Abstract
Bacteria are frequently exposed to changes in environmental conditions, such as fluctuations in temperature, pH or the availability of nutrients. These assaults can be detrimental to cell as they often result in a proteotoxic stress, which can cause the accumulation of unfolded proteins. In order to restore a productive folding environment in the cell, bacteria have evolved a network of proteins, known as the protein quality control (PQC) network, which is composed of both chaperones and AAA+ proteases. These AAA+ proteases form a major part of this PQC network, as they are responsible for the removal of unwanted and damaged proteins. They also play an important role in the turnover of specific regulatory or tagged proteins. In this review, we describe the general features of an AAA+ protease, and using two of the best-characterised AAA+ proteases in Escherichia coli (ClpAP and ClpXP) as a model for all AAA+ proteases, we provide a detailed mechanistic description of how these machines work. Specifically, the review examines the physiological role of these machines, as well as the substrates and the adaptor proteins that modulate their substrate specificity.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
General Introduction
The bacterial cytosol is a complex mixture of macromolecules (proteins, DNA and RNA), which perform a variety of different functions. Given that proteins play a central role in many of these important cellular tasks, their correct maintenance within the cell is critical for cellular viability, not only under normal cellular conditions but also under conditions of stress. As such, a bacterial cell contains a network of molecular chaperones and proteases (often referred to as the protein quality control (PQC) network) dedicated to maintaining homeostasis of protein folding. Chaperones function to protect functional proteins against unfolding and to refold misfolded and aggregated species. The role of proteases is to remove unwanted and hopelessly damaged proteins.
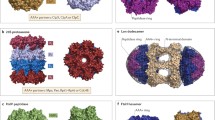
In the bacterial cytosol, protein degradation is performed mainly by a number of different ATP-dependent proteolytic machines. In general these machines are composed of two components, a peptidase and an unfoldase. Invariably, the unfoldase is a member of the AAA+ (ATPase associated with diverse cellular activities) superfamily and as such these molecular machines are commonly referred to as AAA+ proteases [1]. In Gram-negative bacteria, such as Escherichia coli there are generally five different AAA+ proteases (ClpAP, ClpXP, HslUV, Lon (also refereed to as LonA) and FtsH). In contrast most Gram-positive bacteria, such as Bacillus subtilis, contain up to seven different AAA+ protease (ClpCP, ClpEP, ClpXP, HslUV (CodXW), LonA, LonB and FtsH). Interestingly, in bacteria belonging to the Actinobacteria and Nitrospira phyla (e.g. in Mycobacterium tuberculosis (Mtb)) one or more of these AAA+ proteolytic machines is replaced by the proteasome (for a detailed review of this AAA+ machine, and its physiological role in Mtb please refer to Darwin and colleagues [2]). Regardless of their origin, these machines can be divided into two broad groups; those that contain the unfoldase and peptidase components on separate polypeptides (e.g. ClpAP, ClpCP, ClpEP, ClpXP and HslUV (CodXW)), and those that contain both components on a single polypeptide (e.g. LonA, LonB and FtsH) (see Fig. 1.1).
Cartoon representation of the various different AAA+ proteases in bacteria. AAA+ proteases can be separated into two different groups. Two component proteases (e.g. ClpAP, ClpCP, ClpXP, ClpEP and HslUV) contain the unfoldase and peptidase components on separate polypeptides. One component proteases, contain the peptidase and the unfoldase on a single polypeptide (e.g. LonA, LonB and FtsH). The unfoldase component contains one or more AAA+ domains, responsible for ATP-dependent unfolding of the substrate. All unfoldase components also contain at least one accessory domains (e.g. ClpA and ClpC contain a conserved N-terminal domain (N-domain, pink), ClpC and ClpE contain a middle domain (M, grey), ClpE and ClpX contain a Zinc binding domain (ZBD, yellow), HslU contains an accessory domain inserted into the AAA+ domain (I-domain, purple), LonA contains two N-terminal domains unrelated to the N-domain of ClpA and ClpC (N1 and N2, green), while LonB and FtsH both contain a single transmembrane (TM) region), which serve various different functions (see main text for details). In the case of the ClpP-binding unfoldase components, the AAA-2 domain contains an IGF/L loop for interaction with ClpP. The protease components are responsible for cleavage of the unfolded substrate. In the case of ClpP, hydrolysis of the polypeptide is catalysed by the catalytic triad (S, H and D), while FtsH and HslV contain either a conserved HExxH motif or an N-terminal threonine (T) respectively, for peptide bond cleavage
This review will focus on the “two-component” proteolytic machines, primarily those from E. coli (e.g. ClpAP and ClpXP), with a brief comparison to the equivalent machines (e.g. ClpCP and ClpXP) in the model Gram-positive bacterium, B. subtilis. However, for an extensive review on regulatory proteolysis in B. subtilis please refer to [3]. Likewise, for a detailed review on the “single polypeptide” proteases, i.e. Lon and FtsH please refer to [4] and [5], respectively.
Structure and Function of the “ClpP Containing” Proteases (ClpAP, ClpXP and ClpCP)
As mentioned above, bacteria contain a wide variety of different proteolytic machines, of which ClpXP is certainly the best-studied AAA+ protease [1]. ClpXP is known to play a number of critical roles in a wide variety of bacterial species, from the control of different stress response pathways in Gram-positive and Gram-negative bacteria (see [6, 7]) to the regulation of virulence through the degradation of key factors that control virulence (see [8]). ClpXP has also been shown to play an important role in regulating mitochondrial protein homeostasis (proteostasis) in eukaryotes such as worms [9, 10] and plants [11]. Surprisingly however, this proteolytic machine is absent from most fungi including, Saccharomyces cereviseae [12, 13]. For a detailed review of about the role of these AAA+ proteases in regulating mitochondrial function please refer to [14]. Although the AAA+ proteases ClpAP and ClpCP are not as widely conserved as ClpXP, these proteases do, nevertheless, control a number of key proteolytic/regulatory pathways in Gram-negative and Gram-positive bacteria, respectively. Interestingly, ClpCP also appears to play an important role in proteostasis within the plastid of plants (for a recent review see [15, 16]).
Although these machines recognise a variety of different substrates and regulate a range of different physiological processes, each machine shares a common architecture and a similar mode of action. All form barrel-shaped complexes in which the oligomeric AAA+ unfoldase is concentrically aligned with the oligomeric protease component as is best illustrated by the crystal structure of the HslUV complex [17, 18]. Interestingly, the unfoldase component may be located at either or both ends of the peptidase component to form single-headed (1:1) or double-headed (2:1) complexes, respectively. For the ClpAP protease, the symmetric double-headed complexes have been shown to be most efficient at processing substrates [19]. Regardless of whether the complexes are single- or double-headed, both oligomeric components (i.e. the unfoldase and the peptidase) generally exhibit a six-fold symmetry throughout the entire complex. However in the Clp protease complexes (e.g. ClpAP, ClpCP and ClpXP) the machines display a unique symmetry mismatch between the unfoldase and the peptidase. While the AAA+ unfoldase component (i.e. ClpA, ClpC and ClpX) like most AAA+ proteins studied to date, form hexameric ring-shaped oligomers, the peptidase (i.e. ClpP) is composed of two heptameric rings [20]. The two heptameric rings of ClpP stack back-to-back, encapsulating the catalytic (active site) residues of ClpP within a barrel shaped tetradecamer. This symmetry mismatch poses some interesting questions. How do these two rings (the hexameric unfoldase and the heptameric peptidase) interact to form a functional complex, and how many subunits are required for a functional interaction. Regardless of whether the protease complex is symmetric or asymmetric, all AAA+ proteases undergo three basic steps in order to degrade a substrate protein (see Fig. 1.2). In the first step, the substrate is recognised by the unfoldase, although in some cases substrate recognition may be facilitated by an adaptor protein (see later). In bacteria, substrates are usually recognised via short sequence specific motifs (termed degrons), which are often located at the N- or C-terminus of the substrate protein. Following recognition, the substrate is then unfolded in an ATP-dependent fashion (Fig. 1.2, step 2). The unfolded substrate is then translocated into the associated peptidase, where the polypeptide chain is hydrolysed into small peptide fragments (∼3–8 amino acids long), which have been proposed to egress through the holes in the sidewall of the peptidase, although this method of egress remains somewhat controversial (Fig. 1.2, step 3) [21, 22].
Cartoon illustrating the main steps involved in substrate recognition and degradation by AAA+ proteases. The unfoldase (e.g. ClpX) forms a hexameric ring-shaped structure (blue) at one or both ends of the peptidase (e.g. ClpP), which forms two heptameric rings stacked back-to-back (red). The substrate (green) contains a degradation signal (degron) often located at the N- or C-terminus of the protein. The degron is recognised by the unfoldase and the substrate protein unfolded, in an ATP-dependent fashion, then translocated into the peptidase where the protein in cleaved into small peptide fragments, which diffuse through holes in the side-wall of the peptidase
The Peptidase ClpP
The ClpP peptidase is synthesized as a zymogen, containing a N-terminal propeptide [23], which is autocatalytically cleaved upon oligomerization, resulting in the formation of a proteolytically active oligomer. ClpP is a serine protease, composed of a Ser-His-Asp catalytic triad (Fig. 1.1), which exhibits chymotrypsin-like activity, that is, it cleaves peptide bonds mostly after non-polar residues [24, 25]. The active peptidase is a barrel-shaped oligomer composed of two heptameric rings, stacked back-to-back [20], that forms a degradation chamber in which the proteolytic active sites are sequestered away from cytosolic proteins (Fig. 1.3a). Each monomer of ClpP resembles a hatchet and consists of three subdomains: a handle, a globular head and a N-terminal loop. The heptameric ring is formed by the interaction of seven subunits through the head subdomain, and the tetradecamer is formed by the interaction of two heptameric rings through the handle subdomain (Fig. 1.3a). Entry into the catalytic chamber of this serine peptidase is restricted to a narrow entry portal (∼10 Å) at both ends of the barrel-shaped complex. The N-terminal peptides of ClpP flank the axial pore and are proposed to act as a gate for entry into the proteolytic chamber. As a result of this narrow axial entry portal, folded proteins are excluded from entering the catalytic chamber, although small peptides and unfolded proteins can be degraded in an ATPase independent fashion, albeit unfolded proteins are degraded very slowly in the absence of the ATPase [26]. Importantly, the degradation of unfolded substrates can be accelerated by the addition of a cognate unfoldase (i.e. ClpX, ClpA or ClpC), which implies that entry into ClpP is gated and that this gated-entry can be activated by the unfoldase. Indeed, recent cryo-EM reconstructions have shown that binding of ClpA triggers a change in the N-terminal loops of ClpP, from a “down” conformation where they block entry to the catalytic chamber, to an “up” conformation which permits access to the chamber [27]. Consistent with a “gating” role for the N-terminal loops of ClpP, deletion of these loops was shown to accelerate the degradation of short peptides [28]. The cognate AAA+ unfoldase also mediates the degradation of folded substrate proteins by actively unfolding and translocating the substrates through the axial pore and into the proteolytic chamber of ClpP. Indeed, it appears that the oligomeric structure of ClpP has been carefully designed to prevent widespread and indiscriminate degradation of cellular proteins by regulating substrate access to its proteolytic chamber. Consistent with this idea, several recent studies have identified a series of novel antibiotics (e.g. acyldepsipeptides (ADEPs) and ACPs) that activate ClpP (in the absence of its cognate unfoldase) for unregulated protein degradation [29–34]. This activation of ClpP results in the unregulated degradation of nascent polypeptides and unfolded proteins in the cell [34], and in a recent study ADEP was shown to inhibit cell division of Gram positive bacteria, through the ClpP-mediated degradation of FtsZ, a key protein required for septum formation [35].
