Abstract
Rods and cones are highly related, sharing many morphological and functional features. Early lineage studies showed that they are produced by mitotic retinal progenitor cells (RPCs) that produce not only photoreceptor cells, but other retinal cell types as well, even in a terminal division (Holt et al., Neuron 1(1):15–26, 1988; Turner and Cepko, Nature 328 (6126):131–136, 1987; Turner et al., Neuron 4(6):833–845, 1990; Wetts and Fraser, Science 239(4844):1142–1145, 1988). More recent lineage studies have added more depth to the conclusions of these early findings, showing that there are specific and distinct RPCs that produce specific retinal cell types in a terminal division (Godinho et al., Neuron 56(4):597–603, 2007; Hafler et al., Proc Natl Acad Sci USA 109(20):7882–7887, 2012; Rompani and Cepko, Proc Natl Acad Sci USA 105(1):192–197, 2008; Suzuki et al., Proc Natl Acad Sci USA 110(37):15109–15114, 2013). Rather surprisingly, one type of RPC makes cones and horizontal cells (HCs), whereas another makes rods and amacrine cells, and still others make only rods, rods and bipolar cells, or rods and Muller glial cells (Emerson et al., Dev Cell 154(4):928–939, 2013; Hafler et al., Proc Natl Acad Sci USA 109(20):7882–7887, 2012). The genetic networks that are operating in these different types of RPCs have similarities and differences, with one of the differences leading to the cone fate and indirectly repressing the rod fate. The studies that lead to these conclusions are described in this chapter.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
9.1 Introduction
There are a number of fascinating questions concerning the development of photoreceptor cells. One question concerns the cell fate determination mechanisms that direct the irreversible acquisition of the photoreceptor fate, as well as the rod versus cone fate. Another question concerns the intersection between these fate determination events and the patterning machinery that regulates the distribution of rods and cones across the retina: for example, why does the fovea have only cones and no rods? A third question revolves around the mechanistic aspects of the morphogenesis of the highly evolved photoreceptor structure, for example, how is the outer segment built and maintained? Some of these questions are covered within several chapters in this book, and will be of interest for some time, as we know very little about most of these processes, although efforts are being made to learn more. Here, the focus is on the cell fate determination events for rods and cones.
The birthdates of retinal cells are defined as the day when a cell undergoes its last S-phase. Birthdating studies across many vertebrates, including mammals, birds, fish, and amphibians, have shown that the birth order of retinal neurons is conserved (Altshuler et al. 1991). In the mouse, the cone photoreceptors are born early, starting at embryonic day 10 (E10), which is about the same time as the other early-born neurons, the ganglion cells and horizontal cells (HCs) (Fig. 9.1) (Carter-Dawson and LaVail 1979; Young 1985). The last cones are born in the periphery at E18. Rod photoreceptors are born between E13 and postnatal day 7 (P7) and thereby have a period of genesis that overlaps that of cones but extends well beyond it. Rod and cone birthdays in the rat are similar (Rapaport et al. 2004). Carter-Dawson and LaVail quantified the birthdates of rods and cones in the mouse, showing that, within 2 days after the commencement of rod genesis, rods already outnumber cones (Fig. 9.1a). Thus, by E15, there are a greater number of cells fated to be rods in the embryonic mouse retina than there are those fated to be cones. By the end of development, rods comprise 72.3 % of all mouse retinal cells, whereas cones only are 2.2 % (Young 1985). In the chick, which is another useful model of retinal development, there are clearly more cones than rods (Morris and Shorey 1967; Bruhn and Cepko 1996). However, a precise accounting of the frequency of rods and cones is not available and may be difficult to obtain as there is significant variation across the retina in the frequency of rods (Bruhn and Cepko 1996). However, in one estimate cones comprise approximately 80 % of all photoreceptors (Morris and Shorey 1967). Most of the data discussed here are taken from studies of mice and chicks, although many of the observations made in these species have also been made in Xenopus and zebrafish, two other excellent models of retinal development.
Birthdates of rods and cones in the mouse retina. Classical 3H-thymidine birthdating for rods and cones was carried out in the mouse (Carter-Dawson and LaVail 1979). A single injection of 3H-thymidine was given on each day in development in different mice, and it was estimated that the label was available for approximately 30 min. Carter-Dawson and LaVail calculated that this would lead to heavily labeled nuclei for approximately 50 % of the cells born on a given day, as approximately 50 % of the cell cycle is the S-phase (Young 1985). At the termination of development, they quantified the percentage of all rods and all cones that were heavily labeled from the injection on a given day. Using the values of Young (Young 1985) for the fraction of all retinal cells that are rods and cones, 72.3 % and 2.2 %, respectively, the data of Carter-Dawson and LaVail were transformed into the number of rods (red) and cones (blue) born on a given day (a) and the cumulative fraction of rods (red) and cones (blue) born over time in development (b)
9.2 Genes Required for the Genesis of Rods and Cones
The development of photoreceptors can be broken down into several stages, those of cell fate determination, differentiation, and survival. As discussed next, the determination event seems to occur at approximately the point of genesis, or birthday (day of birth), and can be thought of as the decision point to become a photoreceptor or a rod or a cone. Genes required for determination can be defined as those genes whose loss of function leads to a reduction in the number of photoreceptors, with a concomitant increase in another cell type(s), exemplified by loss of Otx2 (Nishida et al. 2003). This definition is meant to allow a distinction between those genes required for determination versus survival, as those required for survival may lead to loss of photoreceptors but they will not lead to an increase in another cell type, such as Neurod1 (Morrow et al. 1999). Differentiation can be defined as the elaboration of phenotype, such as the onset of markers, or morphological changes, that reflect the determination event. Genes that have a major role in the direct regulation of differentiation genes are also referred to as terminal selector genes (Hobert 2011). Loss of a terminal selector gene will lead to a reduction in the expression of photoreceptor markers, but will not lead to an absence of cells that express early markers or other features of photoreceptor cells, such as Crx (Furukawa et al. 1999). Complicating the interpretation of these roles is the possibility that a gene may have more than one role. Removal of gene function at different stages in the development process may enable the appreciation of multiple roles. However, genes with more than one role can present problems of interpretation, if, for example, a gene whose loss leads to a cell fate switch concomitantly leads to a change in proliferation and/or survival.
9.2.1 Notch1
Notch1 is a gene that acts at a very early, perhaps the earliest, point in photoreceptor determination (Jadhav et al. 2006; Yaron et al. 2006). The absence of Notch1 leads to an increase in the number of photoreceptors. Notch1 conditional knockout (CKO) mice have been examined following the introduction of Cre using different Cre strains, or by infection with a retrovirus carrying Cre. Overproduction of cone photoreceptors occurs if Notch1 is removed early, and other cell types are diminished accordingly. It is a bit difficult to do quantitative bookkeeping on this point, however, as there may also be a reduction in proliferation and survival. Nonetheless, one can see a preponderance of cells expressing cone markers, and a reduction in the expression of markers of other early-born cell types, by either microarray, in situ hybridization, or immunohistochemistry, if Notch1 is removed early in development. In the postnatal period, when a retrovirus was used to introduce Cre, there was an increase in rods (Jadhav et al. 2006; Mizeracka et al. 2013a). This experiment was done when proliferation was almost over, and almost every clone in the control was only a single cell, reducing the impact of a change in proliferation. Increases in photoreceptor number also were seen when a chemical inhibitor of gamma secretase, DAPT, which inhibits the enzyme needed to make functional Notch receptor, was added to cultures of the retina (Nelson et al. 2007). Furthermore, when Rbpj, which is a protein that acts in a complex with Notch to regulate transcription, was removed in mice, overproduction of photoreceptors was observed, as well as overproduction of ganglion cells (Riesenberg et al. 2009). It is possible that ganglion cell overproduction occurred because there are multiple Notch genes and all use Rbpj for transcriptional regulation. All of these observations demonstrate the importance of Notch1 in suppressing the photoreceptor fate.
Notch1 is expressed in both mitotic retinal progenitor cells (RPCs) and newly postmitotic cells (Nelson et al. 2006; Trimarchi et al. 2008b; Bao and Cepko 1997; Jadhav et al. 2006; Yaron et al. 2006). Its expression is then extinguished as neurons differentiate, whereas its expression is maintained in Müller glia, which also maintain the expression of many other RPC genes (Blackshaw et al. 2004). The aforementioned studies of the Notch1 CKO were conducted such that Notch1 was removed from mitotic cells. We were interested in whether Notch1 was needed in the newly postmitotic cells to regulate the number of rods. This question was of interest as we wish to understand how RPCs influence the fate of their progeny. It may be there are different types of RPCs and that they determine the fate of their progeny by passing down determinants, such as transcription factors (TFs), microRNAs, and/or chromatin state. These determinants might dictate the fate of progeny, reducing or eliminating the need for extrinsic cues or stochastic processes in the choice of fate within newly postmitotic cells. To this end, we used two methods to remove Notch1 from newly postmitotic cells (Mizeracka et al. 2013a). Almost every cell (~96 %) from which Notch1 was removed at this time became a rod, whereas about 30 % of wild-type cells became bipolar neurons and Müller glia. This study demonstrated that the newborn retinal cells still need Notch1 to escape the rod fate. Interestingly, it is not just the signal from Notch 1 that is required, but new transcription and translation, because the gene itself is required in the postmitotic cells. It is not clear at this time if members of the delta or jagged families of ligands for Notch are required for this function of Notch1 in postmitotic cells. Several ligand genes are expressed at the right time and place to have this role (Bao and Cepko 1997; Nelson and Reh 2008; Rocha et al. 2009), but the role of these ligands in signaling newly postmitotic cells, versus signaling RPCs, has not been analyzed. An excellent candidate is Dll4, which has been shown to be regulated by Foxn4 (Luo et al. 2012), a TF that is required for the genesis of HCs and amacrine cell (ACs) (Li et al. 2004) versus photoreceptors. In addition to identification of the relevant ligand, it will be of interest to determine if the signaling is between siblings, to establish their asymmetry of photoreceptor and nonphotoreceptor fates.
