Abstract
Number of a molecule is discrete by its nature. Therefore, upon better detection sensitivity of analytical methods approaching the single-molecule level, the measured concentration and number of analytes show intrinsic discreteness. An emerging analytical method that makes use of this intrinsic discreteness is digital counting. When an analyte solution is partitioned into many small reaction compartments, such that each molecule is individually encapsulated into a compartment, the analyte quantification is inevitably digitized; each compartment contains none or one molecule of analyte. When a solution of enzyme or enzyme-conjugated molecule is partitioned into such small reactors with fluorogenic substrate, one can count the number of the molecule as that of fluorescent reactors under an optical microscope. We refer to this strategy as “digital counting.” One of the most successful examples of digital counting is digital ELISA, in which target molecules are individually encapsulated in a water-in-oil droplet after bound to enzyme-conjugated antibody. Although the chemistry of digital ELISA; antibody and enzyme is common to conventional ELISA, the detection sensitivity of digital ELISA is higher than that of conventional ELISA by 4–6 orders of magnitude. In this review, we introduce recent achievements in digital ELISA and similar methods.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
An emerging trend in biochemical analysis is digitization. The word “digitization” here does not refer to automatization of instruments nor data analysis. Rather, we use it to describe the conversion of target objects into a temporal and/or spatial distribution of quantized signals. Biochemical substances and their reactions are generally regarded as continuous, but in reality they show discrete and quantized behavior at the single-molecule level. A classic example is the single channel recording reported in 1976 [1]. In this experiment, channel current was quantized by the opening or closing of a single membrane ion channel. In the late twentieth century, single-molecule imaging in aqueous solution demonstrated that the presence or absence of a fluorescence signal correlated with the respective association and dissociation of a fluorescently labeled ligand to a single enzyme molecule [2]. We have conducted single-molecule imaging of rotation of F1-ATPase and demonstrated that discrete 120° stepping rotations are coupled with the single turnover of ATP hydrolysis, where the angular velocity represents on and off, or digital, behavior [3]. These reports are just a few examples that show how single-molecule resolution has revealed an intrinsic discreteness in molecular behavior. At the beginning of this century, the concept of digitalization was extended to the quantification of enzymatic biomolecules by the use of micron-sized (fL volume) reactor chamber systems (hereafter simply called, “reactor chamber”). Successful examples of this approach include digital PCR and digital ELISA [4–6]. In these assays, a sample solution is partitioned into a massive number of reactor chambers and then the number of chambers that show signs of DNA amplification (for digital PCR) or enzymatic activity (for digital ELISA) are counted. We refer to this strategy as “digital counting.” Digital counting requires handling micron-sized aqueous solution, which is now possible because of “Lab-on-a-chip” technology.
2 Lab-on-a-Chip Technology for Biochemical Analysis
Lab-on-a-chip (LOC), also called micro total analysis system (μTAS) or biodevice, has become an invaluable instrument for biochemical analysis. After the first LOC report by Terry and coworkers in 1979 [7], Manz et al. [8] provided a comprehensive review on the concept of LOC technology in the early stage of the field. Over the following quarter-century, LOC technology has provided novel methods for cell biology, molecular biology, and other research fields [9]. Essential to LOC technology is miniaturization, which has benefited from gains in micro/nanofabrication technologies of the semiconductor industry. In the past half century, advancement in semiconductor devices have been driven by a sophistication in highly precise and efficient fabrication such as photolithography and plasma etching. To handle minute sample solutions, a variety of microstructures, including not only microreactors and capillaries, but also micropumps and microvalves, are fabricated onto a chip [10–12]. For easy microfabrication and replication of the device structure, simple fabrication methods, such as molding using silicone rubber poly(dimethylsiloxane) (PDMS), have been developed [13].
What advantageous features does LOC technology bring to biochemistry? These devices allow for the precise control of those factors that regulate microscale biological phenomena. Such factors include laminar flow, electroosmotic flow, the diffusion-limited step, and temperature [8]. Moreover, miniaturization enables a small amount or small size of sample to be treated in parallel [14–16], thus enhancing the throughput of the assay. High-throughput analysis using LOC has proven extremely useful in combinatorial chemistry [17] and directed evolution [18], which require huge libraries, and for epigenome analysis [19] and proteome analysis [20], which exemplify a new trend of biology. Another important feature of LOC is that it reduces the sample consumption, device size, and device cost. These features enable it to be applied to point-of-care testing (POCT) for personalized medicine at home or clinical use in developing countries [21].