Oligomeric structure of ClpP. (a) ClpP (PDB: 1TYF) forms two heptameric ring-shaped oligomers (Top view) stacked back-to-back (Side view) to create a barrel-shaped oligomer. Interactions between adjacent head subdomains drive oligomerisation of the seven-membered ring, while interactions between the handle subdomain of two heptamers are responsible for formation of the tetradecamer. (b) In the absence of the unfoldase, the entry portal into the catalytic chamber of ClpP (PDB: 3KTH) is narrow (∼10 Å), in the presence of chemical activators of ClpP (i.e. ADEPs, ACPs and potentially the unfoldase), the entry portal into the catalytic chamber of ClpP (PDB: 3KTI) is opened (∼30 Å)
Based on a series of biochemical and structural studies, these chemical activators of ClpP dock into a hydrophobic pocket located on the surface of ClpP (Fig. 1.3b). Firstly, and most importantly, ADEP binding to this hydrophobic pocket results in opening of the ClpP pore (from ∼10 Å in the absence of ADEP to ∼21–27 Å in the presence of different forms of ADEP). This “gated-opening” of the ClpP pore, is proposed to be sufficient to allow entry of unfolded proteins into the proteolytic chamber of ClpP (where the catalytic residues are located) and possibly the primary reason for degradation of unfolded substrates. Interestingly, in the case of B. subtilis ClpP, ADEP not only triggers opening of the pore, but also triggers oligomerisation of ClpP from free “inactive” monomers to “active” tetradecamers [32], a step that is normally controlled by the cognate unfoldase, ClpC [36]. Similarly, ADEP activation of human ClpP for unregulated degradation is also likely to result from assembly of the ClpP tetradecamer [37] a process that normally requires the assistance of ClpX [38]. As a consequence, ADEP also appears to be a competitive inhibitor of unfoldase binding to ClpP, preventing the regulated degradation of substrates that would normally be delivered to ClpP by the unfoldase component [32]. As such, the ADEP-bound conformation of ClpP has been proposed to mimic the unfoldase-bound conformation of ClpP. Surprisingly, binding of ClpA to ClpP, as measured from sections of the ClpAP cryo-EM structure, appears to have little effect on the size of the ClpP pore (diameter ∼12 Å) [27] and hence it has been suggested that the size of the pore may vary with translocation of different substrates [39]. Nevertheless, it remains to be seen, if an ordered arrangement of the N-terminal loops on ClpP (as observed in the B. subtilis ClpP-ADEP structure) or a disorder arrangement of the N-terminal loops of ClpP (as observed in the E. coli ClpP-ADEP complex) resembles the unfoldase bound complex.
The Unfoldase Components (ClpX/ClpA/ClpC)
In E. coli, ClpP forms proteolytic complexes with both ClpA and ClpX, while in B. subtilis, ClpP associates with three different unfoldases, ClpC, ClpX and ClpE [3]. Although the overall architecture of the different unfoldase components is similar, each unfoldase contains a unique organisation. While ClpA, ClpC and ClpE each contain two AAA+ domains, ClpX only contains a single AAA+ domain (Fig. 1.1). Regardless of the number of AAA+ domains present, each unfoldase contains one or more accessory domains. In the cases of ClpA and ClpX, a single accessory domain is located at the N-terminus of the protein, while both ClpC and ClpE contain two accessory domains, one at the N-terminus of the protein and the other located between the two AAA+ domains, termed the middle or M-domain (Fig. 1.1). In general, these accessory domains are required for the binding of substrates and/or adaptor proteins. In the case of ClpA, the N-terminal domain is essential for docking of the adaptor protein ClpS [40–42] but also required for the recognition, and hence degradation of some substrates [43, 44]. Similarly, the N-terminal domain of B. subtilis ClpC is essential for the ClpP-mediated degradation of most substrates [45, 46]. However in this case, the N-domain is thought not to be directly involved in substrate recognition but rather plays a crucial role in binding adaptor proteins (i.e. MecA and McsB), which are required for ClpC oligomerisation and/or substrate delivery [36, 47, 48]. Interestingly in the case of B. subtilis ClpC, the second accessory domain (the M-domain) located between the two AAA+ domains, also plays an important role in the recognition of adaptor protein, however the details of substrate delivery by these adaptor proteins is currently unknown [36, 46–48]. For further details regarding the mechanism of action of ClpCP please refer to [3].
In the case of ClpX (and ClpE from Gram-positive bacteria) the N-terminal accessory domain (residues 1–60 in E. coli ClpX) is a C4-type Zinc binding domain (ZBD), which contains four Cysteine residues that coordinate a single Zn atom. In E. coli ClpX, this domain forms a very stable dimer [49], and is responsible for the recognition of several substrates (such as λO and MuA) but not SsrA-tagged proteins [50–52]. This domain is also essential for the recognition of the adaptor proteins, SspB [50, 52, 53] and UmuD [54], discussed in more detail later.
Given that E. coli ClpX is, by far the most extensively characterised Clp-ATPase, this section will focus primarily on the structure and function of ClpX. However, many of the features described here for the AAA+ domain of ClpX are likely to be generally applicable to most AAA+ proteases. At a structural level, the AAA+ domain (∼200–250 a.a.) is composed of two subdomains – a large N-terminal subdomain, which forms an α/β wedge-shaped Rossman fold and a small C-terminal subdomain, which forms a α-helical lid across the nucleotide-binding site [55, 56]. ATP is bound in a cleft between the large and small subdomain of a single subunit and the large subdomain of the adjacent subunit. As such, these interactions provide much of the driving force for formation of the hexamer. To date, several highly conserved sequence motifs have been identified within the AAA+ domain, each of which is responsible for a specific function [57]. The Walker A motif (GXXXXGK [T/S], where X = any amino acid) is required for ATP binding and facilitates oligomerization of the protein into ring-shaped hexamers. The Walker B motif (hhhhDE, where h = any hydrophobic amino acid) is required for hydrolysis of bound ATP and hence drives conformational changes in the protein, mediating substrate binding and translocation. The central pore of the hexamer is comprised of several important motifs and loops (e.g. the pore-1 loop) involved in substrate binding [58–61]. The Sensor 1 and 2 motifs, together with the arginine fingers, are proposed to couple the nucleotide-bound state of the oligomer with conformational changes in the subdomains, which through movement of the substrate-binding loops, results in substrate unfolding and translocation [55, 58]. Despite the broad sequence conservation of AAA+ domains, individual AAA+ domains appear to serve different functions in proteins that contain two or more AAA+ domains (i.e. ClpA or ClpC) [62]. For example, the first AAA+ domain (D1) in ClpA is crucial for oligomerisation while the second AAA+ domain (D2) is primarily responsible for ATP hydrolysis [63]. Interestingly, variants of ClpA lacking ATPase activity in either D1 or D2, are only able to process substrates with “intermediate” or “low” local stability respectively, suggesting that each domain can function independently, at least to a limited extent [64]. However, the ATPase activity of both domains is required for the efficient processing of substrates with “high” local stability [64] indicating that both domains work together to unfold and translocate substrates into ClpP.
As viewed from the top (or ClpP distal face) of the unfoldase, the ClpX hexamer can be divided into six units, each of which was composed of a small AAA+ subdomain from one subunit with a large AAA+ subdomain of the adjacent subunit [55, 56]. Recently, it was shown that the structures of all six of these units were highly superimposable [55] and hence it was proposed that each unit forms a functional rigid body (Fig. 1.4a, b). Despite the high degree of structural similarity between each rigid body unit, the overall shape of the ClpX hexamer is asymmetric, which suggests that the angle of the hinge between the rigid body units (i.e. the angle between the large and the small subdomains within a single subunit of ClpX) varies. This difference in the angle between the rigid body units results in a different ability of each subunit to bind nucleotide. Based on this description, each subunit within the ClpX hexamer can be classified into one of two groups; type 1 subunits, which are able to bind nucleotide (referred to as L, for “loadable”), and type 2 subunits, which are unable to bind nucleotide (referred to as U, for “unloadable”). In the crystal structure of ClpX, the hexamer is composed of four L (or type 1) subunits and two U (or type 2) subunits arranged in the following manner, L-L-U-L-L-U (Fig. 1.4c). Therefore, given that ATP binding and hydrolysis is expected to stabilise the L conformation, while the release of ADP is predicted to result in an transition from the L to the U conformation, it is proposed that the ATPase activity of ClpX will promote domain rotations within a subunit that will propagate around the hexamer and drive transition of the other subunits, in a chain reaction. These ATPase-induced conformational changes are proposed to form an integral part of the mechanism for substrate translocation by ClpX into ClpP (see later).