9.2.2 Rax/Rx
The retinal and anterior homeobox genes identified in mouse as Rax (Furukawa et al. 1997a) and in Xenopus as Rx (Mathers et al. 1997) are the founding members of a group of genes with a paired-type homeobox, an octapeptide domain, and the OAR domain. Alignment of vertebrate and invertebrate genes has revealed a great deal of conservation among the sequences in these domains, and has led to a classification scheme comprising 3 groups (Wu et al. 2009). In the rodent lineage, there is a curious deletion such that they have only one Rax gene, the founding member of the group 1 Rax genes. Gain- and loss-of-function experiments in multiple species have shown multiple roles for Rax/Rx genes (Bailey et al. 2004; Muranishi et al. 2012). The group 2 Rax/Rx genes, which include some members referred to as Rax-L, Rax2, or Rx2 genes, are important in photoreceptor genesis and survival and differentiation. Interference with this type of Rax/Rx gene in chick (Chen and Cepko 2002), Xenopus (Wu et al. 2009), or zebrafish (Nelson et al. 2009) led to a reduction in photoreceptors. In the chick, this was at least in part caused by apoptosis as photoreceptors were being generated. In Xenopus, a clonal analysis following gain and loss of Rax-L indicated photoreceptor number increased or decreased, respectively (Wu et al. 2009), with changes also in amacrine and bipolar cells. The gain of function data from Xenopus is in contrast to gain of function using mouse Rax gene, overexpressed in rat, in which case there was a reduction in the formation of all types of neurons, and an increase in cells that resembled Müller glia/RPCs (Furukawa et al. 2000). Deletion of the sole Rax gene in midembryonic development in mouse using a CKO allele showed a reduction in photoreceptor cells, likely through its role in regulating Otx2 (Muranishi et al. 2011). Of interest was the lack of a requirement for the Rax gene for postnatal Otx2 expression, even though Rax was required for embryonic Otx2 expression (discussed further below). Given the data shown in Fig. 9.1 concerning rod and cone birthdays, it does not appear that the embryonic versus postnatal dependence of Otx2 for Rax is a rod versus cone difference, but rather reflects a difference between embryonic and postnatal RPCs. Rax may also be involved in the direct regulation of some photoreceptor genes in mature photoreceptor cells, through binding to the PCE-1/Ret1 site (Kimura et al. 2000).
9.2.3 Otx2
Temporally downstream of Notch, commencing expression in some cells as they exit the cell cycle, is the Otx2 gene (Muranishi et al. 2012; Trimarchi et al. 2008a; Trimarchi et al. 2008b). Otx2 was found to be required for the production of rods and cones, as a CKO of Otx2 in the mouse was found to have no photoreceptors whereas there was an increase in the number of amacrine cells (Nishida et al. 2003). Although there was cell death in this model, the increase in amacrine fate was quite substantial and likely reflected a change in cell fate. Moreover, misexpression of Otx2 via a retroviral vector delivered to the postnatal mouse retina resulted in clones that contained only rods, whereas wild-type clones normally comprise rods, amacrine cells, bipolar cells, and Müller glia (Nishida et al. 2003). Subsequent to these early studies, it was reported that loss of Otx2 also led to loss of HCs, which do not express Otx2, and also to loss of bipolar cells, which express high levels of Otx2 (Sato et al. 2007).
Given its key role in photoreceptor fate determination, the regulation of Otx2 has been of interest. We used electroporation of chick and mouse retinas to assay for cis-regulatory modules (CRM) that show enhancer activity (Emerson and Cepko 2011). An Otx2 enhancer, Otx2ECR2, was found to drive expression in both chick and mouse retinas in the terminal cell cycle of RPCs that produce photoreceptors. Interestingly, when ECR2 was used to drive Cre, only photoreceptors showed a history of Otx2ECR2 expression, despite the fact that Otx2 is also expressed by bipolar cells, and, as discussed next, transiently in RPCs that produce HCs. Although many of the cells that express Otx2ECR2 are newly postmitotic cells fated to be rods or cones, Otx2ECR2 is expressed by a small number of RPCs in mouse, and these must be restricted to producing only photoreceptors. As clonal analyses have shown that some RPCs produce, for example, bipolar cells and rods in a terminal division (Turner and Cepko 1987; Turner et al. 1990), the restriction of ECR2 to some RPCs that only produce photoreceptors indicates that there are intrinsic differences among RPCs, at least in a terminal division.
Muranishi et al. used transgenic mice to assay for Otx2 enhancer activity (Muranishi et al. 2011). They report an enhancer that is different from Otx2ECR2, which they termed EELPOT. This approximately 500-bp enhancer is located further 5′ relative to ECR2, and had activity in a subset of embryonic cells, but interestingly did not have activity in the postnatal mouse retina. Sequence analysis revealed sites for several families of TFs, including E-boxes for neurogenic basic helix-loop-helix (bHLH genes), N-boxes for bHLH genes of the Hes/Hey type, and binding sites for paired-type homeobox genes, which may include Otx2 or Crx. Rax was shown to transactivate this enhancer in a heterologous in vitro system. Similarly, bHLH genes were tested in this assay and some activity was detected for Ngn2. Hes1, Hes5, and Hey1, which are direct downstream targets of Notch and that bind to N-boxes, were able to reduce expression, even in the presence of Rax. The binding of Rax and Hes 1 to EELPOT in retinal extracts was demonstrated using ChIP. Binding was detected in embryonic, but not postnatal, extracts. Finally, a CKO of Rax was made and examined for Otx2 expression and photoreceptor cell numbers, as already discussed. Otx2 was quite reduced in the CKO and photoreceptor numbers were down. The activity of EELPOT in embryonic but not postnatal cells is reminiscent of the activity of Otx2ECR2 only in a subset of RPCs, and again points to intrinsic differences among the RPCs that produce photoreceptor cells, in this case, differences between embryonic and postnatal RPCs. The link to Notch, as perhaps the most upstream regulator of photoreceptor production, was suggested by these data. It is interesting, however, that Notch regulates rod production in the postnatal retina as well as the embryonic retina but apparently does not work through EELPOT in the postnatal retina.
9.2.4 Basic Helix-Loop-Helix
Multiple bHLH genes are expressed in the right time and place to have a role in photoreceptor development. These genes are heavily interconnected in a network (Kanekar et al. 1997; Hutcheson et al. 2005; Hernandez et al. 2007; Hatakeyama and Kageyama 2004), and are downstream of Notch signaling while also being upstream of the Notch ligands. Functional studies in multiple species have been carried out, showing that they play a role in photoreceptor development. However, it is difficult to assign roles precisely to any particular gene, given redundancy, compensation, and the lack of complementarity between some gain- and loss-of-function assays. However, a short summary of some examples of studies of expression and function is given here, with a focus on the mouse.
We carried out single-cell RNA profiling using microarrays in part to understand the complex patterns of expression of many types of TFs in RPCs, including the bHLH genes (Trimarchi et al. 2008b). A subset of the data regarding bHLH expression (shown in Fig. 9.2) illustrated that there are many patterns of expression of the bHLH genes, including those classified using the clade A, B, and E scheme (Skinner et al. 2010). They are expressed in overlapping patterns in mitotic cells, in newly postmitotic cells, and/or in photoreceptor cells. When double immunohistochemistry has been carried out for several of these proteins, a similar result has been found (Brzezinski et al. 2011). Akagi et al. performed a heroic series of experiments, examining single, double, and triple knockouts (KOs) in mice for effects on retinal development (Akagi et al. 2004). Ngn2, Ascl1 (Mash1), Neurod4 (Math3), and Neurod1, in different KO combinations, led to changes in the complement of retinal cell types. The triple KO for Ascl1, Neurod4, and Neurod1 led to an approximately 66 % reduction in photoreceptors. Removal of only Neurod1 led to a defect only in rod survival (Morrow et al. 1999), whereas removal of Neurod4 along with Ascl1 led to a reduction in bipolar cells (Tomita et al. 2000). On the other hand, overexpression of a single bHLH gene, including either Ascl1 or Neurod4, led to a significant increase in rods (Hatakeyama et al. 2001; Cherry et al. 2011) and a complete loss of Müller glia (Cai et al. 2000). Similarly, overexpression of Ngn1 in the chick led to overproduction of photoreceptor cells and resulted in reduced expression of other bHLH genes (Yan et al. 2009). Overexpression of bHLH genes in Xenopus (Wang and Harris 2005; Kanekar et al. 1997) and in zebrafish (Ochocinska and Hitchcock 2009) similarly led to excess photoreceptor cells.
Expression levels of basic helix-loop-helix (bHLH) genes. Individual single cells were used to prepare cDNA probes, which were amplified and applied to Affymetrix microarrays, with the resulting signal levels shown (Trimarchi et al. 2008b). Each cell is represented on a row and each bHLH gene in a column. The clade A bHLH genes are shown in the first group, clade B genes in the second group, and clade E genes in the third group, with groups demarcated by black boxes in the top row, and with the bHLH clade scheme of Skinner used (Skinner et al. 2010). Additional genes for reference cell types are shown on the right side of the heat map, after the clade E genes. The signal levels were scaled such that maximum red signal is 3,000. Retinal progenitor cells (RPCs) are shown with their corresponding ages, embryonic (E) through postnatal (P) stages. Cells demarcated by an r are immature rods; those with N-r are Notch-negative cells. (From Mizeracka et al. 2013b.) Several more mature cells are included for comparison. (From Cherry et al. 2009; Kim et al. 2008; Trimarchi et al. 2007.) GC, ganglion cell; AC, amacrine cell; BP, bipolar cell; MG, Müller glial cell. Genes that mark these cell types are Fgf15, SFRP2, and cycD1 for RPCs, Nfl1 and Ebf3 for GC, Gad1 and Slc6a9 for AC, Otx2 for rods and BP cells, Prdm1 for newly postmitotic cells fated to be rods, Nrl and Nr2E3 for rods, and ApoE for MG
Given the fact that at least some bHLH genes are negatively regulated by Hes/Hey factors, which are direct targets of Notch1 (Davis and Turner 2001), Notch1 signaling likely keeps the levels of neurogenic bHLHs in check and thus regulates the number of photoreceptors that are produced. Indeed, when we performed microarray analysis on single cells from a Notch1 CKO, one of the most noticeable changes was an increase in Neurod1 and Neurod4 (Mizeracka et al. 2013b). This increase was accompanied by a decrease in the Id factors, Id1 and Id3 (Mizeracka et al. 2013a; Mizeracka et al. 2013b). Ids also limit the activity of neurogenic bHLH factors by direct protein interaction (Benezra et al. 1990), and a functional analysis showed that Ids favor formation of Müeller glia (Mizeracka et al. 2013a). The overall chain of events downstream of Notch signaling, beginning in RPCs that are about to produce postmitotic daughter cells, needs to be elucidated to appreciate this network of TFs and their roles. It is likely that different RPCs, almost all of which can produce photoreceptors, use different bHLH genes, regulated by different upstream regulators, to produce photoreceptors and their distinctive siblings in terminal divisions, as is discussed further below.