Miniaturization especially has important implications for single-molecule assays. The size reduction of the reaction volume magnifies the concentration change of analyte molecules. For enzymatic reactions, single-molecule enzymatic assays can be done using reactor chambers. For example, when two identical enzymes that have a turnover rate of 10 s−1 are separately encapsulated into 1 μL (1 mm3) or 1 fL (1 μm3) reactors, the ensuing reaction product reaches 10 fM and 10 μM, respectively, after 10 min incubation. Note that volume time concentration gives a constant value (quantity of reaction product molecule). While 10 fM is almost undetectable, 10 μM is reliably detectable. Thus, the signal/background ratio is improved to a level that permits the digital counting of enzyme molecules. Large-scale integration of the reactor chambers increases the binary digit (bit) of the measurement, which further improves the sensitivity and dynamic range. Accordingly, we have developed reactor chamber devices for biochemical analysis at the single-molecule level.
3 Femtoliter Droplet Array and Its Application to Single-Molecule Measurements
The Nobel Prize in Chemistry 2014 was awarded to S. W. Hell, E. Betzig, and W. E. Moerner for the development of super-resolved fluorescence microscopy. Among the three laureates, Betzig and Moerner laid the foundation for photoactivated localization microscopy (PALM) using single-molecule fluorescence imaging [22, 23]. Prior to the development of PALM, they separately demonstrated direct optical detection of single-molecules circa 1990 [24, 25]. In contrast, single-molecule detection by enzymatic products was realized using microdroplets as early as the 1960s [26]. This fact underscores the easiness and robustness of single-molecule detection using microdroplets. Nevertheless, this method had not been utilized for digital counting due to heterogeneities in droplet size and shape until LOC technology, which has enabled the generation of monodisperse microdroplets [27, 28]. Recently, such monodisperce microdroplets were employed for digital PCR [29]. For parallel and successive imaging of a massive number of microdroplets, a microdroplet array immobilized on a substrate has been used. Using reactor chamber, we have explored various techniques for the measurement of single biomolecules (Fig. 7.1). As an early example, we encapsulated single F1-ATPase into a reactor chamber and rotated it clockwise using magnetic tweezers. After release from the tweezers, the F1-ATPase rotated counterclockwise, indicating that ATP accumulated in the reactor chamber and was utilized for the autonomous counterclockwise rotation. From the counterclockwise rotation speed, it was revealed that ATP synthesis by F1-ATPase was efficiently coupled with the rotating motion [30]. We also demonstrated digital counting for β-galactosidase (β-gal), which catalyzes hydrolysis of a fluorogenic substrate; fluorescein di-β-d-galactopyranoside (FDG) into fluorescein [31]. Here, the average number of β-gal encapsulated into single reactor chamber was determined from the β-gal concentration and chamber volume. When several pM of β-gal was distributed into the reactor chambers, each chamber randomly showed fluorescence and the ratio of fluorescent chambers increased linearly with β-gal concentration. Additionally, the rate of the fluorescence increase took discretized values of chambers. Thus, the reactor chamber enables measurement of very small, gradual changes in single biomolecules. This advantageous feature of reactor chamber was utilized for efflux activity measurement of single bacterial cell [16].
4 Digital ELISA
Enzyme-linked immunosorbent assay (ELISA) is one of the most widely used methods for diagnostic assays. ELISA that captures a particular target molecule via specific antigen-antibody interaction can detect the target molecule even in a mixture of biomolecules. In a typical procedure of sandwich ELISA, one of the most common types of ELISA, the minimum constituents of assay mixture are; capture antibody that is immobilized on a solid surface, such as a plastic plate or plastic beads, detection antibody that is conjugated to enzyme as a reporter, and reaction substrate for the conjugated enzyme that produces reaction product molecule with different color (colorimetric assay) or fluorescence (fluorogenic assay) from the reaction substrate. The target molecules are at first captured on the capture antibody and then subject to reaction with detection antibody to form the sandwich structure; antibody-antigen-antibody. The quantity of the captured target molecule is determined from the enzymatic activity.