Oligomeric structure of ClpX. In the presence of nucleotide, ClpX forms a hexameric ring-shaped oligomer. (a) Surface representation of the ClpX hexamer (PDB: 3HWS). (b) Cartoon, illustrating the asymmetric organisation of the ClpX hexamer. (c) The asymmetric organisation of the ClpX hexamer results from a differential binding of nucleotide (nuc) within the hexamer. Nucleotides are bound in a cleft formed by the large and small domain of one subunit and the large domain of the adjacent subunit. Depending on the orientation of the small and large domain within a subunit, a subunit can be classified into two types; loadable (L) which are able to bind nucleotide and unloadable (U) which are unable to bind nucleotide. The arrangement of these different subunit types, within the ring gives rise to an asymmetric appearance of the hexamer
The Unfoldase-Peptidase Complex
Given that the AAA+ unfoldase component (i.e. ClpX, ClpA or ClpC) is hexameric and the associated peptidase (e.g. ClpP) is formed by two heptameric rings, the resulting proteolytic machines, ClpXP (ClpAP and ClpCP), exhibit an asymmetry between the two components. This asymmetry, although not unique in biology, poses several interesting questions. How do the two components interact with one-another? How many of these features per hexamer (i.e. how many subunits) are required for formation of a functional complex? Not surprisingly, the formation of the complex is transient, and efficient interaction of the two components is dependent on nucleotide-bound state of the unfoldase. Specifically, formation of the ClpXP complex is only supported by ATP, ATPγS (a slowly hydrolysable analogue of ATP) or a ClpX mutant that is defective in ATP hydrolysis [65]. In contrast, the complex dissociates in the presence of ADP or in the absence of nucleotide [66, 67]. This interaction, (i.e. between the two components), is mediated by two sets of contacts; one at the periphery of the interface and the other near the central pore. The peripheral contact occurs between a flexible loop on ClpX and a hydrophobic pocket on the surface of ClpP, and is important for a strong, nucleotide-independent interaction with ClpP. The flexible loop contains a conserved tripeptide motif ([L/I/V]-G-[F/L]) and as such is often referred to as the IGF/L-loop (Fig. 1.5a). This motif is unique to ClpP-binding unfoldases (i.e. ClpA, ClpC, ClpE and ClpX) and is essential for interaction with ClpP [68, 69]. Consistently, mutation of this motif dramatically reduces the affinity of ClpX to ClpP [67, 68]. The second contact occurs between two loops; one loop (termed the pore-2 loop) protrudes from the axial pore of ClpX, and interacts with the N-terminal loop of ClpP [21, 70, 71]. This interaction, between the two axial loops, appears to be highly dynamic and is dependent on the nucleotide-state of individual subunits of ClpX [71]. Although the ClpXP complex is asymmetric, both sets of loops (the IGF/L-loop, for docking into the hydrophobic pocket on ClpP and the two axial pore loops) appear to be flexible enough that contacts from each subunit of ClpX contribute to the interaction. Indeed loss of a single IGF-loop, within the ClpX hexamer, is sufficient to reduce ClpP binding and activity, while loss of more than one contact per hexamer completely abolishes ClpP binding [71].
ClpP-binding and substrate interaction is mediated by several loops and pockets. (a) Cut-away view of ClpX (blue), highlighting the important interactions that contribute to complex formation with ClpP (red). The IGF/L loops (green) on ClpX form a static interaction with the hydrophobic pocket on ClpP (black). ClpXP complex formation is modulated by the nucleotide state of ClpX, through a set of dynamic interactions, between pore-2 loops of ClpX (red) and the N-terminal loop of ClpP (purple). (b) The substrate is recognised and translocated through the pore via a set of conserved pore loops; RKH (blue), pore-1 (yellow) and pore-2 (red). These loops move up and down the pore of ClpX in a nucleotide-dependent fashion, thereby translocating the substrate into ClpP
Degradation Recognition Motifs (Degrons)
A bacterial cell is composed of thousands of different proteins, the concentration (or copy number) of which varies dramatically (from ∼100 to 105 molecules per cell) [72]. Likewise, the concentration of each individual protein varies in response to changing environmental conditions or stress. As such, in order for the cell to maintain optimal function, not only under normal conditions but also under conditions of stress, the composition and active concentration of its proteins must be monitored and maintained. Hence it is important for the cell to specifically remove unwanted or damaged proteins from the cell when they are no longer required. To achieve this, bacterial proteases need to combine two seemingly incompatible properties, broad recognition of a range of different protein substrates, with a high degree of substrate specificity to prevent the recognition of properly folded or wanted cellular proteins.
A key feature of most, if not all, bacterial protein substrates is the presence of a specific amino acid motif, often referred to as a degradation tag or degron [73]. These degrons are generally located at the N- or C-terminus of the protein, although in some cases they are located internally. Although most degrons are intrinsic to the target protein, a handful of degrons (e.g. the SsrA tag and some N-end rule substrates) are not defined by the primary sequence of the protein, but rather are added (either co- or post-translationaly) to the protein [74, 75]. Often, intrinsic degrons are only revealed (for recognition by the protease) following exposure of the protein to stress (e.g. heat-shock) or processing by an endoprotease [76–79]. This conditional recognition of a protein substrate is ideally suited to the controlled degradation of a key regulatory protein, and forms the basis of controlling several stress response pathways in bacteria (see [6]). In some cases however, a degron may be constitutively exposed under normal conditions, in order to maintain low levels of the protein (e.g. SigmaS) [80].
Trans-translation and the SsrA-Tag: A Specific Protein Tagging System in Bacteria
Messenger RNA molecules normally contain a stop codon at the 3′ end of the transcript, which serves not only to signal the end of translation, but also triggers ribosome dissociation. In some cases however, as a result of truncation of the mRNA or errors during its transcription, the lack of a stop codon in the mRNA sequence caused “stalling” of protein synthesis [81–83]. To overcome this problem, bacteria possess a conserved mechanism, to restart translation and allow ribosome dissociation. This mechanism (illustrated in Fig. 1.6), often referred to as trans-translation, is sensed by an empty A-site and signalled by stalling of the translating ribosome [84]. This signal results in the recruitment of a specialised RNA molecule into the empty A-site of the ribosome. This RNA, encoded by ssrA (small stable RNA gene A) [85] has been termed a tmRNA as it functions both as a tRNA and as an mRNA [84, 86, 87]. The tRNA-like structure can be charged with alanine at its 3′ end, while an extended loop within the same RNA molecule encodes a short open reading frame (ten amino acids in E. coli) that ends in a stop codon. Following docking of the charged tmRNA into the empty A-site, the alanine is transferred to the nascent polypeptide and the open reading frame (encoded by the mRNA portion of the tmRNA) is translated. Noteworthy, trans-translation results in the attachment of a short C-terminal extension (termed the SsrA tag) to the incompletely synthesised protein.
Cartoon, illustrating the process of trans-translation. 1. Truncated mRNA (lacking a stop codon) cause “stalling” of the ribosome. 2. This “stalling” triggers binding of a tmRNA into the empty A-site of the ribosome. 3. Following a transpeptidation reaction, the truncated mRNA is replace with the mRNA from the tmRNA and 4. translation proceeds, resulting in 5. correct termination of protein synthesis 6. rescuing the ribosome and releasing the “tagged” protein for targeted degradation by ClpXP
Importantly, given that SsrA-tagged proteins are produced from aberrant or incomplete mRNA, it is unlikely that they will be able to fold. For this reason, interaction of SsrA-tagged proteins with chaperones is wasteful, as attempts to refold trans-translation products would be futile. Rather, SsrA-tagged proteins are rapidly degraded by proteases. In E. coli, the SsrA tag is 11 amino acids long (AANDENYALAA) and substrates tagged with the sequence are recognised by ClpXP, ClpAP and FtsH [81, 88–90]. Despite the fact that the SsrA tag is recognised by several different proteases in vitro, the in vivo degradation of these substrates is almost exclusively performed by ClpXP [81, 91].
Nevertheless, this tag has been used extensively as a model degron to study the function of both ClpXP and ClpAP. As such, it has proved to be a powerful research tool to study the mechanism of protein recognition and degradation by AAA+ proteases. A major advantage of the SsrA tag, as a research tool to study protein degradation, is that any protein can be converted into a ClpXP (or ClpAP) substrate, simply through the attachment of the SsrA tag to its C-terminus. This has permitted a detailed mechanistic analysis of protein degradation using a range of different substrates with a variety of unique or desired features (i.e. green fluorescent protein (GFP) or the I27 domain of the human titin) to examine unfolding [92–95]. Likewise, it has also served as an excellent tool to study the mechanism of adaptor-mediated substrate delivery (see below).
Other ClpX Recognition Motifs
Apart from the specific recognition of the SsrA-tag, ClpX is also involved in the recognition of several other proteins, including a number of proteins involved in various stress response pathways. In order to determine the complete substrate-binding repertoire of E. coli ClpX, a mutant version of ClpP was used to capture the physiological substrates of ClpXP in vivo [96]. Using this approach, ∼100 putative ClpXP substrate proteins were identified [96, 97]. Following verification of several of these proteins (either by in vitro or in vivo degradation assays) five different ClpX “recognition” motifs were proposed [96]. Of the five different “recognition” motifs, two were located near the C-terminus of the protein and three near the N-terminus of the protein (Fig. 1.7). While both classes of C-terminal motifs (C-motif 1 and 2, Fig. 1.7) shared homology with known ClpXP substrates (i.e. the SsrA-tag and MuA, respectively), only a single N-terminal motif (N-motif 1, Fig. 1.7) had been observed previously (i.e. λO) [98].
Substrate-binding motifs for ClpX and ClpA. In general, AAA+ proteases recognise either the N- or C-terminus of a substrate, as such several motifs have been defined for both ClpX and ClpA. ClpX Substrate recognition by can be divided into five broad groups, three N-terminal motifs (N motif-1, -2 and -3) and two C-terminal motifs (C motif-1 and -2). In contrast only two ClpA recognition motifs have been observed for ClpA (N-degron and C-degron)
Interestingly, the various degradation motifs appear to be recognised by different regions within the unfoldase. Some substrate classes (e.g. N-motif 1) strictly depend on interaction with the N-terminal domain, while other motif classes (e.g. C-motif 1, i.e. SsrA-tagged substrates) do not require this domain for direct recognition [50, 52, 69]. For example, λO (a replication protein of bacteriophage λ) carries an N-terminal degradation motif (N-motif 1, NH2-TNTAKI), which is specifically recognised by the N-terminal domain of ClpX [52, 96, 99]. Indeed deletion of this domain (from ClpX) inhibits the ClpP-mediated degradation of λO [52], which is proposed to result from the low affinity of this class of substrate to the axial loops on ClpX. Tethering of this class of substrate, by the N-terminal domain, is likely to increase the effective concentration of the substrate, near the pore of ClpX. As a result, despite their low affinity to the pore loops, high affinity to the N-terminal domain promotes their engagement by the pore and, consequently, their efficient degradation. The N-terminal domain is also involved in the recognition of the adaptors proteins, SspB and UmuD, and substrate proteins such as MuA (C-motif 2, Fig. 1.7), which appear to share a conserved motif [50, 52, 54]. Importantly however, the adaptor proteins are not degraded by ClpXP, presumably because they are not recognised by the pore-1 motif of ClpX.