9.2.5 Blimp1/PRDM1
Blimp1, or Prdm1, is a gene that is expressed in the period when cells fated to be photoreceptors are exiting mitosis (Hsiau et al. 2007) (Katoh et al. 2010; Brzezinski et al. 2010); this positions Blimp in that critical window where Notch, Otx2, and bHLH genes are expressed and are effecting fate decisions. Loss of Blimp1 was reported to lead to a reduction in rod and cone photoreceptors, with a concomitant gain in bipolar cells and abnormal cells with markers of proliferation (Katoh et al. 2010; Brzezinski et al. 2010). The excess bipolar cells were subsequently lost from cell death, perhaps because of their supernumerary status or perhaps being defective. As with bHLH genes, loss of Blimp1 creates a fate change only in a percentage of photoreceptors, as about 50 % of the normal number of rods remain in the Blimp1 CKO mouse; this may result from redundancy, as there are many members of this gene family and some are expressed in the mouse retina. Alternatively, there may be heterogeneity in the pool of RPCs and/or newly postmitotic daughters, and only some of these rely upon Blimp1 to become photoreceptors.
9.3 Genes That Regulate the Differentiation of Rod and Cone Photoreceptors
The network of genes involved in regulating rod- and cone-specific gene expression in differentiating cells define the specific properties of photoreceptor cells. As well, many of these genes are among the human disease genes that lead to blindness, and an understanding of their roles might lead to a greater understanding of these disease states (Blackshaw et al. 2001)(https://sph.uth.edu/retnet/). A short summary is given here of some of the genes that play a role in directing specific gene expression as photoreceptors differentiate.
9.3.1 Crx
Crx, a gene highly related to Otx2, is expressed in newborn rod and cone photoreceptors (Furukawa et al. 1997b; Chen et al. 1997). We examined the kinetics of expression of this gene and found that it could first be detected in cells that had exited the cell cycle, at about 24 h after the last S-phase (Trimarchi et al. 2008b); this would put Crx temporally later than Otx2, thyroid hormone receptor beta (Thrb) (discussed later), and the aforementioned bHLH genes, in mice and chicks. In zebrafish, Crx is detected in cells that are cycling (Riesenberg et al. 2009), perhaps reflecting the more rapid kinetics of development in the fish relative to mice and chicks. Crx is also detected at lower levels in bipolar cells in several species. The Crx KO mouse has photoreceptors that move to their proper position and turn on some markers of photoreceptor cells (Furukawa et al. 1999). However, the cells fail to express almost every photoreceptor gene and eventually degenerate (Furukawa et al. 1999; Livesey et al. 2000; Morrow et al. 2005), identifying Crx as a terminal selector gene for photoreceptors, necessary for differentiation and survival, but not for determination. In humans, there are also Crx mutations that lead to several forms of photoreceptor disease (Rivolta et al. 2001). Otx2 may regulate Crx directly (Nishida et al. 2003), and both Crx and Otx2 are able to bind to a similar sequence and directly regulate many photoreceptor genes (Chen et al. 1997; Furukawa et al. 1997b; White et al. 2013; Peng and Chen 2005). The combined loss of Otx2 and Crx leads to a phenotype of greater loss of photoreceptor gene expression (Koike et al. 2007).
9.3.2 Nuclear Hormone Receptors
Several members of the nuclear hormone receptor family of TFs play critical roles in photoreceptor differentiation, with some specific roles in opsin regulation clearly identified (Forrest and Swaroop 2012). Thrb has been characterized as the earliest cone-specific marker (Sjoberg et al. 1992; Ng et al. 2001). It was studied in the early chick retina for its kinetics relative to Otx2 and NeuroD1 (Trimarchi et al. 2008a). These three genes are expressed with almost identical kinetics in overlapping patterns, with some cells expressing all three genes and other cells expressing one or two of these genes. They initiate expression in what appears to be the terminal S/G2 phase of the cell cycle. Nr2b3 (RXRg) is also expressed in early cones, although it is not specific to cones, as it is also expressed in cells of the inner nuclear layer (INL) and ganglion cell layer (GCL) (Roberts et al. 2005). Thrb and Nr2b3 have been shown to be important in the regulation of cone opsin genes in mice (Ng et al. 2001). Loss of Thrb led to loss of expression of the medium-wavelength opsin, whereas the short-wavelength opsin was earlier in expression and was maintained in all cones. Nr2b3 is required for proper regulation of short-wavelength opsin (Roberts et al. 2005). Thrb and Nr2b3 regulate, and are themselves the target of, the E3 SUMO ligase, Pias3 (Onishi et al. 2009). Unliganded Thrb2 and liganded RXRγ positively regulate Pias3, which then carries out SUMOylation of Nr1f1, Nr2b3, and Thrb1. These SUMOylated TFs then repress S opsin in M cones and liganded Thrb2 and RXRγ activate M opsin. Interestingly, the Pias-3-mediated SUMOylation of the rod-specific transcription factor, Nr2E3, is required to inhibit S opsin in rods (Onishi et al. 2010). The use of Pias3 in both rods and cones for the inhibition of S opsin may reflect an ancient decision to make S opsin the default opsin type for both rods and cones. Thyroid hormone also plays an important role in opsin regulation within the retinas of salmonid fish. As these fish mature, they undergo a switch from UV to blue opsin-expressing cones. Although originally reported to be the result of a cell death and regeneration mechanism (Allison et al. 2006), it has now been established that this switch is a cone opsin expression switch within single cones (Cheng and Flamarique 2007; Cheng et al. 2009).
Nr2e3(PNR) encodes another nuclear hormone receptor that has been shown to be important in the differentiation of rods versus cones (Chen et al. 2005; Corbo and Cepko 2005; Haider et al. 2009). This gene is completely dependent upon the expression of Nrl, as there is almost no expression of Nr2e3 in the Nrl KO mouse. The rd7 mouse model of the Nr2e3 loss of function has a different phenotype, however, from the Nrl KO. In contrast to the Nrl KO mouse, rd7 rods express rod genes. As in the Nrl KO mouse, however, rd7 shows derepression of cone genes. However, there is a more limited number of cone genes that are derepressed in rods relative to the number seen in the Nrl KO. Nr2e3 is thus required for the repression of a subset of cone genes within rods and is not required for rod gene expression. There are also a small number of additional short-wavelength cones in rd7 (Akhmedov et al. 2000; Haider et al. 2001; Corbo and Cepko 2005), but not nearly the number that one observes in the Nrl KO mouse. These abnormalities likely underlie manifestations of Nr2E3 mutations in the human disease, the enhanced S-cone syndrome, wherein there is increased sensitivity to short wavelengths of light (Haider et al. 2000). As mentioned, the E3 SUMO ligase, Pias3, is involved in the regulation of rod versus cone genes, at least partially through Nr2e3 (Onishi et al. 2009). Nr2e3 works in the realm of differentiation rather than determination, as KO or misexpression leads to misregulation of rod and cone genes, but not a complete transformation of rods or cones into other cell types.
Additional members of the nuclear hormone receptor family have roles to play in the proper regulation of rod and cone genes, as reviewed by Forrest and Swaroop (2012). Some examples include Nr1f1 (RORa) (Fujieda et al. 2009), which is important for the proper levels of cone opsin gene expression within cones. Loss of Nr3b2 (ERRb) leads to the degeneration of rods, and thus it is not a rod determination gene, but a gene important for rod differentiation and survival, in contrast to the role played by Nr1f2 (RORb) whose loss leads to a loss of rod determination (Jia et al. 2009; Srinivas et al. 2006; Montana et al. 2011), as is discussed further.
There is likely a network of nuclear hormone receptors, some of which may be linked through their use of coreceptors, the RXRs. A more in depth view of this network is required to properly interpret the roles of these receptors. However, at this point, all except Nr1f2 appear to be important for the proper regulation of rod and cone genes.
9.3.3 Nrl
In rats and mice, the genesis of cones precedes the genesis of rods, although there is a period of significant overlap in the mid- to late embryonic period (Fig. 9.1). The early-born cones are recognizable by expression of Thrb and Nr2b3. These early-born rods do not have a marker that has been recognized by in situ hybridization or immunohistochemistry, although polymerase chain reaction (PCR) of embryonic mouse tissue reveals expression of Nrl. In addition, an Nrl-GFP transgenic mouse shows green fluorescent protein (GFP) as early at E12 (Akimoto et al. 2006), but this early expression has not been shown to be specific to early-born rods. Nrl is a leucine zipper gene of the Maf family, discovered by the group of Anand Swaroop (Swaroop et al. 1992). They described Nrl as a rod-specific gene in the mouse and report expression of the protein in the cytoplasm of human cones (Swain et al. 2001). A null mutation of Nrl in the mouse showed the very interesting phenotype of loss of rods (Mears et al. 2001). Cells fated to be rods were not dead, however, but were transformed into short-wavelength cones. The cones are very close to bona fide cones, as judged by gene expression, morphology, and physiology (Daniele et al. 2005; Mears et al. 2001; Yoshida et al. 2004). This report led to the model that short-wavelength cones are the default type of photoreceptor; that is, when cells fated to be photoreceptors are first born, they enter the pathway of short-wavelength cones (Fig. 9.3a) (Swaroop et al. 2010). In normal development, many of the default cones are hypothesized to become rods when Nrl is expressed. As Nrl appears to be a critical node, one would like to know how Nrl is regulated. Two groups have examined this question, and report that there are conserved binding sites for Nr1f2 and Otx2/Crx upstream of Nrl (Montana et al. 2011; Kautzmann et al. 2011), in keeping with the loss of Nrl in the Nr1f2 KO mouse (Jia et al. 2009). As pointed out by Montana et al., these regulators cannot explain the specificity for Nrl expression in rods versus cones as both Otx2/Crx and Nr1f2 are expressed in both rods and cones.
Model of rod versus cone determination and the ganglion mother cells (GMCs) that make them (a). A model for photoreceptor development in which all photoreceptors originate from a common photoreceptor precursor, which follows a default pathway to differentiate into a blue cone unless instructed otherwise (Swaroop et al. 2010). Nrl, acting in combination with other transcription factors (TFs), is proposed to induce the rod fate, where it directly activates many rod genes and represses many cone genes. (b) A revised model, based upon lineage data (from Emerson et al. 2013; Emerson and Cepko 2011; Hafler et al. 2012; Suzuki et al. 2013) wherein distinct RPCs produce cones and rods, and there is not a common photoreceptor precursor that is, by default, a blue cone. In mice, some RPCs that produce a cone also produce a horizontal cell (HC) and express Olig2, Oc1, Otx2, Thrb, and Neurod1
The genes regulated by Crx and Nrl have been described in some detail, using expression profiling and CHIP-seq (Hao et al. 2012; Yoshida et al. 2004; Corbo et al. 2007, 2010; Hsiau et al. 2007). They act together at many loci in rods, and both can be considered to be terminal selector genes, directly regulating many of the genes required for rod function. Their importance is underscored by the observation that many of their target genes are human disease genes.