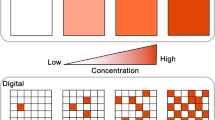
ELISA is a remarkable method, as it can easily obtain high sensitivity and specificity and is widely used in biochemistry and medical studies. However, its sensitivity remains insufficient for the detection of many important biological substances such as the influenza virus in the early phase of infection and biomarkers in early cancer stages. In digital ELISA [4, 6], extremely high sensitivity is achieved by the use of femtoliter reactor chambers. Figure 7.2 shows the schematic image of digital ELISA from Ref. 7, where antigen molecules are reacted with capture antibodies immobilized on microbeads. Because the excess amount of microbeads are mixed with antigen molecules, each microbead carries none or one antigen molecule. After reacted with the detection antibody that is conjugated with β-galactosidase (β-gal), the beads are individually encapsulated in a reactor chamber with the aforementioned fluorogenic substrate of β-gal, FDG. After incubation for a few tens of minutes, some chambers emit distinctive fluorescent signal while the others remain dark, giving clear bimodal peaks in fluorescent histogram of chambers. Thus, when set a threshold to discriminate the fluorescent and nonfluorescent chamber, one can readily count the number of trapped antigen molecules as the number of fluorescent chamber. The probability that a reactor chamber encapsulates two or more antigen molecule in a reactor chamber is negligible due to the extremely low ratio of antigen to beads (typically less than 0.05).
Digital ELISA. a Schematics of digital ELISA. A single sandwich complex is encapsulated into a reactor chamber. The antigen is detected by the fluorescent product of β-gal. The initial concentration of the antigen can be determined by the ratio of fluorescent chambers to the total number of beads. b Digital ELISA for prostate specific antigen (PSA). In the fluorescence images, most bright spots correspond to single PSA molecules. The concentration of PSA was in proportion to the ratio of fluorescent chambers at low concentrations, i.e., less than 200 aM. In this assay, the limit of detection (LOD) was 2 aM, more than one million times lower than that of conventional ELISA. The nonzero fraction at 0 M indicates nonspecific adsorption of the detection antibody
For the proof of concept, digital ELISA for prostate specific antigen (PSA), a biomarker for prostate cancer, was conducted [6]. We determined the number of bright chambers at different PSA concentrations. The fraction of bright chamber showed a good linearity to the PSA concentration from atto- to femto-molar. Thus, digital ELISA of PSA showed a wide dynamic range of 3 digits. The determined limit of detection (LOD) was as low as 2 aM (60 ag/mL), which was a millionfold lower than that of conventional ELISA. It is noteworthy that digital ELISA has no LOD in principle, when any fluorescent chamber is not observed in the absence of target molecule. In the case of digital ELISA for PSA, even in the absence of PSA, 0.2 % of chambers showed fluorescence mainly due to nonspecific absorption of detection antibody on the microbeads.
Digital ELISA would enable early diagnostics of diseases such as influenza, cancer and neurodegenerative diseases. Furthermore, it has the potential to discover new biomarkers that are not currently detectable and to the development of new diagnostic technologies. The reduction of the nonspecific binding is one of the main challenges for improvement of digital ELISA.
5 Future Perspectives
Digital ELISA is a promising analytical method for presymptomatic testing, but several challenges remain for its practical application. The first challenge involves the detection of multiple types of biomarkers in a single assay. Such a feature would improve the specificity of diagnosis and reduce the amount of sample, such as blood, required from a patient. Multicolored digital ELISA, in which multiple types of antibodies are conjugated to distinct enzymes, may be useful for this purpose. Multicolor detection has already been demonstrated with β-gal and alkaline phosphatase, which produces resorufin [32].
The second challenge is the reduction of the assay time. The observation of a massive number of reactor chambers requires wide-field imaging with a low-power objective lens, which weakens the fluorescence incidence on the lens. For example, the digital counting of β-gal using 60 fL chambers and a 10× objective lens takes several tens of minutes even with a scientific CMOS camera due to the slow increase of fluorescence intensity. Moreover, in multicolored digital ELISA, enzymes would function in nonoptimum conditions and take a longer time, because optimum buffer contents and pH are different among enzymes. Thus, a technological advancement that achieves more rapid detection of single enzyme activity is still required. For rapid detection, reactor chambers were further miniaturized to submicron scale and aL volume using nanoimprint technology. We named this aL-droplet array “Nanocell.” In Nanocell, the concentration of fluorescent products rapidly increased and reached detectable levels (Ono et al. unpublished).
The third challenge is to reduce the background noise from nonspecific adsorption to the microbeads. In addition to Poisson noise, nonspecific adsorption of the detection antibody increases background noise and reduces the sensitivity of the assay. Strategies include blocking the microbeads surface with bovine serum albumin or modifying it with polyethylene glycol, but better reductions are still needed.
Finally, the fourth challenge is the miniaturization of the whole ELISA system. In the present digital ELISA system, the reactor chamber chip is as small as several square centimeters, but a conventional microscope is used for the fluorescence detection. To bring POCT to the home or small clinics, a portable imaging unit is needed. We are therefore developing a digital ELISA system that includes a CMOS image sensor behind the chip [33]. Using this system, we have detected single molecule in reactor chambers and expect a convenient and cost-effective diagnostic system will appear in the near future.