Other Degradation Tags
Currently, the substrate recognition motifs for ClpA are only poorly defined. The first ClpAP substrate to be identified was ClpA itself [100]. Interestingly, although the recognition motif within ClpA was originally proposed to be located at the N-terminus of ClpA, it was later shown to be C-terminal [101]. Interestingly, this motif within ClpA shares some similarity with the, well characterised, model degron – the SsrA tag (Fig. 1.7). ClpA has also been shown to recognise proteins via an N-terminal recognition motif but not an internal motif [102, 103]. The N-terminal recognition motifs can be classified into two groups, those that require the adaptor protein (ClpS) – N-end rule substrates, and those that do not. Currently, only a single substrate containing an N-terminal recognition sequence has been identified [104], and consequently a motif has not been defined. In contrast, several N-end rule substrates, both natural and model substrates have been identified and hence a motif for ClpA binding of these substrates has been proposed [74, 78]. The ClpA recognition motif within N-end rule substrates is a dihydrophobic element, located between five and nine residues from the primary destabilising residue at the N-terminus of the protein [74, 105]. Interestingly, one of the N-end rule substrates, Dps (DNA protection during starvation), which protects DNA from reactive oxygen species, contains two N-terminal recognition motifs. One motif is created after endoproteolytic cleavage of the first five residues of Dps, to generate Dps6–167 and is required for recognition by ClpS and ClpA [78, 79], the other N-terminal motif is created following cleavage of the N-terminal Met by methionine aminopeptidase (MetAP), to generate Dps2–167 which contains a ClpX (Nmotif-1) within the first five residues of Dps [96].
Substrate Recognition by AAA+ Proteases (Direct Recognition Versus Indirect or Adaptor Mediated Recognition)
Although the recognition of most protein substrates occurs by direct interaction with the unfoldase, some protein substrates require additional recognition factors to direct them to the protease for degradation. In the following sections, we will describe the molecular details of substrate recognition by the unfoldase and/or delivery by adaptor proteins, using a number of well-characterised examples.
Direct Recognition by ClpX (e.g. Recognition of SsrA Tagged Proteins)
In E. coli, the SsrA tag is composed of 11 amino acids (AANDENYALAA), however recognition of this tag by ClpX, only requires the last two alanines and the C-terminal α-carboxylate [106]. In contrast to some ClpX substrates (e.g. λO), recognition of the SsrA-tag by ClpX, does not involve the N-terminal domain. Consistent with the idea, removal of the N-domain of ClpX, did not alter the ClpP-mediated degradation of SsrA-tagged proteins [50, 52]. Rather, the SsrA-tag is specifically recognised by loops in, or near to, the axial pore of the AAA+ module. Indeed, three sets of pore loops in ClpX (RKH, pore-1 and pore-2, see Fig. 1.5) have been implicated in binding the SsrA tag [71, 107]. The RKH loops, as the name suggests, contains the tripeptide motif (RKH), which surrounds the entrance to the ClpX pore. The positively charged RKH loops are proposed to attract negatively charged sequences (i.e. the charged C-terminal α-carboxylate of the SsrA-tag) to the pore of ClpX [99]. Accordingly, mutations that reduce the positive charge of the RKH loop, reduced binding to SsrA-tagged proteins (or substrates containing a C motif-1), whilst simultaneously improved the binding of substrates containing a positively charged motif [99]. The pore-1 and pore-2 loops, in contrast to the RKH loop, interact with the two last alanine residues of the ssrA-tag [107, 108]. The pore-1 loop of ClpX contains the highly conserved GYVG motif, which plays a central role in substrate translocation across the pore and into the degradation chamber [59–61, 109]. Based on a number of mutations and series of crosslinking experiments, the pore-2 loops were shown to specifically interact with the terminal alanines of the SsrA-tag [108]. Interestingly, neither the RKH nor the pore-2 loops are conserved in human mitochondrial ClpX [108]. As such, human ClpX is unable to recognise proteins tagged with the E. coli SsrA tag. However, a crucial role for these loops in the recognition of SsrA-tagged was elegantly demonstrated by Sauer and colleagues by grafting the E. coli ClpX RKH and pore-2 loops onto human ClpX creating a chimeric ClpX protein [108]. Strikingly, when both the RKH loops and pore-2 loops from E. coli ClpX were grafted onto human ClpX, the resulting chimeric proteins was able, not only to recognize the SsrA-tagged substrates but also to deliver them to ClpP for degradation [108]. Interestingly, grafting of only the RKH or pore-2 loop, was insufficient to promote recognition of the SsrA-tag. Collectively, these results demonstrated the importance of both pore loops in the recognition of SsrA-tagged substrates.
Indirect Recognition (Adaptor Mediated Recognition)
As mentioned above, the recognition of some protein substrates by the unfoldase, either requires or is modulated by an additional component – known as an adaptor protein. In general, an adaptor protein acts as a bridge between the substrate and the unfoldase. As such, adaptor proteins invariably exhibit two separate activities; (i) substrate recognition and (ii) unfoldase docking, however in some cases the adaptor protein is also proposed to activate either the substrate or the unfoldase for delivery to the protease for degradation [42, 110, 111]. Typically, the adaptor protein is released and recycled in this process without being degraded, although in some cases the adaptor protein (e.g. MecA) is also degraded by the protease complex (i.e. ClpCP), which acts a negative feedback loop to control the turnover of the substrates delivered by this adaptor protein. To date, four adaptor proteins have been identified in E. coli, three of which (SspB, UmuD and RssB) deliver specific protein substrates to ClpXP [54, 112, 113] while a single adaptor protein (ClpS) is required for the delivery of a specific class of substrates to ClpAP [40, 114]. SspB increases the affinity of ClpX to SsrA tagged proteins [112]. RssB is essential in bacteria for ClpXP-mediated degradation of the stationary phase sigma factor, σS [113, 115, 116]. Interestingly, four adaptor proteins have also been identified in B. subtilis. However, in contrast to the adaptor proteins from E. coli, the vast majority of B. subtilis adaptor proteins (MecA, McsB and YpbH) function together with ClpC [45, 117–121], and only a single adaptor protein (YjbH) has been identified to function with ClpX [122, 123]. Surprisingly, with the exception of MecA and YpbH, the remaining adaptor proteins share little, to no, sequence homology and hence each adaptor protein is likely to function via a unique mechanism.
SspB (A Multi-functional Adaptor Protein)
SspB is certainly the best characterised ClpX adaptor protein and arguably the best characterised bacterial adaptor protein to be studied. It was first identified, in E. coli as a ribosome-interacting protein that specifically modulates the ClpXP-mediated turnover of SsrA-tagged proteins [112]. Subsequently, SspB was also shown to recognise and deliver another ClpX substrate (i.e. RseA) for ClpP-mediated degradation [76] for a recent review see [6]. Although the distribution of SspB homologs is largely limited to γ- and β-proteobacteria an ortholog of SspB, termed SspBα has also been identified Caulobacter crescentus and other α-proteobacteria [124, 125]. Interestingly, despite the poor sequence homology, the overall fold of SspBα is similar to E. coli SspB [124]. Nevertheless, in contrast to E. coli SspB, C. crescentus SspBα appears to be optimised for binding to the SsrA-tag. In the case of E. coli SspB, the protein is composed of two functional regions separated by a long unstructured segment (∼40–50 residues long). The N-terminal region of SspB (∼110–120 residues long) forms a dimeric module, which is involved in binding of the SsrA-tag [50, 126, 127]. This substrate-binding domain is tethered to ClpX, via a short motif (termed the ClpX-binding region (XBR)), located at the C-terminus of SspB [50, 127]. The XBR of SspB forms an anti-parallel β-sheet with the N-terminal ZBD of ClpX [53]. Indeed, it has been proposed that both XBRs (from the SspB dimer) interact simultaneously with two ZBDs on ClpX – a mode of attachment that places the SspB-bound cargo in an ideal position for interacting with the pore residues in the ClpX hexamer. Hence, SspB tethers the substrate to ClpX thereby increasing the local concentration of SsrA-tagged substrates near the ClpX pore [50, 53, 127–129] (Fig. 1.8). Importantly, both the unfoldase and the adaptor protein recognise exclusive regions within the SsrA tag (AANDENYALAA). The unfoldase recognises the AA motif (C motif-1) at the C-terminus of the SsrA tag (Fig. 1.7), while SspB binds towards the N-terminal end of the SsrA-tag (AANDxxY). In contrast, the ClpA binding motif within the SsrA-tag (AAxxxxxALA) overlaps with the SspB binding and as such SspB inhibits the ClpAP-mediated degradation of SsrA-tagged substrates [50, 130]. As a consequence, SspB-mediated tethering of the SsrA-tag to ClpX results in an increased affinity of the substrate for ClpX, and hence an improved rate of degradation [129]. As such, SspB is likely to play an important role in the delivery of substrates present at low concentrations as tethering to ClpX, effectively increases the local concentration of the substrate. Consistent with this substrate-tethering model, mutation of the ClpX recognition motif within the SsrA-tag (i.e. replacement of LAA with DAS, termed the DAS-tag), significantly reduce the ClpXP-mediated degradation of substrates bearing this tag, while, the addition of SspB improved the recognition and degradation of substrates bearing this modified DAS-tag by more than 100-fold [131].
Adaptor proteins. (a) ClpS (tan) contains a small substrate-binding pocket for the recognition of proteins bearing a primary destabilising residue at their N-terminus. This binding pocket exhibits exquisite specificity and not only forms a number of critical H-bonds with the α-amino group of the N-terminal residue of the substrate (blue), but also forms hydrophobic interactions with the side-chain of the N-terminal residue. The substrate extends away from ClpS and reaches towards the unfoldase, binding to ClpA is proposed to occur through the dihydrophobic (hh) element. (b) SspB (pink) forms a more permisscuous peptide-binding groove, which can accommodate peptides in different orientations. In the case of SsrA-tagged proteins, the substrate extends away from SspB and towards the pore loops of ClpX, which interact with the LAA motif. (c–d) Both ClpS (tan) and MecA (pink) interact with the N-terminal domain of ClpA (blue) and ClpC (light blue), respectively. An α-helix within the adaptor protein, contains a critical Glu residue which projects into the conserved pocket within the appropriate N-domain
RssB
In contrast to SspB, RssB is a dedicated adaptor protein that is uniquely responsible for the recognition of a single substrate – the general stress transcription factor SigmaS (σS, also referred to as RpoS). RssB (also known as SprE (stationary phase regulator) in E. coli or MviA in Salmonella typhimurium) was first identified in E. coli using a genetic screen to discover genes involved in the RpoS expression and/or activity [115, 116]. RssB is a member of the two-component response regulator family and was the first family member to be shown to play a role in protein turnover. As a member of the response regulator family, RssB is phosphorylated on a highly conserved aspartate residue (D58). Phosphorylation of RssB at D58, stimulates binding of σS, resulting in the formation of a stable 1:1 complex [132]. Mutations that inhibit phosphorylation of RssB result in reduced binding to σS, and hence an increased stability of σS, both in vitro and in vivo. In contrast to SspB, which merely enhances the kinetics of substrate recognition, RssB is essential for the recognition of σS by ClpX [113]. Indeed it has been proposed that binding of RssB to σS triggers a conformational change in σS, which exposes a previously concealed ClpX recognition motif, however the mechanistic details of such a model are yet to be confirmed. For further details on the proteolytic control of the general stress response in bacteria refer to [7].