9.3.4 Sall3
Spalt homologue 3 (Sall3), a transcription factor homologous to the Drosophila Spalt, which directly regulates Drosophila opsin expression, plays a role in cone differentiation in mice. It is a positive regulator of the short-wavelength opsin gene, along with additional cone genes, and is required for proper HC development (de Melo et al. 2011). Misexpression studies show that it is sufficient to induce the short-wavelength cone opsin gene in rods, although it is unable to transfate rods or other cell types into cones. Similarly, it plays a role in the intermediate stages of HC differentiation.
9.4 RPCs That Produce Rods and Cones
Almost every clone derived from retroviral marking in the embryonic or postnatal retina or rats and mice contained a photoreceptor (Turner and Cepko 1987; Turner et al. 1990). In addition, many clones, even two-cell clones, contained a photoreceptor and another retinal cell type. These data indicated that almost every RPC has the ability to make a photoreceptor, and that many RPCs are multipotent. However, these data did not address whether all RPCs are equivalent. To probe the nature of RPCs, and to see if they differed from each other, we performed single-cell profiling using microarrays (Trimarchi et al. 2008b). These data showed many differences among RPCs across development as well as at a single time in development. One category of genes that varied was the bHLH category (Fig. 9.2). Olig2, a Clade E bHLH gene, showed variation in expression within RPCs across time. The single-cell microarray data regarding expression of Olig2 were corroborated by in situ hybridization (Hafler et al. 2012) and immunohistochemistry for Olig2 (Nakamura et al. 2006; Shibasaki et al. 2007). All these assays showed that Olig2 was expressed primarily in RPCs, but only in a subset of RPCs at each time in development. Two Cre knock-in lines for Olig2 in mice were available (Schuller et al. 2008; Dessaud et al. 2007). We thus were able to use these lines to analyze the descendants of Olig2+ RPCs using two methods. One method was the classic Cre fate mapping method. By crossing these two different Olig2-Cre strains to three different floxed reporter lines, we could see all the cells with an expression history for Olig2. These fate mapping experiments showed that cells with an Olig2 history comprised primarily rods, cones, HCs, and amacrine cells. Some bipolar cells showed history but Müller glia and RGCs almost never showed any Olig2 history. These results demonstrated that Olig2-expressing RPCs did not behave as totipotent RPCs.
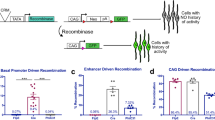
A second method was developed to allow a determination of the types of cells descendant from Olig2-expressing RPCs, at clonal resolution, across time. The classic Cre fating experiment does not provide for temporal resolution, unless used in conjunction with a tamoxifen-regulated allele of Cre, which was not available in this case. The Cre fate mapping method also does not readily provide clonal resolution. Importantly, cells that do not derive from Olig2-expressing RPCs, but rather from expression of Olig2 in postmitotic cells, are mixed in with those cells produced by Olig2-expressing RPCs. As we wished to examine only those cells produced by Olig2-expressing RPCs, and to do so with temporal and clonal resolution, we developed a retroviral marking strategy to accomplish this goal (Beier et al. 2011). This method relies upon expression of the avian retroviral receptor, TVA (Bates et al. 1998).
The TVA receptor is not normally expressed in mammals. However, one can direct expression of TVA to Cre-expressing cells by crossing a Cre line to a floxed TVA line (Beier et al. 2011). Alternatively, one can make a TVA knock-in or transgenic line. Schuller et al. had made a knock-in of both Cre and TVA into the Olig2 locus (Schuller et al. 2008) and had generously given this line to us for our experiments. Infection of TVA-expressing cells can be accomplished using retroviruses that carry on their surface the avian EnvA glycoprotein. By using gamma retroviruses for this analysis, only clones deriving from mitotic cells that express TVA would be produced, as gamma retroviruses are not able to integrate their DNA into postmitotic cells and thus are unable to initiate expression in such cells (Roe et al. 1993). If a gamma retrovirus integrates its genome into a host cell that then exits mitosis, a one-cell clone will be formed. If the integrated cell divides to make two postmitotic daughters, a two-cell clone is formed, etc. We thus infected the Olig2-Cre-IRES-TVA line of mice with a retroviral genome with an EnvA glycoprotein, or used the same virus with a promiscuous glycoprotein that allows infection of any type of RPC, as a control. The clone sizes and compositions from infection at several different ages were then compared.
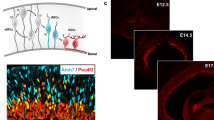
The clonal analysis showed the same trends as the Cre fate mapping experiment but revealed a significant skew in terms of both the size and composition of clones from infection of Olig2-expressing RPCs (Hafler et al. 2012). First, the clones generated by Olig2-expressing RPCs were only 1 or 2 cells, indicating that these RPCs were terminally dividing RPCs. In contrast, the average clone size, for example, for the control group of clones infected by a retrovirus that could infect any RPC, was 32 cells/clone (range, 1–234 cells). Second, there was a striking specificity in the types of cells produced by infection at different times. When infections of Olig2-expressing RPCs were performed at E14.5, only cones and HCs were marked. They comprised single-cell clones of either cell type, or 2-cell clones of 2 cones, 2 HCs, or 1 cone and 1 HC. In contrast, when infections were performed at P0, almost every clone was a single rod, with a few clones comprising an amacrine cell, and a very few 2-cell clones comprising either 2 rods or a rod and an amacrine cell. If marking was initiated at P3, the clones were almost entirely a single rod, although a few single amacrine cells and single bipolar cells were marked as well. Statistical analyses indicated an extreme skew in these data from each infection time point, relative to the cells expected based upon birthdates and the cell types in the control set of clones. Similarly, there was an extreme skew toward small clone size. Results from tamoxifen-regulated Cre fate mapping experiments using history of Ngn2 or Ascl1 also showed an interesting skew in rods and cones (Brzezinski et al. 2011). When marking was initiated at E12.5–E13.5, small “clumps” of cells were labeled, with rods marked preferentially by Ascl1 history, and cones and HCs marked preferentially with Ngn2 history.
The clones produced by Olig2-expressing embryonic RPCs are reminiscent of the clones marked in a study of the chick retina using retroviruses (Rompani and Cepko 2008) and in a study using live imaging of the zebrafish retina (Godinho et al. 2007). The chick has three types of HCs, referred to as HC1, HC2, and HC3, which have different patterns of connectivity to rods and cones. Large clones with many types of retinal cells, and all three types of HCs, were observed following marking near the beginning of chick retinal development. Later marking led to the production of clones with many types of cells, but only one or two HCs. Analysis of the combinations of the types of HCs within clones with two HCs revealed a nonrandom distribution in the types of HCs. Clones with only two HCs had homotypic pairs, either two H1 cells or two H3 cells. No pairs of only H2 cells were seen. Moreover, the clones that had only a single HC were skewed toward H2. When the numbers of each type of HC seen in clones with larger numbers of HC were analyzed, there was a skew toward even numbers of H1 or H3, but not H2. We interpret these data to mean that there is a specific and restricted RPC that divides once to make a pair of H1 cells, and a different RPC that divides once to make a pair of H3 cells. The preponderance toward even numbers suggests that clones contain multiple RPCs of these restricted types. Interestingly, the RPC that makes an H2 does not make a pair of H2 cells. In consideration of the data from the Olig2-expressing RPCs in mouse, we predict that the sibling of the chick H2 cell is a cone photoreceptor. In zebrafish, live imaging showed nonapical divisions of an RPC that produced only HCs (Godinho et al. 2007). The types of HCs produced were not ascertained in this study.
A recent lineage study used live imaging of zebrafish retinas and a fluorescent reporter based upon the Thrb gene (Suzuki et al. 2013). Suzuki et al. found that this reporter marked RPCs that made a terminal division that resulted in L cones, those that express the long-wavelength opsin. Alternatively, RPCs that express this reporter make a terminal division that produces a pair of HCs. A prior division also was seen in three cases where a retinal ganglion cells was produced. If they used a reporter based upon Crx, homotypic pairs of cones were observed, with live imaging in one case showing that the homotypic pairs of UV or M cones were made in terminal divisions.
The finding of restricted RPCs that make limited divisions and specific cell types, along with the observation of larger clones with many cell types, has suggested that the vertebrate retina uses the same strategy as the ventral nerve cord of Drosophila (Baumgardt et al. 2007; Pearson and Doe 2004; Zhong 2003) or the medulla of Drosophila (Li et al. 2013a, b). We have proposed that the terminally dividing RPCs, of the type identified using the Olig2-TVA line and the zebrafish Thrb reporter, are like the ganglion mother cells (GMCs) of the Drosophila ventral nerve cord and medulla. GMCs typically divide only once and make specific types of daughter cells. The types of daughters that are made are different in different segments and different at different times. The temporal order in the nerve cord is reminiscent of the temporal order seen during retinal development. The GMCs are produced by neuroblasts, which make large clones when they are marked, much as we see large clones when we mark with a virus that can mark any RPC. The neuroblasts in each segment rely on a temporal order of TFs to produce their temporal cohorts of GMCs, and even though the same temporal TFs are used in each segment, different types of cells are made in each segment. In the vertebrate retina, there is also expression of paralogues of these temporal TFs, and some of these genes have similar roles in setting up temporal identity in the vertebrate retina (Blackshaw et al. 2004; Brzezinski et al. 2010; Elliott et al. 2008; Trimarchi et al. 2008b; Katoh et al. 2010).
The foregoing studies demonstrate that different RPCs make different daughter cell types in terminal divisions. In the mouse, the E13.5–E14.5 Olig2-expressing RPCs make cones and HCs whereas the postnatal Olig2-RPCs make rods and amacrine cells. The Thrb-expressing RPCs in zebrafish make HCs or L cones and the Crx-expressing RPCs make homotypic pairs of other cone types. The TFs that are responsible for the production of rods versus cones in such RPCs have recently been discovered, as described next.
9.5 Otx2 and Onecut Genes Direct Formation of Cones Versus Rods
The Thrb gene is an early marker of cones. We thus reasoned that the upstream regulators of Thrb would be informative regarding the genesis of cones. We used electroporation (Matsuda and Cepko 2004) to identify a noncoding region of Thrb (“ThrbCRM1”) that directed expression to early cones in the chick and in the mouse. This conserved region was reduced to 40 bp and was found to label early cones, as well as two other cell types. Investigation of these other cell types showed that they were a subset of RPCs, as well as newborn HCs. Reversing the strategy described for the Olig2-RPCs, we used ThrbCRM1 to drive expression of the murine retrovirus receptor, CAT1 (Albritton et al. 1989), in the chick using electroporation. Infecting these electroporated retinas with a retrovirus with the murine glycoprotein gene, gp70, led to the identification of cells produced by the RPCs that expressed ThrbCRM1. The progeny were cones and HCs, providing evidence that, as in the mouse, there is a specific RPC in the chick that makes cones and HCs. The clones comprised almost entirely one cell. Larger clones were not seen, so these RPCs were making terminal divisions.