Aside from digital ELISA, reactor chambers can be used to measure the activity of single-molecule membrane transporters, which are in general very promising drug targets. Active transport by these transporters is quite slow, less than one ten-thousandth of the passive transport rate done by membrane channels. The significant difference in transport rate between active and passive transporters is due to the intrinsic difference of their working principle; in active transport, transporter proteins undergo a large conformational change accompanying main chain rearrangement, to pump substrate molecules one-by-one against substrate concentration gradient or membrane voltage. Such a large conformational change occurs in tens of milliseconds that determines the overall transport rate. In contrast, passive transporter proteins just facilitate substrate diffusion along concentration gradient or voltage by forming nanometer-sized pore. In some ion channel proteins, each ion takes only submicroseconds to travel across the membrane-spanning pore, allowing electrical recording of the ion current by a single ion channel protein. Thus, single-molecule detection of active transporter proteins requires completely different detection principles from single ion channel recording. To address the technical challenge, we developed a new type of femtoliter reactor array in which each reactor is sealed with lipid bilayer [34]. In our arrayed lipid bilayer chamber (ALBiC) system, membrane transporters are reconstituted into a lipid bilayer that covers the upper side of the reactor chamber. When an active transporter protein pumps substrate molecules into the reactor, substrate molecules quickly accumulate in the reactor, enabling the detection of single transporter proteins. We have demonstrated detection of active proton pump activity by single FoF1-ATPase molecules that pump proton, hydrolyzing ATP [34]. This system is now being developed for other transporters and drug screening [35, 36].
6 Conclusion
In our CREST project, we have developed biochemical analysis methods based on digital methodologies for the measurement of biological phenomena at the single-molecule level. More specifically, using our unique LOC devices, we have successfully measured the reaction activity of single F1 motor proteins, enzyme activity and membrane transporter protein. Moreover, we have incorporated digital counting into ELISA format, which has drastically improved the sensitivity (more than one million times higher than that of conventional ELISA). We see these digital methods as the beginnings of a “Digitization Revolution” in biochemical analysis.
References
E. Neher, B. Sakmann, Nature 260, 799 (1976)
T. Funatsu, Y. Harada, M. Tokunaga, K. Saito, T. Yanagida, Nature 374, 555 (1995)
H. Noji, R. Yasuda, M. Yoshida, K. Kinoshita Jr., Nature 386, 299 (1997)
D.M. Rissin, C.W. Kan, T.G. Campbell, S.C. Howes, D.R. Fournier, L. Song, T. Piech, P.P. Patel, L. Chang, A.J. Rivnak, E.P. Ferrell, J.D. Randall, G.K. Provuncher, D.R. Walt, D.C. Duffy, Nat. Biotechnol. 28, 595 (2010)
Y. Men, Y. Fu, Z. Chen, P.A. Sims, W.J. Greenleaf, Y. Huang, Anal. Chem. 84, 4262 (2012)
S.H. Kim, R. Iino, S. Iwai, S. Araki, S. Sakakihara, H. Noji, Lab Chip 12, 4986 (2012)
S.C. Terry, J.H. Jerman, J.B. Angell, IEEE Trans. Electron Device 26, 1880 (1979)
A. Manz, N. Graber, H.M. Widmer, Sens. Actuators B 1, 244 (1990)
E.K. Sackmann, A.L. Fulton, D.J. Beebe, Nature 507, 181 (2014)
T. Thorsen, S.J. Maerkl, S.R. Quake, Science 298, 580 (2002)