UmuD
The third and final, ClpX-specific adaptor protein in E. coli, is UmuD. In response to DNA damage, the first 24 residues of UmuD are auto-catalytically cleaved, in a RecA-dependent fashion. Following cleavage, the resulting protein (termed UmuD′), can form both homo- and hetero-oligomers [133]. As a heterodimer, UmuD/UmuD′ forms a component of the error-prone DNA polymerase V, which is able to bypass DNA lesions in the process of DNA replication and hence facilitates the cells recovery following DNA damage. Since this activity is necessary at times of DNA damage, but toxic under normal growth conditions, it is important that the cellular levels of UmuD′ (and hence UmuD/D′ oligomers) be carefully controlled during and after recovery. Indeed, this is elegantly achieved by the cell, as the N-terminal region of UmuD serves as a ClpX tethering sequence for delivery of UmuD′ to ClpXP when present in an UmuD/D′ complex [134]. Like the XBR of SspB, this region can bind to the ZBD of ClpX, but not as a degradation tag rather as a specific adaptor protein for the delivery of UmuD [54, 134].
Indirect Recognition by ClpA (Recognition of N-End Rule Substrates)
ClpS and the N-End Rule (A Specific ClpAP-Mediated Substrate)
In contrast to ClpX, which uses three different E. coli adaptor proteins, only a single adaptor protein (ClpS) has been identified for ClpA. Although ClpA appears to use only a single adaptor protein, this adaptor protein exhibits broad activity over its cognate unfoldase. Indeed, ClpS is able to regulate ClpA substrate selection, both negatively and positively [40, 114]. Originally identified as an inhibitor of ClpA auto-degradation, both in vitro and in vivo, and a negative regulator of ClpAP-mediated degradation of substrates bearing an SsrA-tag [40], ClpS was also shown to be an essential component of the N-end rule pathway [114].
The N-end rule pathway, originally identified in Saccharomyces cerevisiae, by Alexander Varshavsky’s lab, is a highly conserved protein degradation pathway that is responsible for the recognition and degradation of proteins bearing a specific “destabilising” residue at the N-terminus [135, 136]. This pathway determines the half-life of a protein based on the N-terminal residue of that protein, which may be classified as “stabilising” or “destabilising”. To date, this pathway has been identified in bacteria, plants and mammals and although the details of the various pathways differ, from one organism to the next, each pathway shares a number of common principals [135, 137–139]. In E. coli, like other organisms, the pathway is hierarchical, and destabilising residues can be separated into two classes (primary and secondary) [74]. Primary destabilising residues (L, F, W and Y) are recognised directly by the bacterial N-recognin, ClpS [114], while secondary destabilising residues (R, K and M) must first be converted to primary destabilising residues by the enzyme Leu/Phe-tRNA-protein transfersase (LFTR) before they are recognised by the adaptor protein [78, 140]. Interestingly, the first clue for a role of ClpS in the N-end rule pathway came from the structure of ClpS and comparison to the secondary structure of the human N-end rule recognition component (N-recognin), the E3-ligase, UBR1 [42, 141]. From this bioinformatic analysis, despite very low sequence homology, Lupas and colleagues proposed that ClpS was involved in the N-end rule pathway in bacteria [141]. Consistently, the crystal structure of ClpS (in complex with the N domain of ClpA) identified two conserved regions, one for interaction with the N-domain of ClpA and the other proposed to be involved in a substrate interaction [41, 42]. Subsequent biochemical and structural analysis confirmed that ClpS was indeed essential for the recognition of N-end rule substrates and that the second conserved region within ClpS was the N-degron binding site [142–144].
ClpS, like most characterised adaptor proteins is a small protein composed of two regions. The C-terminal domain of ClpS is the “workhorse” of the protein, it is responsible, not only for recognition of the substrate but also for docking to the N-terminal domain of ClpA [40–42, 114]. Despite both of these functions being located on the C-terminal domain of ClpS, this domain alone is neither sufficient for the inhibition of substrates bearing an SsrA-tag nor the delivery of N-end rule substrates [41, 110], suggesting that the N-terminal region of ClpS plays a crucial role in activation of ClpA. Hence in contrast to SspB, which merely modulates the affinity of ClpX for recognition of the SsrA-tag, the adaptor protein ClpS alters the substrate specificity of ClpA, by activating the unfoldase for recognition of N-degron bearing substrates. In summary, a substrate bearing an N-terminal primary destabilising residue, is bound by a small hydrophobic pocket on the surface of ClpS (Fig. 1.8). Importantly, this pocket exhibits exquisite specificity – it forms a number of important hydrogen bonds with both the α-amino group of the N-terminal residue and the carbonyl oxygen of the peptide bond, as well as several hydrophobic interactions with the side chain of the N-terminal amino acid [142–144]. Following recognition of the substrate by the adaptor protein, the substrate-ClpS complex docks to the N-terminal domain of ClpA [40–42]. Next, the N-terminal region is proposed to activate, an as yet undefined region of ClpA, for recognition of the N-degron bearing substrate [110]. The unfoldase (ClpA), then recognises a hydrophobic region in the substrate approximately ∼5–9 residues downstream of the primary destabilising residue (Fig. 1.7) [74, 78, 105].
MecA
Of the three known ClpC-adaptor proteins, MecA is currently the best characterised. It was first discovered in a genetic screen for repressors of competence development (which is a physiological state that permits B. subtilis cells to actively import DNA). In non-competent cells, the “competence” transcription factor (ComK) is recognised by MecA and targeted for degradation by ClpCP. Competence is triggered by the accumulation of a small peptide (ComS), which binds to MecA and thereby inhibits the MecA-dependent degradation of ComK by ClpCP [145]. Interestingly, MecA is not only involved in the development of competence through the regulated degradation of ComK but has also been proposed to be involved in general protein quality control, through the ClpCP-mediated degradation of misfolded and aggregated proteins [120]. Similar to most other adaptor proteins, MecA is composed of two regions an N-terminal domain, which is responsible for substrate recognition (i.e. ComK and ComS) and a C-terminal domain, which is required for docking to the unfoldase [146]. Interestingly, in contrast to other characterised adaptor proteins, docking of MecA (and hence substrate delivery to the protease) requires both the N-domain and the M-domain of ClpC [36, 48]. Despite the additional requirement for MecA binding to ClpC (i.e. to the M-domain), the mode of docking of MecA to the N-domain is strikingly similar to that of ClpS with the N-domain of ClpA [41, 42]. Indeed both adaptor proteins (ClpS and MecA) use a single α-helix to interact with the same region of the N-domain, and stabilise the complex by the formation of several H-bonds (Fig. 1.8). Interestingly, MecA is absent in cyanobacteria and ClpC was shown to cooperate with the adaptor protein ClpS [147]. Consistently, the distribution of MecA and ClpS appears to be mutually exclusive throughout evolution.
Substrate Processing by AAA+ Proteins
Substrate translocation is a basic mechanical process of all AAA+ proteins. This process is performed solely by the AAA+ module of the unfoldase, and like substrate binding has been extensively studied using both ClpA and ClpX as a model AAA+ protein. In recent years however, there have been many advances in this area of research by several researchers, including numerous contributions by the laboratory of Robert Sauer and Tania Baker to study the basic mechanism of action of ClpX.
Substrate Unfolding and Translocation
In order for a folded protein to enter the degradation chamber of ClpP, it must first be unfolded by the ATPase component. Although the pore of ClpX is large enough to simultaneously accommodate two or three peptide chains, most folded proteins are too large to enter [148]. As such, the narrow size of the ClpX pore prevents diffusion of folded proteins through ClpX and hence prevents the uncontrolled degradation of folded proteins by the ClpXP protease. Therefore protein substrates must first be unfolded, to enter the proteolytic chamber of ClpP. To achieve this, the unfoldase component converts the energy released from the binding and hydrolysis of ATP into a pulling force. This pulling force is responsible for the global unfolding of the substrate by threading it through the unfoldase pore, and into the degradation chamber of ClpP in a vectorial manner [95, 149].
The pulling force, generated by nucleotide-driven changes in the structure of ClpX is proposed to be transmitted to the substrate via movement of the pore-1 loops (Fig. 1.7). Indeed, the location of each pore-1 loop, within the central pore of the ClpX hexamer was shown to vary depending on the nucleotide bound state of the subunit [55]. As such, it has been proposed that ATP binding and hydrolysis drives conformational changes in the unfoldase, which result in movement of the pore-1 loops up and down the central pore [107, 150]. As these loops (in particular the highly conserved tyrosine residue) interact with the substrate’s polypeptide chain, their movement along the pore provides the pulling force that is necessary for substrate unfolding and translocation. Do the subunits in a ClpX hexamer have to operate in a concerted fashion to promote successful unfolding and translocation of protein substrates? To examine this question Sauer and colleagues employed a method first used by the lab of Art Horwich, to study the role of individual subunits in GroEL [151]. In this case however, Sauer and colleagues created a single-chain hexamer of ClpX (lacking the N-terminal ZBD) by fusing six copies of clpX (lacking the sequence coding for the N-terminal ZBD) into a single gene [152]. This elegant experimental setup allowed the incorporation of specific mutations into various different ClpX subunits within the hexamer. Specifically, mutations in either the Walker B motif (E185Q) – which prevents ATP hydrolysis, or the sensor-2 motif (R370K) – which prevents both ATP hydrolysis and uncouples conformational changes linked to ClpP- and substrate-binding, were combined with wild type subunits and the degradation of SsrA-tagged substrates was examined [152]. Consequently, ClpX hexamers were created which contained either a single wild type subunit, or two wild type subunits and so on. Remarkably, the degradation rate increased linearly with the amount of the wild type subunits in a hexamer. For example, a ClpX hexamer that contained one wild type subunit led to degradation of an SsrA-tagged substrate 17% as fast as a wild type hexamer. In accordance, two wild type subunits in a hexamer resulted in a degradation rate that was 30% of that observed for a single-chain hexamer that was constructed of six wild type subunits. Moreover, in all cases, degradation was performed at a similar energetic cost (i.e. the amount of ATP hydrolyzed per substrate). These results indicated that even a single active subunit in a hexamer can promote efficient unfolding and translocation of substrates by ClpX and that concerted or sequential activity of multiple subunits is not essential for degradation.