Investigation of the TFs that could bind and activate the 40-bp ThrbCRM1 led to the identification of Otx2 and Onecut1 (Oc1) as both necessary and sufficient, not only for activation of this enhancer, but also the endogenous Thrb gene in both chick and mouse. Chromatin immunoprecipitation experiments using retinal extracts confirmed that these two proteins bind ThrbCRM1. Otx2 and Oc1 were found to overlap in expression with Olig2, in a subset of RPC cells, as predicted by the activity of ThrbCRM1 in a subset of RPCs, and the previous observations from infection of Olig2-TVA in the mouse.
It was of interest to determine if Otx2 and Oc1 regulate the production of cones, HCs, and potentially, rod photoreceptors. To this end, gain and loss of function experiments were carried out in mice and chicks. Misexpression of Oc1 in the postnatal mouse retina, where Otx2 is expressed, could induce the formation of immature cones. This induction was dependent on Otx2 as removal of a conditional allele of Otx2 prevented this induction. The induced cones did not progress to fully differentiated cones. We speculate that this might be caused by two factors. One is that expression of the Oc1 gene is normally reduced as cones mature, and the misexpression construct was constitutive. Second, a gene such as Sall3 might need to be upregulated at the proper time and proper level, and misexpression of Oc1 did not lead to this. Interestingly, in addition to induction of cones, cells with markers of HCs were produced from introduction of Oc1 into the postnatal mouse retina.
Two methods were used to examine the necessity of Oc1. Electroporation of the chick retina with a construct in which the Engrailed repressor domain was fused to Oc1 led to a reduction in Thrb reporter expression. This construct also led to an upregulation of MafA, the chick homologue of Nrl (Ochi et al. 2004), and to expression of a rhodopsin promoter construct. These findings suggest an induction of rod genesis. In embryonic Oc1 KO mice, a reduction in Thrb RNA and an upregulation in Nrl RNA were seen. These data all point to a role of the Oc1 gene in regulating the rod versus cone decision. These data suggest a revised model for rod and cone genesis (Fig. 9.3b).
9.6 Model of Rod Versus Cone Determination
In keeping with the notion of GMCs and neuroblasts in retinal development, we propose that there are specific GMCs that produce rods, cones, HCs, and likely other retinal cell types as well (Fig. 9.4). At least some of the types of GMCs that produce cones also produce HCs, and the GMCs that make cones are not the same ones that make rods. Rods are proposed to be produced by multiple types of GMCs. We propose multiple GMCs for rod production for two reasons. One reason is the nature of two-cell clones that are produced by viral infection in the postnatal period of rats and mice (Turner and Cepko 1987; Turner et al. 1990). Here, we see clones in which there are two rods, a rod and amacrine cell, a rod and a bipolar cell, or a rod and a Müller glial cell. Although one could model these clone types as deriving from a single type of rod GMC with competence to make all four cell types (Gomes et al. 2011), the Olig2 lineage data argue against this idea. Olig2-expressing RPCs in the postnatal period make either two rods or a rod and amacrine cell, whereas Olig2-negative RPCs make rod and bipolar and rod and Müller glial clones. Interestingly, the newly postmitotic daughter cells that would normally take on the non-rod fates still rely on Notch1 (Mizeracka et al. 2013a) and Numb (Kechad et al. 2012) to escape the rod fate. This finding argues against GMCs passing on irreversible fate decisions to their daughters. Rather, it is likely that determinants are passed to daughter cells from GMCs (Kechad et al. 2012), and although different determinants are passed from different GMCs, the newly postmitotic daughter cells must work out their fate using at least some other cue, such as the Notch signal, to adopt the proper fate. Because deletion of the Notch gene in the newly postmitotic cell prevented the acquisition of the non-rod fate, Notch transcription and translation in the newly postmitotic cell are needed to generate Notch signals (Mizeracka et al. 2013a). This Notch dependence has also been seen in the Drosophila ventral nerve cord in the newly postmitotic daughters of the GMCs (Spana and Doe 1996) as well as in vertebrate neurons that are produced as asymmetrical pairs (Del Barrio et al. 2007). Regarding the rod versus cone fate, the network operating in cone GMCs includes, at least in part, Otx2 and Oc1. The daughters of these GMCs then differentially regulate the Oc1 and Otx2 genes, as HCs upregulate Oc genes (Wu et al. 2012) and downregulate Otx2, and cones do the opposite. Additional levels of regulation downstream then can lead to photoreceptors with rod gene expression, as interference with Oc1 function leads to cells with Nrl or MafA, and thus likely will become rods. Because Nrl was upregulated following introduction of an Oc1-EnR allele, it is likely that Oc1 positively regulates a repressor of Nrl expression.
Distinct RPCs that make terminal divisions to produce at least one rod or one cone, based upon marker expression and lineage tracing experiments, are shown (Emerson and Cepko 2011; Hafler et al. 2012; Rompani and Cepko 2008; Turner and Cepko 1987; Turner et al. 1990); (Suzuki et al. 2013). These are modeled to behave as GMCs similar to those seen in Drosophila. Three types of two cell clones are produced at E13.5–E14.5 in the mouse from Olig2-expressing GMCs. At least two of these clone types include a cone, as shown, and one type produces two HCs (not shown). The GMC that produces only cones likely uses the Otx2ECR2 to regulate Otx2 (Emerson and Cepko 2011). In zebrafish, a reporter based upon Thrb makes a pair of L cones from a terminal division (Suzuki et al. 2013). Other Thrb-expressing RPCs divide to make a pair of HCs (not shown). Crx-expressing RPCs produce homotypic pairs with respect to cone opsin type, with live imaging showing that these are likely the result of terminal divisions, as shown. GMCs that produce rods, but not cones, exist later in development in mice. They do not express Oc1 or Thrb and some of them express Olig2. At least some of those that make only rods express Otx2ECR2. HC, horizontal cell; BP, bipolar cell; AC, amacrine cell; MG, Müller glial cell
It is worth considering the phenotypes of the KO mice for Nrl, Nr1f2, and Nr2e3 in terms of the foregoing model. The fact that rods are proposed to be produced by RPCs that are distinct from the GMCs that make cones render the idea of a default blue cone fate unlikely. The lack of a blue cone default state is further supported by the lack of an upregulation of the early cone genes in the embryonic Nrl KO retina (Emerson et al. 2013). Rather than being a reflection of a normal developmental state, expression of blue cone genes in the aforementioned KO strains likely results from dysregulation of rod and cone genes in maturing photoreceptors. The blue cone gene expression program may be the program that is set up by expression of the common TFs in photoreceptors, including TFs such as Crx/Otx2 and Nr1f2. The differential expression of specific TFs, such as Nrl and Nr2e3, and their cofactors and regulators, such as Pias3, then defines the specific gene expression programs for rods and the different types of cones. The difference in these models is not merely a semantic one, as the blue cone default model invokes the blue cone fate as the state into which a newly postmitotic cell fated to be a photoreceptor enters, no matter which RPC produces it. Instead, we propose that the newly postmitotic cells fated to be photoreceptors are already instructed by their GMCs to be a cone or a rod, by virtue of differences within the GMCs that make these cells. We have defined the Oc1 gene as one of the critical differences between rod and cone GMCs. There are undoubtedly others, and the networks of which they are part need to be elucidated to understand the actual mechanisms that guide, and then lock, each type of photoreceptor into its final state. These networks must also be understood in terms of the spatial patterning across the retina that govern the frequency of the different types of photoreceptors, as exemplified by the fovea and similar specialized areas in different organisms. The final fate may also be dependent upon chromatin changes to make the changes irreversible.
Finally, it is interesting to note the similarities of the genes discussed herein to those used in the development of Drosophila photoreceptors. The discovery of the determination network that defines the eye field in disparate organisms has been well noted and discussed (Fernald 2006; Gehring 2011). In addition to these early genes, the Drosophila homologue of Otx/Crx, orthodenticle (Otd) (Vandendries et al. 1996), and the Drosophila homologue of Sall3, Spalt (Domingos et al. 2004), are required for Drosophila S cone regulation. The Drosophila Onecut gene likely also plays a role in photoreceptor differentiation, as it is able to bind to a conserved enhancer upstream of the Rh1 gene, and a dominant negative allele disrupts photoreceptor differentiation (Nguyen et al. 2000). These genes have some common expression patterns, as well as some shared functions in Drosophila and mammals; for example, Crx and Otd are functionally similar in developing photoreceptors (Ranade et al. 2008; Tahayato et al. 2003). However, we are now proposing that the strategy used by the vertebrate retina for the production of the diverse set of cell types does not follow that of the Drosophila retina but rather follows that of the Drosophila ventral nerve cord and medulla (Pearson and Doe 2004). The terminally dividing RPCs have the same properties as GMCs, and there is support for both expression and function for the homologues of the Drosophila temporal TFs (Elliott et al. 2008). The strategy used for diversification of cells during development may be an overarching theme in many tissues, with specific cohorts of TFs acting to specify and differentiate the distinct cell types in tissues. Photoreceptors may have enough deep homology that they use some of the same TFs used by Drosophila photoreceptors for development, evolving different roles over time.
Finally, in keeping with the ventral nerve cord and medulla strategy, it may be that there are lineages in the vertebrate retina that are distinct; that is, specific and distinct RPCs may be upstream of specific GMCs. There is some evidence for this, in that RPCs that express Cad6 preferentially make RGCs that express Cad6, as well as other retinal cell types (De la Huerta et al. 2012). This observation could mean that there are some limited types of daughter cells produced by Cad6-expressing RPCs, or perhaps there is one division that produces a Cad6 RGC, with a sibling being a more generic type of RPC. Similarly, analysis of clones with many HCs in large chick clones shows biases toward the types of HCs that they contain, perhaps implying differences among very early RPCs toward one type of HC (Rompani and Cepko, unpublished data). Future lineage and molecular studies are needed to determine the full set of RPCs and GMCs and the mechanisms that reliably lead to the production of such a beautifully complex tissue.