H.T.G. van Lintel, F.C.M. van De Pol, S. Bouwstra, Sens. Actuators 15, 153 (1988)
M. Esahsi, S. Shoji, A. Nakano, Sens. Actuators 20, 163 (1989)
D.C. Duffy, J.C. McDonald, O.J.A. Schueller, G.M. Whitesides, Anal. Chem. 70, 4974 (1998)
D.D. Carlo, L.Y. Wu, L.P. Lee, Lab Chip 6, 1445 (2006)
L. Cai, N. Friedman, X.S. Xie, Nature 440, 358 (2006)
R. Iino, K. Hayama, H. Amezawa, S. Sakakihara, S.H. Kim, Y. Matsumono, K. Nishino, A. Yamaguchi, H. Noji, Lab Chip 12, 3923 (2012)
M.C. Mitchell, V. Spikmans, A. Manz, A.J. de Mello, J. Chem. Soc. Perkin Trans. 15, 514 (2001)
J.J. Agresti, E. Antipov, A.R. Abate, K. Ahn, A.C. Rowat, J.-C. Baret, M. Marquez, A.M. Klibanov, A.D. Griffiths, D.A. Weitz, Proc. Natl. Acad. Sci. USA 107, 4004 (2010)
J. Clarke, H.-C. Wu, L. Jayasinghe, A. Patel, S. Reid, H. Bayley, Nat. Nanotechnol. 4, 265 (2009)
Y. Taniguchi, P.J. Choi, G.-W. Li, H. Chen, M. Babu, J. Hearn, A. Emili, X.S. Xie, Science 329, 553 (2010)
C.D. Chin, T. Laksanasopin, Y.K. Cheung, D. Steinmiller, V. Linder, H. Parsa, J. Wang, H. Moore, R. Rouse, G. Umviligihozo, E. Karita, L. Mwambarangwe, S.L. Braunstein, J. van de Wijgert, R. Sahabo, J.E. Justman, W. El-Sadr, S.K. Sia, Nat. Med. 17, 1015 (2011)
R.M. Dickson, A.B. Cubitt, R.Y. Tsien, W.E. Moerner, Nature 388, 355 (1997)
E. Betzig, G.H. Patterson, R. Sougrat, O.W. Lindwasser, S. Olenych, J.S. Bonifacino, M.W. Davidson, J. Lippincott-Schwartz, H.F. Hess, Science 313, 1642 (2006)
W.E. Moerner, L. Kador, Phys. Rev. Lett. 62, 2535 (1989)
E. Betzig, R.J. Chichester, Science 262, 1422 (1993)
B. Rotman, Proc. Natl. Acad. Sci. USA 47, 1981 (1961)
P.B. Umbanhowar, V. Prasad, D.A. Weitz, Langmuir 16, 347 (2000)
T. Thorsen, R.W. Roberts, F.H. Arnold, S.R. Quake, Phys. Rev. Lett. 86, 4163 (2001)
B.J. Hindson, K.D. Ness, D.A. Masquelier, P. Belgrader, N.J. Heredia, A.J. Makarewicz, I.J. Bright, M.Y. Lucero, A.L. Hiddessen, T.C. Legler, T.K. Kitano, M.R. Hodel, J.F. Petersen, P.W. Wyatt, E.R. Steenblock, P.H. Shah, L.J. Bousse, C.B. Troup, J.C. Mellen, D.K. Wittmann, N.G. Erndt, T.H. Cauley, R.T. Koehler, A.P. So, S. Dube, K.A. Rose, L. Montesclaros, S. Wang, D.P. Stumbo, S.P. Hodges, S. Romine, F.P. Milanovich, H.E. White, J.F. Regan, G.A. Karlin-Neumann, C.M. Hindson, S. Saxonov, B.W. Colston, Anal. Chem. 83, 8604 (2011)
Y. Rondelez, G. Tresset, T. Nakashima, Y. Kato-Yamada, H. Fujita, S. Takeuchi, H. Noji, Nature 433, 773 (2005)
S. Sakakihara, S. Araki, R. Iino, H. Noji, Lab Chip 10, 3355 (2010)
Y. Obayashi, R. Iino, H. Noji, Analyst 140, 5065 (2015)
K. Sasagawa, K. Ando, T. Kobayashi, T. Noda, T. Tokuda, S.H. Kim, R. Iino, H. Noji, J. Ohta, Jpn. J. Appl. Phys. 51, 02BL01 (2012)
R. Watanabe, N. Soga, D. Fujita, K.V. Tabata, L. Yamauchi, S.H. Kim, D. Asanuma, M. Kamiya, Y. Urano, H. Suga, H. Noji, Nat. Commun. 5, 4519 (2014)
R. Watanabe, N. Soga, T. Yamanaka, H. Noji, Sci. Rep. 4, 7076 (2014)
N. Soga, R. Watanabe, H. Noji, Sci. Rep. 5, 11025 (2015)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Japan
About this chapter
Cite this chapter
Ono, T., Noji, H. (2016). Digital Bioassay with Femtoliter Reactor Array. In: Sone, J., Tsuji, S. (eds) Intelligent Nanosystems for Energy, Information and Biological Technologies. Springer, Tokyo. https://doi.org/10.1007/978-4-431-56429-4_7
Download citation
DOI: https://doi.org/10.1007/978-4-431-56429-4_7
Published:
Publisher Name: Springer, Tokyo
Print ISBN: 978-4-431-56427-0
Online ISBN: 978-4-431-56429-4
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)