Degradation of folded substrates proceeds much slower than degradation of unfolded substrates, suggesting that substrate unfolding is a rate-limiting step for proteolysis by AAA+ proteases. This principle was elegantly demonstrated using the I27 domain of human titin as a substrate [92]. Titin-I27 was converted into a ClpX substrate by creating a genetic fusion of titin-I27 with SsrA. The I27 domain of titin is extremely resistant to mechanical unfolding [153] and consistent with this, its degradation by ClpXP is relatively slow [92, 94]. Remarkably, an unfolded variant of titin-I27-SsrA can be obtained, simply by carboxymethylation of its two cysteine residues [92]. This simple chemical modification completely unfolds titin-I27 without altering its solubility. This permitted a direct comparison of the degradation kinetics of the substrate with respect to its folded state (i.e. either stably folded or unfolded). Interestingly, both the folded and unfolded substrates had a K M of ∼1 μM, similar to that observed for other SsrA tagged proteins. By contrast, the energetic cost for degradation of different substrates by ClpXP varied dramatically. For instance, the degradation of native titin-I27 required ∼600 molecules of ATP, while the degradation of an unfolded mutant of titin-I27 only required ∼100 molecules of ATP, suggesting that the cost of titin-I27 unfolding is ∼500 ATP. Interestingly however, the rates of degradation do not correlate with the global thermodyamic stability of a substrate but rather seem to depend on the local stability of the region to which the recognition tag is attached [94, 154, 155]. In summary, the current model suggests that following binding, ClpX pulls on the degradation tag in an attempt to unfold the substrate. In some cases unfolding of the substrate may require multiple rounds of ATP hydrolysis until a power stroke of ClpX coincides with transient unfolding of a structural element near the degradation tag. When this occurs, ClpX can initiate substrate translocation and complete substrate unfolding very rapidly and with a high degree of cooperativity. As mentioned previously substrates may be recognised from either an internal site or from the N- or C-terminus of the protein [46]. Not surprisingly, substrate translocation may occur in either direction (i.e. from N-terminus to C-terminus or visa versa). Strikingly, single molecule experiments indicate that substrate unfolding eventually results from a single ClpX power stroke [154]. Following the initial unfolding event, substrate translocation proceeds rapidly and without considerable specificity [156]. Indeed, ClpX was shown to efficiently translocate a variety of different polymers, including homopolymeric tracts of glycine, proline and lysine non-amino-acid aliphatic chains. In addition, ClpX can carry out translocation of a polypeptide from the N-terminus to the C-terminus as efficiently as in the opposite direction.
References
Sauer RT, Baker TA (2011) AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem 80:587–612
Samanovic M, Li H, Darwin KH (2013) The Pup-proteasome system of Mycobacterium tuberculosis. In: Dougan DA (ed) Regulated proteolysis in microorganisms. Springer, Subcell Biochem 66:267–295
Molière N, Turgay K (2013) General and regulatory proteolysis in Bacillus subtilis. In: Dougan DA (ed) Regulated proteolysis in microorganisms. Springer, Subcell Biochem 66:73–103
Gur E (2013) The Lon AAA+ protease. In: Dougan DA (ed) Regulated proteolysis in microorganisms. Springer, Subcell Biochem 66:35–51
Okuno T, Ogura T (2013) FtsH protease-mediated regulation of various cellular functions. In: Dougan DA (ed) Regulated proteolysis in microorganisms. Springer, Subcell Biochem 66:53–69
Barchinger SE, Ades SE (2013) Regulated proteolysis: control of the Escherichia coli σE-dependent cell envelope stress response. In: Dougan DA (ed) Regulated proteolysis in microorganisms. Springer, Subcell Biochem 66:129–160
Micevski D, Dougan DA (2013) Proteolytic regulation of stress response pathways in Escherichia coli. In: Dougan DA (ed) Regulated proteolysis in microorganisms. Springer, Subcell Biochem 66:105–128
Frees D, Brøndsted L, Ingmer H (2013) Bacterial proteases and virulence. In: Dougan DA (ed) Regulated proteolysis in microorganisms. Springer, Subcell Biochem 66:161–192
Kirstein-Miles J, Morimoto RI (2010) Caenorhabditis elegans as a model system to study intercompartmental proteostasis: interrelation of mitochondrial function, longevity, and neurodegenerative diseases. Dev Dyn 239(5):1529–1538
Truscott KN, Bezawork-Geleta A, Dougan DA (2011) Unfolded protein responses in bacteria and mitochondria: a central role for the ClpXP machine. IUBMB Life 63(11):955–963
Kwasniak M, Pogorzelec L, Migdal I, Smakowska E et al (2012) Proteolytic system of plant mitochondria. Physiol Plant 145(1):187–195
Truscott KN, Lowth BR, Strack PR, Dougan DA (2010) Diverse functions of mitochondrial AAA+ proteins: protein activation, disaggregation, and degradation. Biochem Cell Biol 88(1):97–108
van Dyck L, Dembowski M, Neupert W, Langer T (1998) Mcx1p, a ClpX homologue in mitochondria of Saccharomyces cerevisiae. FEBS Lett 438(3):250–254
Voos W, Ward L, Truscott KN (2013) The role of AAA+ proteases in mitochondrial protein biogenesis, homeostasis and activity control. In: Dougan DA (ed) Regulated proteolysis in microorganisms. Springer, Subcell Biochem 66:223–263
Clarke AK (2012) The chloroplast ATP-dependent Clp protease in vascular plants – new dimensions and future challenges. Physiol Plant 145(1):235–244
Olinares PD, Kim J, van Wijk KJ (2011) The Clp protease system; a central component of the chloroplast protease network. Biochim Biophys Acta 1807(8):999–1011
Bochtler M, Hartmann C, Song HK, Bourenkov GP et al (2000) The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature 403(6771):800–805
Sousa MC, Trame CB, Tsuruta H, Wilbanks SM et al (2000) Crystal and solution structures of an HslUV protease-chaperone complex. Cell 103(4):633–643
Maglica Z, Kolygo K, Weber-Ban E (2009) Optimal efficiency of ClpAP and ClpXP chaperone-proteases is achieved by architectural symmetry. Structure 17(4):508–516
Wang J, Hartling JA, Flanagan JM (1997) The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis. Cell 91(4):447–456
Gribun A, Kimber MS, Ching R, Sprangers R et al (2005) The ClpP double ring tetradecameric protease exhibits plastic ring-ring interactions, and the N termini of its subunits form flexible loops that are essential for ClpXP and ClpAP complex formation. J Biol Chem 280(16):16185–16196
Sprangers R, Gribun A, Hwang PM, Houry WA et al (2005) Quantitative NMR spectroscopy of supramolecular complexes: dynamic side pores in ClpP are important for product release. Proc Natl Acad Sci U S A 102(46):16678–16683
Maurizi MR, Clark WP, Katayama Y, Rudikoff S et al (1990) Sequence and structure of Clp P, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J Biol Chem 265(21):12536–12545
Arribas J, Castano JG (1993) A comparative study of the chymotrypsin-like activity of the rat liver multicatalytic proteinase and the ClpP from Escherichia coli. J Biol Chem 268(28):21165–21171
Thompson MW, Maurizi MR (1994) Activity and specificity of Escherichia coli ClpAP protease in cleaving model peptide substrates. J Biol Chem 269(27):18201–18208
Jennings LD, Lun DS, Medard M, Licht S (2008) ClpP hydrolyzes a protein substrate processively in the absence of the ClpA ATPase: mechanistic studies of ATP-independent proteolysis. Biochemistry 47(44):11536–11546
Effantin G, Ishikawa T, De Donatis GM, Maurizi MR et al (2010) Local and global mobility in the ClpA AAA+ chaperone detected by cryo-electron microscopy: functional connotations. Structure 18(5):553–562
Jennings LD, Bohon J, Chance MR, Licht S (2008) The ClpP N-terminus coordinates substrate access with protease active site reactivity. Biochemistry 47(42):11031–11040
Li DH, Chung YS, Gloyd M, Joseph E et al (2010) Acyldepsipeptide antibiotics induce the formation of a structured axial channel in ClpP: a model for the ClpX/ClpA-bound state of ClpP. Chem Biol 17(9):959–969
Leung E, Datti A, Cossette M, Goodreid J et al (2011) Activators of cylindrical proteases as antimicrobials: identification and development of small molecule activators of ClpP protease. Chem Biol 18(9):1167–1178
Lee BG, Park EY, Lee KE, Jeon H et al (2010) Structures of ClpP in complex with acyldepsipeptide antibiotics reveal its activation mechanism. Nat Struct Mol Biol 17(4):471–478
Kirstein J, Hoffmann A, Lilie H, Schmidt R et al (2009) The antibiotic ADEP reprogrammes ClpP, switching it from a regulated to an uncontrolled protease. EMBO Mol Med 1(1):37–49
Dougan DA (2011) Chemical activators of ClpP: turning Jekyll into Hyde. Chem Biol 18(9):1072–1074
Brotz-Oesterhelt H, Beyer D, Kroll HP, Endermann R et al (2005) Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat Med 11(10):1082–1087
Sass P, Josten M, Famulla K, Schiffer G et al (2011) Antibiotic acyldepsipeptides activate ClpP peptidase to degrade the cell division protein FtsZ. Proc Natl Acad Sci U S A 108(42):17474–17479
Kirstein J, Schlothauer T, Dougan DA, Lilie H et al (2006) Adaptor protein controlled oligomerization activates the AAA+ protein ClpC. EMBO J 25(7):1481–1491
Lowth BR, Kirstein-Miles J, Saiyed T, Brotz-Oesterhelt H et al (2012) Substrate recognition and processing by a Walker B mutant of the human mitochondrial AAA+ protein CLPX. J Struct Biol 179(2):193–201
Kang SG, Dimitrova MN, Ortega J, Ginsburg A et al (2005) Human mitochondrial ClpP is a stable heptamer that assembles into a tetradecamer in the presence of ClpX. J Biol Chem 280(42):35424–35432