References
Akagi T, Inoue T, Miyoshi G, Bessho Y, Takahashi M, Lee JE, Guillemot F, Kageyama R (2004) Requirement of multiple basic helix-loop-helix genes for retinal neuronal subtype specification. J Biol Chem 279(27):28492–28498. doi:10.1074/jbc.M400871200
Akhmedov NB, Piriev NI, Chang B, Rapoport AL, Hawes NL, Nishina PM, Nusinowitz S, Heckenlively JR, Roderick TH, Kozak CA, Danciger M, Davisson MT, Farber DB (2000) A deletion in a photoreceptor-specific nuclear receptor mRNA causes retinal degeneration in the rd7 mouse. Proc Natl Acad Sci USA 97(10):5551–5556
Akimoto M, Cheng H, Zhu D, Brzezinski JA, Khanna R, Filippova E, Oh EC, Jing Y, Linares JL, Brooks M, Zareparsi S, Mears AJ, Hero A, Glaser T, Swaroop A (2006) Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc Natl Acad Sci USA 103(10):3890–3895. doi:10.1073/pnas.0508214103
Albritton LM, Tseng L, Scadden D, Cunningham JM (1989) A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57(4):659–666
Allison WT, Dann SG, Veldhoen KM, Hawryshyn CW (2006) Degeneration and regeneration of ultraviolet cone photoreceptors during development in rainbow trout. J Comp Neurol 499(5):702–715. doi:10.1002/cne.21164
Altshuler D, Lo Turco JJ, Cepko C (1991) Specification of cell type in the vertebrate retina. In: Lam M-K, Shatz C (eds) Development of the visual system. MIT Press, Cambridge, pp 37–58
Bailey TJ, El-Hodiri H, Zhang L, Shah R, Mathers PH, Jamrich M (2004) Regulation of vertebrate eye development by Rx genes. Int J Dev Biol 48(8-9):761–770. doi:10.1387/ijdb.041878tb
Bao ZZ, Cepko CL (1997) The expression and function of Notch pathway genes in the developing rat eye. J Neurosci 17(4):1425–1434
Bates P, Rong L, Varmus HE, Young JA, Crittenden LB (1998) Genetic mapping of the cloned subgroup A avian sarcoma and leukosis virus receptor gene to the TVA locus. J Virol 72(3):2505–2508
Baumgardt M, Miguel-Aliaga I, Karlsson D, Ekman H, Thor S (2007) Specification of neuronal identities by feedforward combinatorial coding. PLoS Biol 5(2):e37. doi:10.1371/journal.pbio.0050037
Beier KT, Samson ME, Matsuda T, Cepko CL (2011) Conditional expression of the TVA receptor allows clonal analysis of descendents from Cre-expressing progenitor cells. Dev Biol 353(2):309–320. doi:10.1016/j.ydbio.2011.03.004
Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H (1990) The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61(1):49–59
Blackshaw S, Fraioli RE, Furukawa T, Cepko CL (2001) Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell 107(5):579–589
Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, Weber G, Lee K, Fraioli RE, Cho SH, Yung R, Asch E, Ohno-Machado L, Wong WH, Cepko CL (2004) Genomic analysis of mouse retinal development. PLoS Biol 2(9):E247
Bruhn SL, Cepko CL (1996) Development of the pattern of photoreceptors in the chick retina. J Neurosci 16(4):1430–1439
Brzezinski JA, Lamba DA, Reh TA (2010) Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development (Camb) 137(4):619–629. doi:10.1242/dev.043968
Brzezinski JA, Kim EJ, Johnson JE, Reh TA (2011) Ascl1 expression defines a subpopulation of lineage-restricted progenitors in the mammalian retina. Development (Camb) 138(16):3519–3531. doi:10.1242/dev.064006
Cai L, Morrow EM, Cepko CL (2000) Misexpression of basic helix-loop-helix genes in the murine cerebral cortex affects cell fate choices and neuronal survival. Development (Camb) 127(14):3021–3030
Carter-Dawson LD, LaVail MM (1979) Rods and cones in the mouse retina. II. Autoradiographic analysis of cell generation using tritiated thymidine. J Comp Neurol 188(2):263–272. doi:10.1002/cne.901880205
Chen CM, Cepko CL (2002) The chicken RaxL gene plays a role in the initiation of photoreceptor differentiation. Development (Camb) 129(23):5363–5375
Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ (1997) Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron 19(5):1017–1030
Chen J, Rattner A, Nathans J (2005) The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J Neurosci 25(1):118–129. doi:10.1523/JNEUROSCI.3571-04.2005
Cheng CL, Flamarique IN (2007) Chromatic organization of cone photoreceptors in the retina of rainbow trout: single cones irreversibly switch from UV (SWS1) to blue (SWS2) light sensitive opsin during natural development. J Exp Biol 210(pt 23):4123–4135. doi:10.1242/jeb.009217
Cheng CL, Gan KJ, Flamarique IN (2009) Thyroid hormone induces a time-dependent opsin switch in the retina of salmonid fishes. Invest Ophthalmol Vis Sci 50(6):3024–3032. doi:10.1167/iovs.08-2713
Cherry TJ, Trimarchi JM, Stadler MB, Cepko CL (2009) Development and diversification of retinal amacrine interneurons at single cell resolution. Proc Natl Acad Sci USA 106(23):9495–9500. doi:10.1073/pnas.0903264106
Cherry TJ, Wang S, Bormuth I, Schwab M, Olson J, Cepko CL (2011) NeuroD factors regulate cell fate and neurite stratification in the developing retina. J Neurosci 31(20):7365–7379. doi:10.1523/JNEUROSCI.2555-10.2011
Corbo JC, Cepko CL (2005) A hybrid photoreceptor expressing both rod and cone genes in a mouse model of enhanced S-cone syndrome. PLoS Genet 1(2):e11. doi:10.1371/journal.pgen.0010011
Corbo JC, Myers CA, Lawrence KA, Jadhav AP, Cepko CL (2007) A typology of photoreceptor gene expression patterns in the mouse. Proc Natl Acad Sci USA 104(29):12069–12074. doi:10.1073/pnas.0705465104
Corbo JC, Lawrence KA, Karlstetter M, Myers CA, Abdelaziz M, Dirkes W, Weigelt K, Seifert M, Benes V, Fritsche LG, Weber BH, Langmann T (2010) CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors. Genome Res 20(11):1512–1525. doi:10.1101/gr.109405.110
Daniele LL, Lillo C, Lyubarsky AL, Nikonov SS, Philp N, Mears AJ, Swaroop A, Williams DS, Pugh EN Jr (2005) Cone-like morphological, molecular, and electrophysiological features of the photoreceptors of the Nrl knockout mouse. Invest Ophthalmol Vis Sci 46(6):2156–2167. doi:10.1167/iovs.04-1427
Davis RL, Turner DL (2001) Vertebrate hairy and Enhancer of split related proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene 20(58):8342–8357. doi:10.1038/sj.onc.1205094
De la Huerta I, Kim IJ, Voinescu PE, Sanes JR (2012) Direction-selective retinal ganglion cells arise from molecularly specified multipotential progenitors. Proc Natl Acad Sci USA 109(43):17663–17668. doi:10.1073/pnas.1215806109
de Melo J, Peng GH, Chen S, Blackshaw S (2011) The Spalt family transcription factor Sall3 regulates the development of cone photoreceptors and retinal horizontal interneurons. Development (Camb) 138(11):2325–2336. doi:10.1242/dev.061846
Del Barrio MG, Taveira-Marques R, Muroyama Y, Yuk DI, Li S, Wines-Samuelson M, Shen J, Smith HK, Xiang M, Rowitch D, Richardson WD (2007) A regulatory network involving Foxn4, Mash1 and delta-like 4/Notch1 generates V2a and V2b spinal interneurons from a common progenitor pool. Development (Camb) 134(19):3427–3436. doi:10.1242/dev.005868
Dessaud E, Yang LL, Hill K, Cox B, Ulloa F, Ribeiro A, Mynett A, Novitch BG, Briscoe J (2007) Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature (Lond) 450(7170):717–720. doi:10.1038/nature06347
Domingos PM, Brown S, Barrio R, Ratnakumar K, Frankfort BJ, Mardon G, Steller H, Mollereau B (2004) Regulation of R7 and R8 differentiation by the spalt genes. Dev Biol 273(1):121–133. doi:10.1016/j.ydbio.2004.05.026
Elliott J, Jolicoeur C, Ramamurthy V, Cayouette M (2008) Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron 60(1):26–39. doi:10.1016/j.neuron.2008.08.008
Emerson MM, Cepko CL (2011) Identification of a retina-specific Otx2 enhancer element active in immature developing photoreceptors. Dev Biol 360(1):241–255. doi:10.1016/j.ydbio.2011.09.012
Emerson MM, Surzenko N, Goetz JJ, Trimarchi J, Cepko CL (2013) The Otx2 and Onecut factors promote cone photoreceptor and horizontal cell genesis over rod photoreceptors. Dev Cell 154(4):928–939
Fernald RD (2006) Casting a genetic light on the evolution of eyes. Science 313(5795):1914–1918. doi:10.1126/science.1127889
Forrest D, Swaroop A (2012) Minireview: the role of nuclear receptors in photoreceptor differentiation and disease. Mol Endocrinol 26(6):905–915. doi:10.1210/me.2012-1010
Fujieda H, Bremner R, Mears AJ, Sasaki H (2009) Retinoic acid receptor-related orphan receptor alpha regulates a subset of cone genes during mouse retinal development. J Neurochem 108(1):91–101. doi:10.1111/j.1471-4159.2008.05739.x
Furukawa T, Kozak CA, Cepko CL (1997a) rax, a novel paired-type homeobox gene, shows expression in the anterior neural fold and developing retina. Proc Natl Acad Sci USA 94(7):3088–3093
Furukawa T, Morrow EM, Cepko CL (1997b) Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell 91(4):531–541
Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL (1999) Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat Genet 23(4):466–470. doi:10.1038/70591
Furukawa T, Mukherjee S, Bao ZZ, Morrow EM, Cepko CL (2000) rax, Hes1, and notch1 promote the formation of Müller glia by postnatal retinal progenitor cells. Neuron 26(2):383–394
Gehring WJ (2011) Chance and necessity in eye evolution. Genome Biol Evol 3:1053–1066. doi:10.1093/gbe/evr061
Godinho L, Williams PR, Claassen Y, Provost E, Leach SD, Kamermans M, Wong RO (2007) Nonapical symmetric divisions underlie horizontal cell layer formation in the developing retina in vivo. Neuron 56(4):597–603. doi:10.1016/j.neuron.2007.09.036
