholt, B.: Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation 5(3–4), 161–174 (1994)
van Beilen, J.B., Panke, S., Lucchini, S., Franchini, A.G., Röthlisberger, M., Witholt, B.: Analysis of Pseudomonas putida alkane-degradation gene clusters and flanking insertion sequences: evolution and regulation of the alk genes. SGM Microbiol. 147, 1621–1630 (2001)
Van Elsas, J.D., Fry, J., Hirsch, P., Molin, S.: Ecology of plasmid transfer and spread. In: Thomas, C.M. (ed.) The Horizontal Gene Pool. Bacterial Plasmids and Gene Spread, p. 419. Harwood Academic Publishers, Amsterdam (2000)
Williams, P.A., Murray, K.: Metabolism of benzoate and the methylbenzoates by Pseudomonas putida (arvilla) mt-2: evidence for the existence of a TOL plasmid. J. Bacteriol. 120, 416–423 (1974)
Yano, H., Miyakoshi, M., Ohshima, K., Tabata, M., Nagata, Y., Hattori, M., Tsuda, M.: Complete nucleotide sequence of TOL plasmid pDK1 provides evidence for evolutionary history of IncP-7 catabolic plasmids. J. Bacteriol. 192, 4337–4347 (2010)
Zhao, H., Chen, D., Li, Y., Cai, B.: Overexpression, purification and characterization of a new salicylate hydroxylase from naphthalene-degrading Pseudomonas sp. strain ND6. Microbiol. Res. 160, 307–313 (2005)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Boronin, A.M., Kosheleva, I.A. (2014). The Role of Catabolic Plasmids in Biodegradation of Petroleum Hydrocarbons. In: Cao, G., Orrù, R. (eds) Current Environmental Issues and Challenges. Springer, Dordrecht. https://doi.org/10.1007/978-94-017-8777-2_9
Download citation
DOI: https://doi.org/10.1007/978-94-017-8777-2_9
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-017-8776-5
Online ISBN: 978-94-017-8777-2
eBook Packages: Earth and Environmental ScienceEarth and Environmental Science (R0)