Alexopoulos JA, Guarne A, Ortega J (2012) ClpP: a structurally dynamic protease regulated by AAA+ proteins. J Struct Biol 179(2):202–210
Dougan DA, Reid BG, Horwich AL, Bukau B (2002) ClpS, a substrate modulator of the ClpAP machine. Mol Cell 9(3):673–683
Guo F, Esser L, Singh SK, Maurizi MR et al (2002) Crystal structure of the heterodimeric complex of the adaptor, ClpS, with the N-domain of the AAA+ chaperone, ClpA. J Biol Chem 277(48):46753–46762
Zeth K, Ravelli RB, Paal K, Cusack S et al (2002) Structural analysis of the adaptor protein ClpS in complex with the N-terminal domain of ClpA. Nat Struct Biol 9(12):906–911
Erbse AH, Wagner JN, Truscott KN, Spall SK et al (2008) Conserved residues in the N-domain of the AAA+ chaperone ClpA regulate substrate recognition and unfolding. FEBS J 275(7):1400–1410
Lo JH, Baker TA, Sauer RT (2001) Characterization of the N-terminal repeat domain of Escherichia coli ClpA-A class I Clp/HSP100 ATPase. Protein Sci 10(3):551–559
Kirstein J, Dougan DA, Gerth U, Hecker M et al (2007) The tyrosine kinase McsB is a regulated adaptor protein for ClpCP. EMBO J 26(8):2061–2070
Kirstein J, Moliere N, Dougan DA, Turgay K (2009) Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat Rev Microbiol 7(8):589–599
Kojetin DJ, McLaughlin PD, Thompson RJ, Dubnau D et al (2009) Structural and motional contributions of the Bacillus subtilis ClpC N-domain to adaptor protein interactions. J Mol Biol 387(3):639–652
Wang F, Mei Z, Qi Y, Yan C et al (2011) Structure and mechanism of the hexameric MecA-ClpC molecular machine. Nature 471(7338):331–335
Donaldson LW, Wojtyra U, Houry WA (2003) Solution structure of the dimeric zinc binding domain of the chaperone ClpX. J Biol Chem 278(49):48991–48996
Dougan DA, Weber-Ban E, Bukau B (2003) Targeted delivery of an ssrA-tagged substrate by the adaptor protein SspB to its cognate AAA+ protein ClpX. Mol Cell 12(2):373–380
Thibault G, Houry WA (2012) Role of the N-terminal domain of the chaperone ClpX in the recognition and degradation of lambda phage protein O. J Phys Chem B 116(23):6717–6724
Wojtyra UA, Thibault G, Tuite A, Houry WA (2003) The N-terminal zinc binding domain of ClpX is a dimerization domain that modulates the chaperone function. J Biol Chem 278(49):48981–48990
Park EY, Lee BG, Hong SB, Kim HW et al (2007) Structural basis of SspB-tail recognition by the zinc binding domain of ClpX. J Mol Biol 367(2):514–526
Neher SB, Sauer RT, Baker TA (2003) Distinct peptide signals in the UmuD and UmuD′ subunits of UmuD/D′ mediate tethering and substrate processing by the ClpXP protease. Proc Natl Acad Sci U S A 100(23):13219–13224
Glynn SE, Martin A, Nager AR, Baker TA et al (2009) Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell 139(4):744–756
Kim DY, Kim KK (2003) Crystal structure of ClpX molecular chaperone from Helicobacter pylori. J Biol Chem 278(50):50664–50670
Neuwald AF, Aravind L, Spouge JL, Koonin EV (1999) AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res 9(1):27–43
Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW et al (2005) Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell 121(7):1029–1041
Okuno T, Yamanaka K, Ogura T (2006) Characterization of mutants of the Escherichia coli AAA protease, FtsH, carrying a mutation in the central pore region. J Struct Biol 156(1):109–114
Schlieker C, Weibezahn J, Patzelt H, Tessarz P et al (2004) Substrate recognition by the AAA+ chaperone ClpB. Nat Struct Mol Biol 11(7):607–615
Yamada-Inagawa T, Okuno T, Karata K, Yamanaka K et al (2003) Conserved pore residues in the AAA protease FtsH are important for proteolysis and its coupling to ATP hydrolysis. J Biol Chem 278(50):50182–50187
Turgay K, Persuh M, Hahn J, Dubnau D (2001) Roles of the two ClpC ATP binding sites in the regulation of competence and the stress response. Mol Microbiol 42(3):717–727
Singh SK, Maurizi MR (1994) Mutational analysis demonstrates different functional roles for the two ATP-binding sites in ClpAP protease from Escherichia coli. J Biol Chem 269(47):29537–29545
Kress W, Mutschler H, Weber-Ban E (2009) Both ATPase domains of ClpA are critical for processing of stable protein structures. J Biol Chem 284(45):31441–31452
Hersch GL, Burton RE, Bolon DN, Baker TA et al (2005) Asymmetric interactions of ATP with the AAA+ ClpX6 unfoldase: allosteric control of a protein machine. Cell 121(7):1017–1027
Grimaud R, Kessel M, Beuron F, Steven AC et al (1998) Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J Biol Chem 273(20):12476–12481
Joshi SA, Hersch GL, Baker TA, Sauer RT (2004) Communication between ClpX and ClpP during substrate processing and degradation. Nat Struct Mol Biol 11(5):404–411
Kim YI, Levchenko I, Fraczkowska K, Woodruff RV et al (2001) Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat Struct Biol 8(3):230–233
Singh SK, Rozycki J, Ortega J, Ishikawa T et al (2001) Functional domains of the ClpA and ClpX molecular chaperones identified by limited proteolysis and deletion analysis. J Biol Chem 276(31):29420–29429
Bewley MC, Graziano V, Griffin K, Flanagan JM (2006) The asymmetry in the mature amino-terminus of ClpP facilitates a local symmetry match in ClpAP and ClpXP complexes. J Struct Biol 153(2):113–128
Martin A, Baker TA, Sauer RT (2007) Distinct static and dynamic interactions control ATPase-peptidase communication in a AAA+ protease. Mol Cell 27(1):41–52
Ishihama Y, Schmidt T, Rappsilber J, Mann M et al (2008) Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics 9:102
Schrader EK, Harstad KG, Matouschek A (2009) Targeting proteins for degradation. Nat Chem Biol 5(11):815–822
Dougan DA, Truscott KN, Zeth K (2010) The bacterial N-end rule pathway: expect the unexpected. Mol Microbiol 76(3):545–558
Karzai AW, Roche ED, Sauer RT (2000) The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat Struct Biol 7(6):449–455
Flynn JM, Levchenko I, Sauer RT, Baker TA (2004) Modulating substrate choice: the SspB adaptor delivers a regulator of the extracytoplasmic-stress response to the AAA+ protease ClpXP for degradation. Genes Dev 18(18):2292–2301
Neher SB, Flynn JM, Sauer RT, Baker TA (2003) Latent ClpX-recognition signals ensure LexA destruction after DNA damage. Genes Dev 17(9):1084–1089
Ninnis RL, Spall SK, Talbo GH, Truscott KN et al (2009) Modification of PATase by L/F-transferase generates a ClpS-dependent N-end rule substrate in Escherichia coli. EMBO J 28(12):1732–1744
Schmidt R, Zahn R, Bukau B, Mogk A (2009) ClpS is the recognition component for Escherichia coli substrates of the N-end rule degradation pathway. Mol Microbiol 72(2):506–517
Lange R, Hengge-Aronis R (1994) The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev 8(13):1600–1612
Lies M, Maurizi MR (2008) Turnover of endogenous SsrA-tagged proteins mediated by ATP-dependent proteases in Escherichia coli. J Biol Chem 283(34):22918–22929
Moore SD, Sauer RT (2005) Ribosome rescue: tmRNA tagging activity and capacity in Escherichia coli. Mol Microbiol 58(2):456–466
Moore SD, Sauer RT (2007) The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem 76:101–124
Keiler KC, Waller PR, Sauer RT (1996) Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271(5251):990–993
Oh BK, Chauhan AK, Isono K, Apirion D (1990) Location of a gene (ssrA) for a small, stable RNA (10Sa RNA) in the Escherichia coli chromosome. J Bacteriol 172(8):4708–4709
Komine Y, Kitabatake M, Yokogawa T, Nishikawa K et al (1994) A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc Natl Acad Sci U S A 91(20):9223–9227
Tu GF, Reid GE, Zhang JG, Moritz RL et al (1995) C-terminal extension of truncated recombinant proteins in Escherichia coli with a 10Sa RNA decapeptide. J Biol Chem 270(16):9322–9326
Gottesman S, Roche E, Zhou Y, Sauer RT (1998) The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev 12(9):1338–1347
Gur E, Sauer RT (2008) Evolution of the ssrA degradation tag in Mycoplasma: specificity switch to a different protease. Proc Natl Acad Sci U S A 105(42):16113–16118
Herman C, Prakash S, Lu CZ, Matouschek A et al (2003) Lack of a robust unfoldase activity confers a unique level of substrate specificity to the universal AAA protease FtsH. Mol Cell 11(3):659–669
Farrell CM, Grossman AD, Sauer RT (2005) Cytoplasmic degradation of ssrA-tagged proteins. Mol Microbiol 57(6):1750–1761
Kenniston JA, Baker TA, Fernandez JM, Sauer RT (2003) Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell 114(4):511–520
Kenniston JA, Baker TA, Sauer RT (2005) Partitioning between unfolding and release of native domains during ClpXP degradation determines substrate selectivity and partial processing. Proc Natl Acad Sci U S A 102(5):1390–1395
Kenniston JA, Burton RE, Siddiqui SM, Baker TA et al (2004) Effects of local protein stability and the geometric position of the substrate degradation tag on the efficiency of ClpXP denaturation and degradation. J Struct Biol 146(1–2):130–140
Weber-Ban EU, Reid BG, Miranker AD, Horwich AL (1999) Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature 401(6748):90–93
Flynn JM, Neher SB, Kim YI, Sauer RT et al (2003) Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell 11(3):671–683
Neher SB, Villen J, Oakes EC, Bakalarski CE et al (2006) Proteomic profiling of ClpXP substrates after DNA damage reveals extensive instability within SOS regulon. Mol Cell 22(2):193–204
Gonciarz-Swiatek M, Wawrzynow A, Um SJ, Learn BA et al (1999) Recognition, targeting, and hydrolysis of the lambda O replication protein by the ClpP/ClpX protease. J Biol Chem 274(20):13999–14005
Farrell CM, Baker TA, Sauer RT (2007) Altered specificity of a AAA+ protease. Mol Cell 25(1):161–166