Gomes FL, Zhang G, Carbonell F, Correa JA, Harris WA, Simons BD, Cayouette M (2011) Reconstruction of rat retinal progenitor cell lineages in vitro reveals a surprising degree of stochasticity in cell fate decisions. Development (Camb) 138(2):227–235. doi:10.1242/dev.059683
Hafler BP, Surzenko N, Beier KT, Punzo C, Trimarchi JM, Kong JH, Cepko CL (2012) Transcription factor Olig2 defines subpopulations of retinal progenitor cells biased toward specific cell fates. Proc Natl Acad Sci USA 109(20):7882–7887. doi:10.1073/pnas.1203138109
Haider NB, Jacobson SG, Cideciyan AV, Swiderski R, Streb LM, Searby C, Beck G, Hockey R, Hanna DB, Gorman S, Duhl D, Carmi R, Bennett J, Weleber RG, Fishman GA, Wright AF, Stone EM, Sheffield VC (2000) Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat Genet 24(2):127–131. doi:10.1038/72777
Haider NB, Naggert JK, Nishina PM (2001) Excess cone cell proliferation due to lack of a functional NR2E3 causes retinal dysplasia and degeneration in rd7/rd7 mice. Hum Mol Genet 10(16):1619–1626
Haider NB, Mollema N, Gaule M, Yuan Y, Sachs AJ, Nystuen AM, Naggert JK, Nishina PM (2009) Nr2e3-directed transcriptional regulation of genes involved in photoreceptor development and cell-type specific phototransduction. Exp Eye Res 89(3):365–372. doi:10.1016/j.exer.2009.04.006
Hao H, Kim DS, Klocke B, Johnson KR, Cui K, Gotoh N, Zang C, Gregorski J, Gieser L, Peng W, Fann Y, Seifert M, Zhao K, Swaroop A (2012) Transcriptional regulation of rod photoreceptor homeostasis revealed by in vivo NRL targetome analysis. PLoS Genet 8(4):e1002649. doi:10.1371/journal.pgen.1002649
Hatakeyama J, Kageyama R (2004) Retinal cell fate determination and bHLH factors. Semin Cell Dev Biol 15(1):83–89. doi:10.1016/j.semcdb.2003.09.005
Hatakeyama J, Tomita K, Inoue T, Kageyama R (2001) Roles of homeobox and bHLH genes in specification of a retinal cell type. Development (Camb) 128(8):1313–1322
Hernandez J, Matter-Sadzinski L, Skowronska-Krawczyk D, Chiodini F, Alliod C, Ballivet M, Matter JM (2007) Highly conserved sequences mediate the dynamic interplay of basic helix-loop-helix proteins regulating retinogenesis. J Biol Chem 282(52):37894–37905. doi:10.1074/jbc.M703616200
Hobert O (2011) Regulation of terminal differentiation programs in the nervous system. Annu Rev Cell Dev Biol 27:681–696. doi:10.1146/annurev-cellbio-092910-154226
Holt CE, Bertsch TW, Ellis HM, Harris WA (1988) Cellular determination in the Xenopus retina is independent of lineage and birth date. Neuron 1(1):15–26
Hsiau TH, Diaconu C, Myers CA, Lee J, Cepko CL, Corbo JC (2007) The cis-regulatory logic of the mammalian photoreceptor transcriptional network. PloS One 2(7):e643. doi:10.1371/journal.pone.0000643
Hutcheson DA, Hanson MI, Moore KB, Le TT, Brown NL, Vetter ML (2005) bHLH-dependent and -independent modes of Ath5 gene regulation during retinal development. Development (Camb) 132(4):829–839. doi:10.1242/dev.01653
Jadhav AP, Mason HA, Cepko CL (2006) Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development (Camb) 133(5):913–923. doi:10.1242/dev.02245
Jia L, Oh EC, Ng L, Srinivas M, Brooks M, Swaroop A, Forrest D (2009) Retinoid-related orphan nuclear receptor RORbeta is an early-acting factor in rod photoreceptor development. Proc Natl Acad Sci USA 106(41):17534–17539. doi:10.1073/pnas.0902425106
Kanekar S, Perron M, Dorsky R, Harris WA, Jan LY, Jan YN, Vetter ML (1997) Xath5 participates in a network of bHLH genes in the developing Xenopus retina. Neuron 19(5):981–994
Katoh K, Omori Y, Onishi A, Sato S, Kondo M, Furukawa T (2010) Blimp1 suppresses Chx10 expression in differentiating retinal photoreceptor precursors to ensure proper photoreceptor development. J Neurosci 30(19):6515–6526. doi:10.1523/JNEUROSCI.0771-10.2010
Kautzmann M-AI, Kim DS, Felder-Schmittbuhl M-P, Swaroop A (2011) Combinatorial regulation of photoreceptor differentiation factor, neural retina leucine zipper gene NRL, revealed by in vivo promoter analysis. J Biol Chem 286(32):28247–28255. doi:10.1074/jbc.M111.257246
Kechad A, Jolicoeur C, Tufford A, Mattar P, Chow RW, Harris WA, Cayouette M (2012) Numb is required for the production of terminal asymmetric cell divisions in the developing mouse retina. J Neurosci 32(48):17197–17210. doi:10.1523/JNEUROSCI.4127-12.2012
Kim DS, Ross SE, Trimarchi JM, Aach J, Greenberg ME, Cepko CL (2008) Identification of molecular markers of bipolar cells in the murine retina. J Comp Neurol 507(5):1795–1810. doi:10.1002/cne.21639
Kimura A, Singh D, Wawrousek EF, Kikuchi M, Nakamura M, Shinohara T (2000) Both PCE-1/RX and OTX/CRX interactions are necessary for photoreceptor-specific gene expression. J Biol Chem 275(2):1152–1160
Koike C, Nishida A, Ueno S, Saito H, Sanuki R, Sato S, Furukawa A, Aizawa S, Matsuo I, Suzuki N, Kondo M, Furukawa T (2007) Functional roles of Otx2 transcription factor in postnatal mouse retinal development. Mol Cell Biol 27(23):8318–8329. doi:10.1128/MCB.01209-07
Li S, Mo Z, Yang X, Price SM, Shen MM, Xiang M (2004) Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron 43(6):795–807. doi:10.1016/j.neuron.2004.08.041
Li X, Chen Z, Desplan C (2013a) Temporal patterning of neural progenitors in Drosophila. Curr Top Dev Biol 105:69–96. doi:10.1016/B978-0-12-396968-2.00003-8
Li X, Erclik T, Bertet C, Chen Z, Voutev R, Venkatesh S, Morante J, Celik A, Desplan C (2013b) Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature (Lond) 498(7455):456–462. doi:10.1038/nature12319
Livesey FJ, Furukawa T, Steffen MA, Church GM, Cepko CL (2000) Microarray analysis of the transcriptional network controlled by the photoreceptor homeobox gene Crx. Curr Biol 10(6):301–310
Luo H, Jin K, Xie Z, Qiu F, Li S, Zou M, Cai L, Hozumi K, Shima DT, Xiang M (2012) Forkhead box N4 (Foxn4) activates Dll4-Notch signaling to suppress photoreceptor cell fates of early retinal progenitors. Proc Natl Acad Sci USA 109(9):E553–562. doi:10.1073/pnas.1115767109
Mathers PH, Grinberg A, Mahon KA, Jamrich M (1997) The Rx homeobox gene is essential for vertebrate eye development. Nature 387(6633):603–607. doi:10.1038/42475
Matsuda T, Cepko CL (2004) Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci USA 101(1):16–22
Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A (2001) Nrl is required for rod photoreceptor development. Nat Genet 29(4):447–452. doi:10.1038/ng774
Mizeracka K, Demaso CR, Cepko CL (2013a) Notch1 is required in newly postmitotic cells to inhibit the rod photoreceptor fate. Development (Camb). doi:10.1242/dev.090696
Mizeracka K, Trimarchi JM, Stadler MB, Cepko CL (2013b) Analysis of gene expression in wild type and Notch1 mutant retinal cells by single cell profiling. Dev Dyn. doi:10.1002/dvdy.24006
Montana CL, Lawrence KA, Williams NL, Tran NM, Peng GH, Chen S, Corbo JC (2011) Transcriptional regulation of neural retina leucine zipper (Nrl), a photoreceptor cell fate determinant. J Biol Chem 286(42):36921–36931. doi:10.1074/jbc.M111.279026
Morris VB, Shorey CD (1967) An electron microscope study of types of receptor in the chick retina. J Comp Neurol 129(4):313–340. doi:10.1002/cne.901290404
Morrow EM, Furukawa T, Lee JE, Cepko CL (1999) NeuroD regulates multiple functions in the developing neural retina in rodent. Development (Camb) 126(1):23–36
Morrow EM, Furukawa T, Raviola E, Cepko CL (2005) Synaptogenesis and outer segment formation are perturbed in the neural retina of Crx mutant mice. BMC Neurosci 6:5. doi:10.1186/1471-2202-6-5
Muranishi Y, Terada K, Inoue T, Katoh K, Tsujii T, Sanuki R, Kurokawa D, Aizawa S, Tamaki Y, Furukawa T (2011) An essential role for RAX homeoprotein and NOTCH-HES signaling in Otx2 expression in embryonic retinal photoreceptor cell fate determination. J Neurosci 31(46):16792–16807. doi:10.1523/JNEUROSCI.3109-11.2011
Muranishi Y, Terada K, Furukawa T (2012) An essential role for Rax in retina and neuroendocrine system development. Dev Growth Differ 54(3):341–348. doi:10.1111/j.1440-169X.2012.01337.x
Nakamura K, Harada C, Namekata K, Harada T (2006) Expression of olig2 in retinal progenitor cells. Neuroreport 17(4):345–349. doi:10.1097/01.wnr.0000203352.44998.6b
Nelson BR, Reh TA (2008) Relationship between Delta-like and proneural bHLH genes during chick retinal development. Dev Dyn 237(6):1565–1580. doi:10.1002/dvdy.21550
Nelson BR, Gumuscu B, Hartman BH, Reh TA (2006) Notch activity is downregulated just prior to retinal ganglion cell differentiation. Dev Neurosci 28(1-2):128–141. doi:10.1159/000090759
Nelson BR, Hartman BH, Georgi SA, Lan MS, Reh TA (2007) Transient inactivation of Notch signaling synchronizes differentiation of neural progenitor cells. Dev Biol 304(2):479–498. doi:10.1016/j.ydbio.2007.01.001
Nelson SM, Park L, Stenkamp DL (2009) Retinal homeobox 1 is required for retinal neurogenesis and photoreceptor differentiation in embryonic zebrafish. Dev Biol 328(1):24–39. doi:10.1016/j.ydbio.2008.12.040
Ng L, Hurley JB, Dierks B, Srinivas M, Salto C, Vennstrom B, Reh TA, Forrest D (2001) A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat Genet 27(1):94–98. doi:10.1038/83829
Nguyen DN, Rohrbaugh M, Lai Z (2000) The Drosophila homolog of Onecut homeodomain proteins is a neural-specific transcriptional activator with a potential role in regulating neural differentiation. Mech Dev 97(1-2):57–72
Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, Furukawa T (2003) Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci 6(12):1255–1263. doi:10.1038/nn1155
Ochi H, Sakagami K, Ishii A, Morita N, Nishiuchi M, Ogino H, Yasuda K (2004) Temporal expression of L-Maf and RaxL in developing chicken retina are arranged into mosaic pattern. Gene Expr Patterns 4(5):489–494. doi:10.1016/j.modgep.2004.03.005
Ochocinska MJ, Hitchcock PF (2009) NeuroD regulates proliferation of photoreceptor progenitors in the retina of the zebrafish. Mech Dev 126(3-4):128–141. doi:10.1016/j.mod.2008.11.009
Onishi A, Peng GH, Hsu C, Alexis U, Chen S, Blackshaw S (2009) Pias3-dependent SUMOylation directs rod photoreceptor development. Neuron 61(2):234–246. doi:10.1016/j.neuron.2008.12.006
Onishi A, Peng GH, Poth EM, Lee DA, Chen J, Alexis U, de Melo J, Chen S, Blackshaw S (2010) The orphan nuclear hormone receptor ERRbeta controls rod photoreceptor survival. Proc Natl Acad Sci USA 107(25):11579–11584. doi:10.1073/pnas.1000102107
Pearson BJ, Doe CQ (2004) Specification of temporal identity in the developing nervous system. Annu Rev Cell Dev Biol 20:619–647. doi:10.1146/annurev.cellbio.19.111301.115142
Peng GH, Chen S (2005) Chromatin immunoprecipitation identifies photoreceptor transcription factor targets in mouse models of retinal degeneration: new findings and challenges. Vis Neurosci 22(5):575–586. doi:10.1017/S0952523805225063
Ranade SS, Yang-Zhou D, Kong SW, McDonald EC, Cook TA, Pignoni F (2008) Analysis of the Otd-dependent transcriptome supports the evolutionary conservation of CRX/OTX/OTD functions in flies and vertebrates. Dev Biol 315(2):521–534. doi:10.1016/j.ydbio.2007.12.017
Rapaport DH, Wong LL, Wood ED, Yasumura D, LaVail MM (2004) Timing and topography of cell genesis in the rat retina. J Comp Neurol 474(2):304–324
Riesenberg AN, Liu Z, Kopan R, Brown NL (2009) Rbpj cell autonomous regulation of retinal ganglion cell and cone photoreceptor fates in the mouse retina. J Neurosci 29(41):12865–12877. doi:10.1523/JNEUROSCI.3382-09.2009
Rivolta C, Berson EL, Dryja TP (2001) Dominant Leber congenital amaurosis, cone-rod degeneration, and retinitis pigmentosa caused by mutant versions of the transcription factor CRX. Hum Mutat 18(6):488–498. doi:10.1002/humu.1226
Roberts MR, Hendrickson A, McGuire CR, Reh TA (2005) Retinoid X receptor (gamma) is necessary to establish the S-opsin gradient in cone photoreceptors of the developing mouse retina. Invest Ophthalmol Vis Sci 46(8):2897–2904. doi:10.1167/iovs.05-0093
Rocha SF, Lopes SS, Gossler A, Henrique D (2009) Dll1 and Dll4 function sequentially in the retina and pV2 domain of the spinal cord to regulate neurogenesis and create cell diversity. Dev Biol 328(1):54–65. doi:10.1016/j.ydbio.2009.01.011
Roe T, Reynolds TC, Yu G, Brown PO (1993) Integration of murine leukemia virus DNA depends on mitosis. EMBO J 12(5):2099–2108
Rompani SB, Cepko CL (2008) Retinal progenitor cells can produce restricted subsets of horizontal cells. Proc Natl Acad Sci USA 105(1):192–197. doi:10.1073/pnas.0709979104
Sato S, Inoue T, Terada K, Matsuo I, Aizawa S, Tano Y, Fujikado T, Furukawa T (2007) Dkk3-Cre BAC transgenic mouse line: a tool for highly efficient gene deletion in retinal progenitor cells. Genesis 45(8):502–507. doi:10.1002/dvg.20318
Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, Huillard E, Sun T, Ligon AH, Qian Y, Ma Q, Alvarez-Buylla A, McMahon AP, Rowitch DH, Ligon KL (2008) Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell 14(2):123–134. doi:10.1016/j.ccr.2008.07.005
Shibasaki K, Takebayashi H, Ikenaka K, Feng L, Gan L (2007) Expression of the basic helix-loop-factor Olig2 in the developing retina: Olig2 as a new marker for retinal progenitors and late-born cells. Gene Expr Patterns 7(1–2):57–65. doi:10.1016/j.modgep.2006.05.008
Sjoberg M, Vennstrom B, Forrest D (1992) Thyroid hormone receptors in chick retinal development: differential expression of mRNAs for alpha and N-terminal variant beta receptors. Development (Camb) 114(1):39–47
Skinner MK, Rawls A, Wilson-Rawls J, Roalson EH (2010) Basic helix-loop-helix transcription factor gene family phylogenetics and nomenclature. Differentiation 80(1):1–8. doi:10.1016/j.diff.2010.02.003
Spana EP, Doe CQ (1996) Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron 17(1):21–26
Srinivas M, Ng L, Liu H, Jia L, Forrest D (2006) Activation of the blue opsin gene in cone photoreceptor development by retinoid-related orphan receptor beta. Mol Endocrinol 20(8):1728–1741. doi:10.1210/me.2005-0505
Suzuki SC, Bleckert A, Williams PR, Takechi M, Kawamura S, Wong RO (2013) Cone photoreceptor types in zebrafish are generated by symmetric terminal divisions of dedicated precursors. Proc Natl Acad Sci USA 110(37):15109–15114. doi:10.1073/pnas.1303551110
Swain PK, Hicks D, Mears AJ, Apel IJ, Smith JE, John SK, Hendrickson A, Milam AH, Swaroop A (2001) Multiple phosphorylated isoforms of NRL are expressed in rod photoreceptors. J Biol Chem 276(39):36824–36830. doi:10.1074/jbc.M105855200
Swaroop A, Xu JZ, Pawar H, Jackson A, Skolnick C, Agarwal N (1992) A conserved retina-specific gene encodes a basic motif/leucine zipper domain. Proc Natl Acad Sci USA 89(1):266–270
Swaroop A, Kim D, Forrest D (2010) Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci 11(8):563–576. doi:10.1038/nrn2880
Tahayato A, Sonneville R, Pichaud F, Wernet MF, Papatsenko D, Beaufils P, Cook T, Desplan C (2003) Otd/Crx, a dual regulator for the specification of ommatidia subtypes in the Drosophila retina. Dev Cell 5(3):391–402
Tomita K, Moriyoshi K, Nakanishi S, Guillemot F, Kageyama R (2000) Mammalian achaete-scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system. EMBO J 19(20):5460–5472. doi:10.1093/emboj/19.20.5460
Trimarchi JM, Stadler MB, Roska B, Billings N, Sun B, Bartch B, Cepko CL (2007) Molecular heterogeneity of developing retinal ganglion and amacrine cells revealed through single cell gene expression profiling. J Comp Neurol 502(6):1047–1065. doi:10.1002/cne.21368
Trimarchi JM, Harpavat S, Billings NA, Cepko CL (2008a) Thyroid hormone components are expressed in three sequential waves during development of the chick retina. BMC Dev Biol 8:101. doi:10.1186/1471-213X-8-101
Trimarchi JM, Stadler MB, Cepko CL (2008b) Individual retinal progenitor cells display extensive heterogeneity of gene expression. PloS One 3(2):e1588. doi:10.1371/journal.pone.0001588
Turner DL, Cepko CL (1987) A common progenitor for neurons and glia persists in rat retina late in development. Nature (Lond) 328(6126):131–136
Turner DL, Snyder EY, Cepko CL (1990) Lineage-independent determination of cell type in the embryonic mouse retina. Neuron 4(6):833–845
Vandendries ER, Johnson D, Reinke R (1996) orthodenticle is required for photoreceptor cell development in the Drosophila eye. Dev Biol 173(1):243–255. doi:10.1006/dbio.1996.0020
Wang JC, Harris WA (2005) The role of combinational coding by homeodomain and bHLH transcription factors in retinal cell fate specification. Dev Biol 285(1):101–115. doi:10.1016/j.ydbio.2005.05.041
Wetts R, Fraser SE (1988) Multipotent precursors can give rise to all major cell types of the frog retina. Science 239(4844):1142–1145
White MA, Myers CA, Corbo JC, Cohen BA (2013) Massively parallel in vivo enhancer assay reveals that highly local features determine the cis-regulatory function of ChIP-seq peaks. Proc Natl Acad Sci USA. doi:10.1073/pnas.1307449110
Wu HY, Perron M, Hollemann T (2009) The role of Xenopus Rx-L in photoreceptor cell determination. Dev Biol 327(2):352–365. doi:10.1016/j.ydbio.2008.12.017
Wu F, Sapkota D, Li R, Mu X (2012) Onecut 1 and Onecut 2 are potential regulators of mouse retinal development. J Comp Neurol 520(5):952–969. doi:10.1002/cne.22741
Yan RT, He L, Wang SZ (2009) Pro-photoreceptor activity of chick neurogenin 1. Invest Ophthalmol Vis Sci 50(12):5567–5576. doi:10.1167/iovs.09-3647
Yaron O, Farhy C, Marquardt T, Applebury M, Ashery-Padan R (2006) Notch1 functions to suppress cone-photoreceptor fate specification in the developing mouse retina. Development (Camb) 133(7):1367–1378. doi:10.1242/dev.02311
Yoshida S, Mears AJ, Friedman JS, Carter T, He S, Oh E, Jing Y, Farjo R, Fleury G, Barlow C, Hero AO, Swaroop A (2004) Expression profiling of the developing and mature Nrl-/- mouse retina: identification of retinal disease candidates and transcriptional regulatory targets of Nrl. Hum Mol Genet 13(14):1487–1503. doi:10.1093/hmg/ddh160
Young RW (1985) Cell differentiation in the retina of the mouse. Anat Rec 212(2):199–205. doi:10.1002/ar.1092120215
Zhong W (2003) Diversifying neural cells through order of birth and asymmetry of division. Neuron 37(1):11–14
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Japan
About this chapter
Cite this chapter
Cepko, C. (2014). Cell Fate Determination of Photoreceptor Cells. In: Furukawa, T., Hurley, J., Kawamura, S. (eds) Vertebrate Photoreceptors. Springer, Tokyo. https://doi.org/10.1007/978-4-431-54880-5_9
Download citation
DOI: https://doi.org/10.1007/978-4-431-54880-5_9
Published:
Publisher Name: Springer, Tokyo
Print ISBN: 978-4-431-54879-9
Online ISBN: 978-4-431-54880-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)