Gottesman S, Clark WP, Maurizi MR (1990) The ATP-dependent Clp protease of Escherichia coli. Sequence of clpA and identification of a Clp-specific substrate. J Biol Chem 265(14):7886–7893
Maglica Z, Striebel F, Weber-Ban E (2008) An intrinsic degradation tag on the ClpA C-terminus regulates the balance of ClpAP complexes with different substrate specificity. J Mol Biol 384(2):503–511
Hoskins JR, Singh SK, Maurizi MR, Wickner S (2000) Protein binding and unfolding by the chaperone ClpA and degradation by the protease ClpAP. Proc Natl Acad Sci U S A 97(16):8892–8897
Hoskins JR, Wickner S (2006) Two peptide sequences can function cooperatively to facilitate binding and unfolding by ClpA and degradation by ClpAP. Proc Natl Acad Sci U S A 103(4):909–914
Hoskins JR, Kim SY, Wickner S (2000) Substrate recognition by the ClpA chaperone component of ClpAP protease. J Biol Chem 275(45):35361–35367
Wang KH, Oakes ES, Sauer RT, Baker TA (2008) Tuning the strength of a bacterial N-end rule degradation signal. J Biol Chem 283(36):24600–24607
Kim YI, Burton RE, Burton BM, Sauer RT et al (2000) Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol Cell 5(4):639–648
Martin A, Baker TA, Sauer RT (2008) Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat Struct Mol Biol 15(11):1147–1151
Martin A, Baker TA, Sauer RT (2008) Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. Mol Cell 29(4):441–450
Park E, Rho YM, Koh OJ, Ahn SW et al (2005) Role of the GYVG pore motif of HslU ATPase in protein unfolding and translocation for degradation by HslV peptidase. J Biol Chem 280(24):22892–22898
Hou JY, Sauer RT, Baker TA (2008) Distinct structural elements of the adaptor ClpS are required for regulating degradation by ClpAP. Nat Struct Mol Biol 15(3):288–294
Roman-Hernandez G, Hou JY, Grant RA, Sauer RT et al (2011) The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease. Mol Cell 43(2):217–228
Levchenko I, Seidel M, Sauer RT, Baker TA (2000) A specificity-enhancing factor for the ClpXP degradation machine. Science 289(5488):2354–2356
Zhou Y, Gottesman S, Hoskins JR, Maurizi MR et al (2001) The RssB response regulator directly targets sigma(S) for degradation by ClpXP. Genes Dev 15(5):627–637
Erbse A, Schmidt R, Bornemann T, Schneider-Mergener J et al (2006) ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature 439(7077):753–756
Muffler A, Fischer D, Altuvia S, Storz G et al (1996) The response regulator RssB controls stability of the sigma(S) subunit of RNA polymerase in Escherichia coli. EMBO J 15(6):1333–1339
Pratt LA, Silhavy TJ (1996) The response regulator SprE controls the stability of RpoS. Proc Natl Acad Sci U S A 93(6):2488–2492
Elsholz AK, Turgay K, Michalik S, Hessling B et al (2012) Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis. Proc Natl Acad Sci U S A 109(19):7451–7456
Fuhrmann J, Schmidt A, Spiess S, Lehner A et al (2009) McsB is a protein arginine kinase that phosphorylates and inhibits the heat-shock regulator CtsR. Science 324(5932):1323–1327
Persuh M, Mandic-Mulec I, Dubnau D (2002) A MecA paralog, YpbH, binds ClpC, affecting both competence and sporulation. J Bacteriol 184(8):2310–2313
Schlothauer T, Mogk A, Dougan DA, Bukau B et al (2003) MecA, an adaptor protein necessary for ClpC chaperone activity. Proc Natl Acad Sci U S A 100(5):2306–2311
Turgay K, Hahn J, Burghoorn J, Dubnau D (1998) Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J 17(22):6730–6738
Chan CM, Garg S, Lin AA, Zuber P (2012) Geobacillus thermodenitrificans YjbH recognizes the C-terminal end of Bacillus subtilis Spx to accelerate Spx proteolysis by ClpXP. Microbiology 158(Pt 5):1268–1278
Nakano MM, Nakano S, Zuber P (2002) Spx (YjbD), a negative effector of competence in Bacillus subtilis, enhances ClpC-MecA-ComK interaction. Mol Microbiol 44(5):1341–1349
Chien P, Grant RA, Sauer RT, Baker TA (2007) Structure and substrate specificity of an SspB ortholog: design implications for AAA+ adaptors. Structure 15(10):1296–1305
Lessner FH, Venters BJ, Keiler KC (2007) Proteolytic adaptor for transfer-messenger RNA-tagged proteins from alpha-proteobacteria. J Bacteriol 189(1):272–275
Levchenko I, Grant RA, Wah DA, Sauer RT et al (2003) Structure of a delivery protein for an AAA+ protease in complex with a peptide degradation tag. Mol Cell 12(2):365–372
Wah DA, Levchenko I, Rieckhof GE, Bolon DN et al (2003) Flexible linkers leash the substrate binding domain of SspB to a peptide module that stabilizes delivery complexes with the AAA+ ClpXP protease. Mol Cell 12(2):355–363
Bolon DN, Grant RA, Baker TA, Sauer RT (2004) Nucleotide-dependent substrate handoff from the SspB adaptor to the AAA+ ClpXP protease. Mol Cell 16(3):343–350
Bolon DN, Wah DA, Hersch GL, Baker TA et al (2004) Bivalent tethering of SspB to ClpXP is required for efficient substrate delivery: a protein-design study. Mol Cell 13(3):443–449
Flynn JM, Levchenko I, Seidel M, Wickner SH et al (2001) Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc Natl Acad Sci U S A 98(19):10584–10589
McGinness KE, Baker TA, Sauer RT (2006) Engineering controllable protein degradation. Mol Cell 22(5):701–707
Becker G, Klauck E, Hengge-Aronis R (1999) Regulation of RpoS proteolysis in Escherichia coli: the response regulator RssB is a recognition factor that interacts with the turnover element in RpoS. Proc Natl Acad Sci U S A 96(11):6439–6444
Jonczyk P, Nowicka A (1996) Specific in vivo protein-protein interactions between Escherichia coli SOS mutagenesis proteins. J Bacteriol 178(9):2580–2585
Gonzalez M, Rasulova F, Maurizi MR, Woodgate R (2000) Subunit-specific degradation of the UmuD/D′ heterodimer by the ClpXP protease: the role of trans recognition in UmuD′ stability. EMBO J 19(19):5251–5258
Varshavsky A (1996) The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci U S A 93(22):12142–12149
Varshavsky A (2011) The N-end rule pathway and regulation by proteolysis. Protein Sci 20(8):1298–1345
Dougan DA, Micevski D, Truscott KN (2012) The N-end rule pathway: from recognition by N-recognins, to destruction by AAA+ proteases. Biochim Biophys Acta 1823(1):83–91
Graciet E, Wellmer F (2010) The plant N-end rule pathway: structure and functions. Trends Plant Sci 15(8):447–453
Mogk A, Schmidt R, Bukau B (2007) The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell Biol 17(4):165–172
Shrader TE, Tobias JW, Varshavsky A (1993) The N-end rule in Escherichia coli: cloning and analysis of the leucyl, phenylalanyl-tRNA-protein transferase gene aat. J Bacteriol 175(14):4364–4374
Lupas AN, Koretke KK (2003) Bioinformatic analysis of ClpS, a protein module involved in prokaryotic and eukaryotic protein degradation. J Struct Biol 141(1):77–83
Roman-Hernandez G, Grant RA, Sauer RT, Baker TA (2009) Molecular basis of substrate selection by the N-end rule adaptor protein ClpS. Proc Natl Acad Sci U S A 106(22):8888–8893
Schuenemann VJ, Kralik SM, Albrecht R, Spall SK et al (2009) Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS. EMBO Rep 10(5):508–514
Wang KH, Roman-Hernandez G, Grant RA, Sauer RT et al (2008) The molecular basis of N-end rule recognition. Mol Cell 32(3):406–414
Turgay K, Hamoen LW, Venema G, Dubnau D (1997) Biochemical characterization of a molecular switch involving the heat shock protein ClpC, which controls the activity of ComK, the competence transcription factor of Bacillus subtilis. Genes Dev 11(1):119–128
Persuh M, Turgay K, Mandic-Mulec I, Dubnau D (1999) The N- and C-terminal domains of MecA recognize different partners in the competence molecular switch. Mol Microbiol 33(4):886–894
Andersson FI, Blakytny R, Kirstein J, Turgay K et al (2006) Cyanobacterial ClpC/HSP100 protein displays intrinsic chaperone activity. J Biol Chem 281(9):5468–5475
Burton RE, Siddiqui SM, Kim YI, Baker TA et al (2001) Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine. EMBO J 20(12):3092–3100
Reid BG, Fenton WA, Horwich AL, Weber-Ban EU (2001) ClpA mediates directional translocation of substrate proteins into the ClpP protease. Proc Natl Acad Sci U S A 98(7):3768–3772
Wang J, Song JJ, Seong IS, Franklin MC et al (2001) Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU. Structure 9(11):1107–1116
Farr GW, Furtak K, Rowland MB, Ranson NA et al (2000) Multivalent binding of nonnative substrate proteins by the chaperonin GroEL. Cell 100(5):561–573
Martin A, Baker TA, Sauer RT (2005) Rebuilt AAA+ motors reveal operating principles for ATP-fuelled machines. Nature 437(7062):1115–1120
Li H, Carrion-Vazquez M, Oberhauser AF, Marszalek PE et al (2000) Point mutations alter the mechanical stability of immunoglobulin modules. Nat Struct Biol 7(12):1117–1120
Aubin-Tam ME, Olivares AO, Sauer RT, Baker TA et al (2011) Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell 145(2):257–267
Lee C, Schwartz MP, Prakash S, Iwakura M et al (2001) ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell 7(3):627–637
Barkow SR, Levchenko I, Baker TA, Sauer RT (2009) Polypeptide translocation by the AAA+ ClpXP protease machine. Chem Biol 16(6):605–612
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Gur, E., Ottofueling, R., Dougan, D.A. (2013). Machines of Destruction – AAA+ Proteases and the Adaptors That Control Them. In: Dougan, D. (eds) Regulated Proteolysis in Microorganisms. Subcellular Biochemistry, vol 66. Springer, Dordrecht. https://doi.org/10.1007/978-94-007-5940-4_1
Download citation
DOI: https://doi.org/10.1007/978-94-007-5940-4_1
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-007-5939-8
Online ISBN: 978-94-007-5940-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)